Abstract
In an attempt to further increase transgene expression levels in plants over and above the enhancement obtained with a 5′ untranslated leader intron, three different maize introns were inserted at three different positions within the coding sequence of the luciferase reporter gene. Constructs were transformed into maize (Black Mexican Sweet) cells and protoplasts, and their activity determined. Although all introns tested were correctly spliced, only one of them in a particular position was able to enhance gene expression. Correct splicing sites were used for intron removal and the quantity of luciferase mRNA produced did not differ significantly. These data indicate that both the position and the sequence of an intron have marked effects on expression levels, suggesting that nuclear processing of the pre-mRNA determines final expression levels through the structure of the mRNP.
INTRODUCTION
Introns are intervening sequences present in the pre-mRNA but absent in the mature RNA following excision by a precise splicing mechanism. In both animals and plants numerous studies involving introns have focused on their effects when inserted between the promoter region and the coding sequence of the gene (Callis et al., 1987; Matsumoto et al., 1998). Introns inserted inside the coding region of the reporter gene were shown to enhance gene expression (Tanaka et al., 1990). Enhancement of gene expression by introns is not a general phenomenon as some naturally occurring genes do not contain introns and are expressed efficiently, while some intron insertions into transgenes fail to enhance expression (Buchman and Berg, 1988; Mascarenhas et al., 1990). In plants enhancement of gene expression is observed only in the more GC-rich (monocot) genomes (Taylor et al., 1993; Vain et al., 1996). Moreover, it has been reported that variation in expression levels can even be found in different tissues within a single plant (Fennell and Hauptmann, 1992; Gallie and Young, 1994) and that intron-mediated enhancement may be a gene dependent process (Rethmeier et al., 1997). We investigated the affects of adding additional introns into the coding sequence of the luciferase reporter gene to determine whether they would enhance expression over and above the expression of constructs with a 5′ leader intron. Three introns, from the maize nuclear RpoT gene, which encodes one of the maize T3/T7 like DNA dependent RNA polymerases (Young et al., 1998), were, independently and in combination, inserted into the coding sequence of the firefly luciferase reporter gene. Only one out of the three maize introns tested (RpoT-i4) inserted at nucleotide +165, position 1, resulted in a significant and reproducible increase in luciferase expression. The two other introns or intron i4, at other positions or in combination with one of the other introns, in any one of three positions, strongly inhibited luciferase expression. Missplicing was not detected and there was no significant difference between the quantities of luciferase mRNA produced with the different constructs. The results suggest that the reduction in luciferase expression observed was due to a post-transcriptional level mechanism, which although not reducing the mRNA levels, significantly reduced the quantity of active luciferase protein over several orders of magnitude.
RESULTS AND DISCUSSION
Relative luciferase activities of the constructs
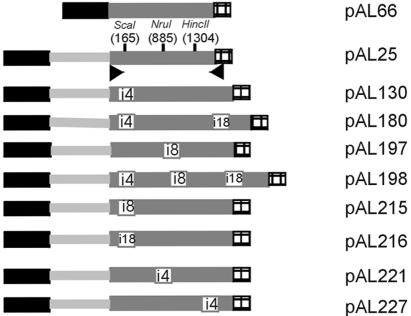
The different plasmids (Figure 1) were mixed with the p2920 plasmid (GUS reporter gene) in a 1:1 ratio and cotransformed by bombardment into maize BMS cells, and by electroporation into protoplasts. All constructs with the exception of pAL66 contained the maize ubiquitin leader intron. In each set of experiments pAL25 was included. Its luciferase activity relative to the level of GUS expression (p2920) was normalized to one. The luciferase/GUS ratios for all other plasmids were determined and expressed as a value of pAL25. The resulting luciferase activities of each of the constructs relative to that of pAL25 are given in Figure 2.
Fig. 1. Chimeric luciferase constructs. The maize ubiquitin promoter (black box) plus leader intron (grey line) is fused to the Luc+ reporter gene (grey box) in presence of the Nos terminator (black squares). White boxes represent maize introns and their corresponding number is indicated. Restriction sites used to introduce the different introns in the luciferase cDNA are indicated as well as their position in the coding region. Arrowheads indicate primers used for the PCR amplifications.
