Abstract
We have recently isolated and characterized a novel protein associated with Escherichia coli ribosomes and named protein Y (pY). Here we show that the ribosomes from bacterial cells growing at a normal physiological temperature contain no pY, whereas a temperature downshift results in the appearance of the protein in ribosomes. The protein also appears in the ribosomes of those cells that reached the stationary phase of growth at a physiological temperature. Our experiments with cell-free translation systems demonstrate that the protein inhibits translation at the elongation stage by blocking the binding of aminoacyl-tRNA to the ribosomal A site. The function of the protein in adaptation of cells to environmental stress is discussed.
INTRODUCTION
The involvement of ribosomes in low temperature adaptation of bacteria was implied a long time ago (Das and Goldstein, 1968; Friedman et al., 1969). It was reported that the ribosomal fraction of the bacterial cell is responsible for the arrest of translation at low temperatures (Das and Goldstein, 1968). Some antibiotics specifically affecting bacterial ribosomes were shown to mimic cold shock or heat shock response, and so ribosomes were claimed to be sensors of cold and heat shock in bacteria (VanBogelen and Neidhard, 1990). More recently, a ribosome-binding factor, RbfA, was discovered (Dammel and Noller, 1995) that proved to be a cold shock protein (Jones and Inouye, 1996). RbfA was shown to bind with 30S ribosomal subunit and suppress a cold-sensitive mutation in 16S ribosomal RNA (Dammel and Noller, 1995). Another cold shock protein, CsdA, was also characterized as a ribosome-associated protein, and its capacity to unwind double-stranded RNA was observed (Jones et al., 1996). The major cold shock protein of Escherichia coli, CspA (Goldstein et al., 1990), was reported to be an RNA-binding protein and qualified as an RNA chaperone (Jiang et al., 1997). A number of other cold shock proteins were discovered in bacteria (Graumann and Marahiel, 1996; Jones and Inouye, 1994). In no case, however, were the mechanisms and effects of cold shock protein on protein synthesis and cell growth clarified.
RESULTS AND DISCUSSION
The presence of the recently discovered ribosome-associated protein pY in ribosomes (Agafonov et al., 1999) was analysed depending on the conditions of the E. coli culture growth. To our surprise, the protein was not found in the ribosomal fraction of cells grown at the temperature for physiological activity in E. coli (37°C). The protein appeared in the ribosomal fraction when the cultivated cells were chilled rapidly to 15°C followed by incubation at the same temperature, or cooled gradually to 4°C. Figure 1 presents the area of the two-dimensional electrophoresis slab where pY migrates among ribosomal proteins. It can be seen that the proteins extracted from ribosomes of cells grown under normal physiological conditions contain very little, if any, pY (Figure 1A), whereas a significant amount of the protein is detected in the ribosomal fraction from cells subjected to a temperature downshift prior to harvesting (Figure 1B). The appearance pY in some of the preparations of ribosomes from cells grown at 37°C (Agafonov et al., 1999) seems to be the result of the cooling of cell culture during the harvesting of bacteria from the large-volume fermentation medium.
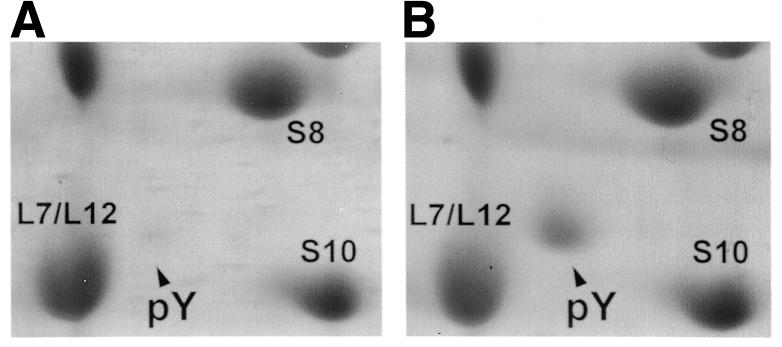
Fig. 1. Cold-induced synthesis of pY in E. coli cells. Shown are fragments of gel slabs stained with Coomassie Blue G 250. (A) Proteins from ribosomes of cells grown at 37°C. (B) Proteins from ribosomes of cells subjected to a temperature downshift.
It is noteworthy that the protein became detectable in the ribosomal fraction within the first hour of cold shock response and remained associated with ribosomes during the subsequent 4 h of the growth arrest phase. When growth was resumed the protein disappeared from ribosomes. At the same time it should be noted that the ribosomes from cells grown to stationary phase at the physiological temperature (37°C) contained the same amount of pY as ribosomes from cells subjected to temperature downshift (our data not shown). Hence, the protein expression or its binding to ribosomes was induced by environmental stress such as temperature downshift or excessive cell culture density. Comparing the intensities of protein spots in electrophoretic slabs it can be concluded that the molar ratio of pY to ribosomes varied from 1:10 to 1:3 depending on conditions.