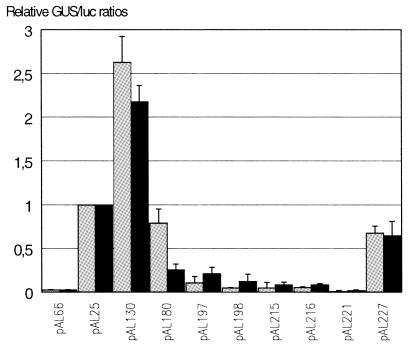
Fig. 2. Luciferase activity in transformed maize BMS cells and protoplasts. Average luciferase activities from 11 bombardments of BMS cells (in grey) and three electroporations of BMS protoplasts (in black) are given as the ratio of luciferase units to GUS units. Luciferase activity of the plasmid pAL25 was set to one.
The inclusion of the ubiquitin 5′ leader intron enhances expression of the luciferase gene by 3800% (pAL66 -v- pAL25). This enhancement is also obtained when the ubiquitin promoter is replaced with either a wheat actin promoter (our unpublished results) or with a wheat FKBP promoter (Kurek et al., 1999). Intron mediated enhancement of translation is therefore promoter independent.
The plasmid pAL130 contains the maize intron i4 in position 1 of the Luc+ cDNA, this was the only construct to show a significant and reproducible enhancement of luciferase activity, averaging 240%, when compared to the plasmid pAL25. Insertion of intron i4 in either position 2 or position 3 (pAL221 and pAL227) resulted in reductions of luciferase expression, 99 and 35%, respectively, when compared to pAL25 (Figure 2). Severe reductions in luciferase activity were also obtained when intron i8 or i18 were placed in position 1 (pAL215 and pAL216); both producing a 93% decrease when compared to pAL25. Similarly, insertion of i8 into position 2 (pAL197) resulted in an 83% decrease in expression.
Only two constructs with combinations of introns were made and tested: pAL180 (i4 and i8) and pAL198 (i4, i8 and i18). Both resulted in reduced expression levels relative to pAL25; reductions of 44 and 92%, respectively.
The data demonstrates that with the exception of i4 in position 1 (ScaI site), one or more introns inserted into the coding sequence severely reduce expression levels. This effect appears to be position dependent (i4: pAL130, pAL221 and pAL227) and intron dependent (i4 v i8 and i18: pAL130, pAL215 and pAL216).
Maize introns in the luciferase transgene are correctly spliced in maize BMS cells
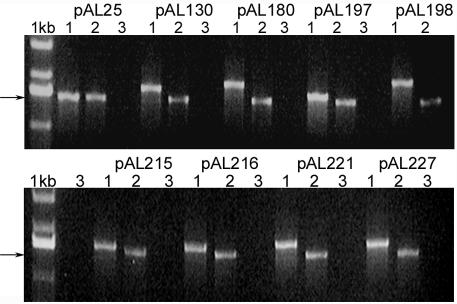
Missplicing was a possible explanation for the decrease in luciferase activity observed in constructs pAL180 to pAL227 (Figure 2). cDNA was synthesized from mRNA extracted from bombarded cells. Six different combinations of primers were used on each cDNA. Primer pairs were used to amplify the sequences flanking intron positions 1, 2 and 3, independently; the sequence from 116 bp to 1037 bp (intron positions 1 and 2) and 682 bp to 1645 bp (intron positions 2 and 3), and lastly the entire cDNA from 116 bp to 1645 bp. Plasmid DNAs were amplified in parallel to provide the sizes of the intron containing amplicons. PCR amplicons were analysed on 1% agarose gels. In each case, a single band corresponding to the expected size of the spliced product, amplified from pAL25, was observed. The result of the amplification of the entire cDNA (primer positions shown in Figure 1) is given in Figure 3. The sizes of the PCR products correspond to a non-spliced version of the luciferase reporter gene. Only one band of 1.5 kb, corresponding to a correctly spliced luciferase RNA, was ever detected on the gel for all nine cDNAs. The absence of bands in lanes corresponding to the RT–PCR negative control, mRNA sample, confirms that amplicons derive entirely from cDNA and not from contaminating plasmids. We concluded that the maize introns inserted into the luciferase transgene are correctly spliced in maize BMS cells.