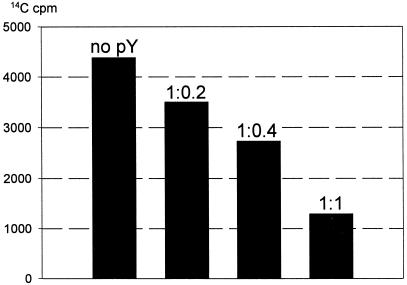
The isolation of pY, its selective binding to the 30S ribosomal subunit of the ribosome and its stabilizing effect on the ribosomal subunits association has been described in our previous communication (Agafonov et al., 1999). In order to test possible functional consequences of its physical interaction with the ribosome, cell-free translation experiments in the presence of increasing amounts of purified pY were performed. Figure 2 demonstrates that pY strongly inhibits cell-free translation of green fluorescent protein (GFP) mRNA in bacterial (E. coli) extract. The inhibition is dose-dependent and proportional to the amount of pY added, being ∼75–80% at the equimolar ratio relative to ribosomes.
Fig. 2. Effect of pY on cell-free translation of GFP mRNA. Molar ratio of ribosomes to pY is indicated above bars. The amount of newly synthesized protein was determined from the radioactivity of hot 10% trichloroacetic acid precipitate.
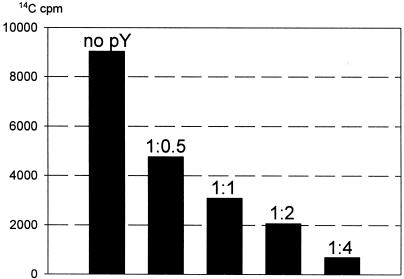
Translation of a natural mRNA consists of three main stages: initiation, elongation and termination. On the other hand, translation of a synthetic polynucleotide, such as polyuridylic acid [poly(U)], does not involve the mechanisms of initiation and termination, and is regarded as involving only elongation. Figure 3 shows the effect of pY on poly(U)-directed elongation of polyphenylalanine in a cell-free translation system. It can be seen that pY effectively inhibits elongation. The extent of inhibition is found to be approximately the same as in the case of the three-stage translation of natural mRNA: ∼65% inhibition is observed at equimolar amounts of pY and ribosomes, and the 4-fold excess of pY over ribosomes in the translation system leads to >90% inhibition. Hence, we can conclude that pY inhibits translation mainly at the elongation stage.
Fig. 3. Inhibition of poly(U)-directed polyphenylalanine elongation by pY. The molar ratio of ribosomes to pY is indicated above bars.
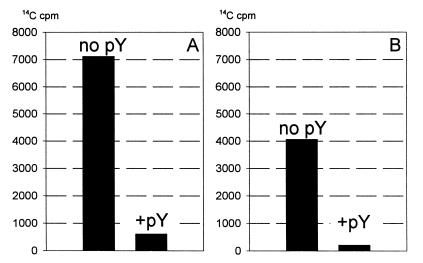
The elongation stage of translation is a sequence of elongation cycles, each of which can be divided into three consecutive steps—aminoacyl-tRNA binding, transpeptidation and translocation. Thus, each elongation cycle of the ribosome results in the selection of one aminoacyl-tRNA from the milieu, the formation of one peptide bond and the read-out of one codon of message. The question then arises, which of the partial steps of the elongation cycle is inhibited by pY? Figure 4 shows the results of testing the codon-cognate aminoacyl-tRNA binding with poly(U)-programmed 70S ribosomes in the absence and in the presence of pY. Both elongation factor Tu (EF-Tu)-promoted (‘enzymatic’) and ‘non-enzymatic’ binding of phenylalanyl-tRNA were tested. The ternary complex [14C]phenylalanyl-tRNAPhe–EF-Tu–GTP formed as described in the Methods was incubated with poly(U)-programmed 70S ribosomes, and the resultant enzymatic binding of [14C]phenylalanyl-tRNAPhe to the ribosome was measured. As seen in Figure 4A, the presence of pY strongly inhibits the enzymatic binding of the codon-cognate aminoacyl-tRNA to the A site of the 70S ribosome. Figure 4B shows that pY also suppresses the non-enzymatic binding of [14C]phenylalanyl-tRNAPhe with the 70S ribosome, to approximately the same extent as in the case of the enzymatic binding experiment.
Fig. 4. Blocking of aminoacyl-tRNA binding to the ribosomal A site by pY. (A) Binding of [14C]phenylalanyl-tRNAPhe–EF-Tu–GTP ternary complex to 70S ribosomes. (B) Binding of [14C]phenylalanyl-tRNAPhe to 70S ribosomes.