Fig. 3. PCR amplification of the spliced products from mRNA isolated from bombarded maize BMS cells. Lane 1, plasmid amplicons; lane 2, RT–PCR amplicons; lane 3, mRNA controls from RT–PCR. The size of the expected spliced product (1.5 kb) is indicated with arrows.
Sequencing across splice junctions
All six spliced junctions of the luciferase transcripts were sequenced to check whether the correct sites were recognized in the transgenes. Two selected amplified DNA fragments corresponding to the 1.5 kb luciferase cDNA of pAL180 and pAL198 were subcloned in a pKS(+) plasmid and the entire cDNA sequenced. Two independent clones from each RT–PCR were sequenced. The sequences demonstrated that the correct splicing sites were used for intron removal and a precise excision of each intron using the predicted GT/AG recreated the correct splicing site. As a consequence, the spliced products were identical to the intron-less luciferase cDNA of pAL25. The decrease in the luciferase activity found in maize BMS cells could, therefore, not be attributed to any defect in the splicing mechanism. The introns could conceivably target the pre-mRNA to degradation pathways within the nucleus resulting in RNA instability and differences in the steady-state level of cytoplasmic RNA.
Determination of the steady-state level of mature mRNAs
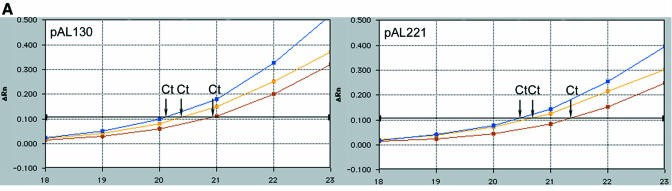
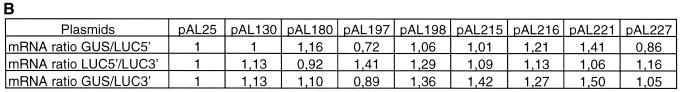
In order to determine whether the level of mRNA was affected by the presence of introns in the luciferase-coding region, quantitative RT–PCR was performed using the TaqMan (Perkin-Elmer 7700). The different plasmids shown in Figure 1 were mixed with the GUS reporter plasmid (p2920) in a 1:1 ratio and electroporated into maize BMS protoplasts. cDNA was prepared from mRNA isolated from samples of electroporated cells which had been assayed for luciferase and GUS (Figure 2). Two different luciferase 5′-carboxyfluorescein (FAM) probes hybridizing to the 5′ and 3′ ends of the cDNA were used to detect the entire luciferase mRNA. The GUS mRNA was used as an internal standard to quantify the luciferase mRNA. An example of one TaqMan experiment is illustrated in Figure 4A. Ratios between the GUS and the 5′ and 3′ luciferase mRNA were calculated from two separate electroporations and average values are given in Figure 4B. Relative to the level of GUS mRNA the levels of luciferase mRNA, whether it be 5′ or 3′, exhibited little variation from experiment to experiment. The low variation observed in these ratios and more specifically between the luciferase expression values corresponding to pAL130, showing a 240% increase in luciferase expression, and pAL221, showing a drastic 99% reduction in expression (Figure 2), cannot be explained by the levels of steady-state mRNA. Additionally the results from using 5′ and 3′ FAM probes reveal that the luciferase mRNA produced is not destabilized by the intervening sequences. As with the RT–PCR (Figure 3) no signal was detected using mRNA as negative controls demonstrating that the signals observed derived only from cDNA. No signal was obtained in the controls without nucleic acids.
Fig. 4. Quantification of the luciferase mRNA in maize BMS protoplasts. (A) Example of two cDNA amplifications using GUS and both LUC probes. Only the PCR cycles in which the reporter dye emission crosses the default threshold (horizontal line) are shown, corresponding Ct values for both cDNA with each probe (GUS in blue, 5′LUC in red and 3′LUC in yellow) are indicated. (B) Comparison of mRNA ratios between GUS and 5′Luc, GUS and 3′Luc, and 5′Luc and 3′Luc. Ratios are set to pAL25 values.
In plants introns are characterized by their AT content (60%) and their consensus splicing sites (Brown, 1986; Lorkovic et al., 2000). The AT content of the three maize introns used in this study were close to the optimal AT content of maize introns (between 54 and 63%). Comparison of the plant consensus splice site (5′-AAG:GTAAGT//TTGCAG:GT) with the maize introns shows that introns i8 and i18 differ more from the consensus (from 41 to 53% identity depending on the insertion site) than those of intron i4 (between 70 and 76% identity). Again, these introns derived from a natural and functional maize gene (Young et al., 1998) and are spliced correctly. It seems unlikely that the intron flanking sequences and the intron phase (position 1, 0; position 2, 1; position 3, 2) relative to the coding frame (Logsdon, 1998) have any affect on pre-mRNA processing as the steady-state mature mRNA levels are similar.
Similar arguments can be applied to the sequences surrounding the introns as it has already been suggested that these sequences are important for efficiency of intron splicing (Matsumoto et al., 1998; Luehrsen and Walbot, 1991). Our results tend to suggest otherwise. No unspliced RT–PCR products were ever detected for any of the introns in positions 1 to 3. So if intron splicing efficiency were reduced, it was at an undetectable level and could have no observable impact on the mature mRNA levels.
Enhancement of gene expression has been correlated to an increased level of mRNA (Tanaka et al., 1990). However, Matsumoto et al. (1998) demonstrated that even though the addition of a 5′ leader intron to a CAT transgene enhanced mRNA levels, the increase could not account for the increase in CAT activity suggesting that the ‘quality’ of the mRNA was important. In this study we have shown that either a 240% increase or a 99% decrease in luciferase expression did not correlate with the level of steady-state mRNA. As neither the splicing mechanism or the quantity of mRNA produced is affected, we envisage that a post-transcriptional mechanism is responsible for the variation in luciferase expression found with the constructs tested, excluding pAL66. This hypothesis correlates with observations made earlier in animal cells using the CAT reporter gene where the inhibitory effect of the intron was detectable only at the protein level (Evans and Scarpulla, 1989).
Speculation
Recently, Mascarenhas et al. (1990) demonstrated that in animal cells the presence of an intron in the 3′ UTR of the mRNA was a major determinant of translational efficiency in the absence of any change in the steady-state mRNA level. They concluded that it was the ‘nuclear history’ of the mRNA that was the major determinant of expression levels. The results that we have obtained clearly demonstrate that introns, even within the coding sequence, are a major determinant of transgene expression levels. Their sequence and position within any open reading frame could determine how and with what efficiency the ribosomes recruit the mRNA once the mRNA has been exported from the nucleus. Transacting factors binding during intron excision have been shown to form a stable mRNP with proteins remaining at the exon–exon junctions after intron excision (Le Hir et al., 2000). If the factors recruited to excise introns and those left at the exon–exon boundaries are determined not only by the intron but also by its position as a result of interactions with the intron flanking sequences then the mature mRNP particles could have significantly different structures and compositions. Our results indicate that, in the absence of any detectable splicing defects or changes in mature mRNA levels, the formation of the mature mRNP and its ability to be recruited by the translational machinery is a consequence of the position and sequence of the introns. In other words, the processed mRNA is imprinted with intron and intron position information which, when exported from the nucleus, governs its functionality.
We can speculate that the expression of genes, the majority of which contain introns, is primarily determined by the introns they contain. Introns effectively modulate gene expression in both a positive and negative sense. Missplicing of introns, as exemplified by the maize RpoT gene where it occurs at intron i12 (Young et al., 1998), produces a full-length transcript which accounts for 25% of the steady-state mRNA. As the remaining six introns now reside in the 3′ UTR, this could effectively be a signal to downregulate expression of this misspliced transcript (Evans and Scarpulla, 1989; Mascarenhas et al., 1990). Alternative splicing is another common phenomenon in eukaryotes, our results suggest that its consequences may dictate the expression levels of each alternatively spliced product based on the combination of intron signatures remaining in the spliced and mature mRNA.
A number of the plasmids used in this study have been introduced into wheat to investigate whether the intron effects observed in transient experiments are the same in transgenic plants and to examine the association of the transcripts with polysomes that could provide information on the mechanism of this phenomenon.
METHODS
Luciferase constructs. All plasmids are derived from pAL25 (Lonsdale et al., 1998), which contains the maize ubiquitin promoter plus a leader intron fused to the firefly luciferase cDNA (Luc+, Promega; DDBJ/EMBL/GenBank accession No. U47122). Three introns, i4 (234 bp, 54% AT), i8 (103 bp, 56% AT) and i18 (117 bp, 63% AT), from the maize RpoT gene (Young et al., 1998; DDBJ/EMBL/GenBank accession No. AJ005343) were selected on the basis that their 5′ and 3′ sequences corresponded to the 3′ and 5′ halves of SnaBI and PvuII sites, respectively. These introns were inserted into the three unique blunt end restriction sites of the Luc+ cDNA: ScaI site (position 1, nucleotide +165), NruI site (position 2, +885) and HincII (position 3, +1304). A silent C→G transversion of nucleotide +887 using PCR mutagenesis and subsequent sequencing of the modified cDNA created the NruI site. Based on the abundance of GCC and GCG codons no deleterious effects on expression were predicted. Plasmid p2920 containing the maize ubiquitin promoter plus leader intron fused to the uidA reporter gene (GUS; kindly provided by Paul Christou, John Innes Centre, Norwich, UK) was used as an internal standard.
Transient assays. Maize BMS suspension cells were bombarded with the luciferase and GUS constructs (1:1 ratio using a DuPont He1000 particle gun, 1100 rupture disc). Eleven independent experiments were performed with 3–5 bombardments per construct. Luciferase and GUS activities were determined 2 days after bombardment (Lonsdale et al., 1998). mRNA was isolated from parallel bombardments for RT–PCR experiments.
Maize BMS protoplasts were prepared and electroporated with the luciferase and GUS constructs (Fromm et al., 1987). Three electroporations were performed. One day after electroporation cells were divided, part was assayed for GUS and luciferase activities and mRNA was extracted from the remainder.
Quantification and analyses of the cDNA. Poly(A)+ mRNA was isolated from maize BMS cells and protoplasts two days and one day after transformation, respectively, using the Dynabeads mRNA Direct kit (Dynal, Oslo, Norway). Reverse transcription was performed for 1 h at 50°C with 1 µg of mRNA using a poly-dT primer and the Superscript Reverse Transcriptase (Gibco-BRL, Paisley, UK). A negative control of reverse transcription without enzyme was performed in parallel for each mRNA isolation for use in TaqMan and RT–PCR experiments.
A fraction of 1/20 of the synthesized cDNA was used as a template to quantify the mRNA in the TaqMan 7700 (Perkin-Elmer) using LUC and GUS specific primers and FAM-labelled oligo probes. The TaqMan 5′ luciferase probe, 5′-ACGTTCGTCACATCTCATCTACCTCCCG was used to detect a 115 bp luciferase amplicon, beginning at +443 bp. The 3′ luciferase probe, 5′-CGCCAGTCAAGTAACAACCGCGAAAA was used to detect a 74 bp luciferase amplicon, beginning at +1478 bp. The GUS probe, 5′-AACGTGCTGATGGTGCACGACCAC was used to detect a 69 bp amplicon beginning at +899 bp. TaqMan runs were performed from two independent electroporations and each cDNA was amplified in triplicate. In each run, the ratio of GUS mRNA to Luc mRNA or Luc5′ mRNA to Luc3′ mRNA in the control pAL25 plasmid was set to one and other ratios were calculated accordingly. Controls without nucleic acids were included for each probe. Probes and primers were designed using Primer Express (Perkin-Elmer).
Nested PCR was used to analyse the fidelity of intron splicing. 1/1000 of the poly-dT primed cDNA was used as a template. In order to amplify the luciferase cDNA, a primer specific from the 5′ end of the luciferase coding region (5′-CGATCTTTCCGCCCTTCTTGG-3′) was used in combination with primers 3′ to each intron and with a primer located in the Nos terminator (5′-CGCAAGACCGGCAACAGG-3′) 5′ to the poly(A) addition site. Amplified fragments were loaded on a 1% agarose gel. PCR products were cloned into a pKS(+) plasmid (Stratagene, Amsterdam, The Netherlands) and sequenced with the 373 DNA Sequencer (Perkin-Elmer). Sequence comparisons were performed using Pileup in the GCG program (Devereux et al., 1984).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Bill Martin (Technical University of Braunsweig, Germany) for his views on our results and Allan Wickham for his technical assistance. This work was supported by MAFF funding (CSA 4786). We would also like to acknowledge the support of the John Innes Centre, Norwich, UK.
References
- Brown J.W.S. (1986) A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res., 14, 9549–9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A.R. and Berg, P. (1988) Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol., 8, 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J., Fromm, M. and Walbot, V. (1987) Introns increase gene expression in cultured maize cells. Genes Dev., 1, 1183–1200. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli, P. and Smithies, O. (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res., 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J. and Scarpulla, R.C. (1989) Introns in the 3′-untranslated region can inhibit chimeric CAT and β-galactosidase gene expression. Gene, 84, 135–142. [DOI] [PubMed] [Google Scholar]
- Fennell A. and Hauptmann, R. (1992) Electroporation and PEG delivery of DNA into maize microspores. Plant Cell Rep., 11, 567–570. [DOI] [PubMed] [Google Scholar]
- Fromm M., Callis, J., Taylor, L.P. and Walbot, V. (1987) Electroporation of DNA and RNA into plant protoplasts. Methods Enzymol., 153, 351–366. [Google Scholar]
- Gallie D.R. and Young, T.E. (1994) The regulation of gene expression in transformed maize aleurone and endosperm protoplasts. Analysis of promoter activity, intron enhancement, and mRNA untranslated regions on expression. Plant Physiol., 106, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore, M.J. and Maquat, L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Kurek I., Harvey, A.J., Lonsdale, D. and Breiman, A. (1999) Isolation and characterization of the wheat prolyl isomerase FK506 binding protein (FKBP) 73 promoter. Plant Mol. Biol., 42, 489–497. [DOI] [PubMed] [Google Scholar]
- Logsdon J.M. Jr, (1998) The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev., 8, 637–648. [DOI] [PubMed] [Google Scholar]
- Lonsdale D.M., Moisan, L.J. and Harvey, A.J. (1998) The effect of altered codon usage on luciferase activity in tobacco, maize and wheat. Plant Cell Rep., 17, 396–399. [DOI] [PubMed] [Google Scholar]
- Lorkovic Z.J., Wieczorek Kirk, D.A., Lambermon, M.H.L. and Filipowicz, W. (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci., 5, 160–167. [DOI] [PubMed] [Google Scholar]
- Luehrsen K.R. and Walbot, V. (1991) Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol. Gen. Genet., 225, 81–93. [DOI] [PubMed] [Google Scholar]
- Mascarenhas D., Mettler, I.J., Pierce, D.A. and Lowe, H.W. (1990) Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol. Biol., 15, 913–920. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman, K.M. and Wolffe, A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethmeier N., Seurinck, J., Van Montagu, M. and Corelissen, M. (1997) Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J., 12, 895–899. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Mita, S., Ohta, S., Kyozuka, J., Shimamoto, K. and Nakamura, K. (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res., 18, 6767–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.G., Vasil, V. and Vasil, I.K. (1993) Enhanced GUS gene expression in cereal/grass cell suspensions and immature embryos using the maize ubiquitin-based plasmid pAHC25. Plant Cell Rep., 12, 491–495. [DOI] [PubMed] [Google Scholar]
- Vain P., Finer, K.R., Engler, D.E., Pratt, R.C. and Finer, J.J. (1996) Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep., 15, 489–494. [DOI] [PubMed] [Google Scholar]
- Young D.A., Allen, R.L., Harvey, A.J. and Lonsdale, D.M. (1998) Characterization of a gene encoding a single-subunit bacteriophage-type RNA polymerase from maize which is alternatively spliced. Mol. Gen. Genet., 260, 30–37. [DOI] [PubMed] [Google Scholar]