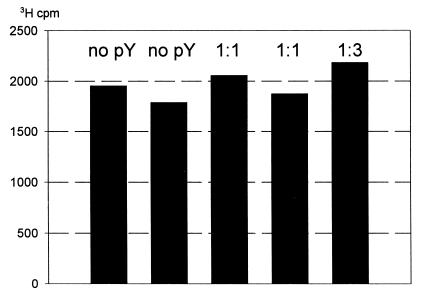
At the same time, experiments with radioactive tetracycline, a specific inhibitor of the ribosomal A site, have demonstrated that the antibiotic and pY bind to the ribosome independently. Tetracycline binding to the ribosome remained unaffected in the presence of excessive amounts of pY (Figure 5). As judged by ultrafiltration testing (Agafonov et al., 1999), the interaction of the protein with the ribosome was also shown to be resistant against teteracycline (see Supplementary data). Hence, both the protein and tetracycline do not interfere with each other when binding to the ribosome, thus implying their non-overlapping binding within the A site.
Fig. 5. Binding of [3H]tetracycline to 70S ribosomes in the presence of pY. The molar ratio of ribosomes to pY is indicated above bars.
The poly(U)-directed cell-free translation system can be made in such a way that either the catalyst of aminoacyl-tRNA binding, EF-Tu or the catalyst of translocation, elongation factor G (EF-G), are absent from the translation mixture. The result is that either aminoacyl-tRNA binding or translocation, respectively, pass via the non-enzymatic (factor-free) pathways (Gavrilova et al., 1976) and, hence, become the rate-limiting steps of the entire elongation process. In our experiments with the EF-Tu-promoted system, where translocation was the rate-limiting step, the addition of pY had no influence on the elongation rate. At the same time the presence of pY in the EF-G-catalysed system, where the aminoacyl-tRNA-binding step limited the rate of translation, inhibited elongation to the same extent as in the case of the full translation system (data not shown). Thus, the results confirm the conclusion that pY specifically affects the aminoacyl-tRNA-binding step, rather than other steps of the elongation cycle.
The observation that pY, a novel ribosome-associated stress protein, inhibits protein synthesis at the elongation stage of translation in cell-free systems is in agreement with the fact that cold shock leads to the decline of protein synthesis in vivo and to the arrest of bacterial growth (Das and Goldstein, 1968; Friedman et al., 1969). It also correlates well with the growth inhibition observed at the stationary phase of cell culture. It seems to be important that the protein appears in ribosomes at the early stage of cold shock response, mediating the overall adaptation to lower temperature. It should be noted that the protein appears in the ribosomal fraction only when cell growth is arrested, either during cold shock response or at the stationary phase of cell culture. When growth is resumed the protein disappears from ribosomes. It seems plausible that the protein function is to arrest translation in response to an environmental stress.
The results obtained are the first evidence of the direct functional interplay between the ribosome and a protein involved in cold shock response. The mechanism of pY action via the blocking of the ribosomal A site is in agreement with the assumption that the occupation of the A site triggers cold and heat response in bacteria (VanBogelen and Neidhard, 1990). The A site blocking function of pY is also consistent with the localization of pY at the ribosomal interface (Agafonov et al., 1999) since the intersubunit position of the ribosome-bound aminoacyl-tRNA is clearly revealed by recent crystallographic studies of ribosome functional complexes (Cate et al., 1999).
From the results reported here we propose to name the protein Y as ribosome associated inhibitor A (RaiA) and accordingly rename its open reading frame (yfia) to RaiA.
Genes homologous to the RaiA (pY) gene of E. coli can be found in various species of bacteria (Agafonov et al., 1999). The wide occurrence of RaiA homologues suggests that the protein may be important generally for survival under stress conditions. In any case, this novel protein seems to represent a direct link between ribosomes and adaptation to environmental stress.
METHODS
Bacterial strain and growth conditions
Cells of E. coli, strain MRE-600, were grown at 37°C in Luria-Bertani medium to a density of A590 0.5 (mid-log phase), subjected to a temperature downshift from 37 to 15°C and incubated for 1 h. Cell growth was then inhibited with antibiotics (rifampicin plus chloramphenicol) and sodium azide. Control cells received the inhibitors at 37°C and were pelleted immediately. Protoplasts were prepared from both the control and cold shocked cells, and lysates were obtained from protoplasts (Flessel et al., 1967). Ribosomes were pelleted from the lysates by centrifugation for 90 min at 430 000 g, and proteins of the ribosome fraction analysed by two-dimensional gel electrophoresis (Agafonov et al., 1999).
Cell-free translation of GFP mRNA
Translation in E. coli RNA-free S100 extract (Gold and Schweiger, 1971) was carried out at 30°C for 1 h in the presence of purified pY. The final concentration of the cell extract in the reaction mixtures was 0.6 mg/ml total extract protein. The reaction mixtures contained 1.5 mM ATP, 0.2 mM GTP, 0.6 mg/ml total E. coli tRNA, 0.01 mg/ml leucovorin, 5 mM phosphoenolpyruvate, 0.025 mg/ml pyruvate kinase and 18 µM amino acid each (except phenylalanine and leucine) in buffer A (8 mM MgCl2, 85 mM NH4Cl, 20 mM Tris–HCl pH 7.6, 0.1 mM EDTA and 1 mM DTT). Other components were 16 µM [14C]leucine and 16 µM [14C]phenylalanine, 0.11 µM 30S and 0.11 µM 50S ribosomal subunits, and 0.28 µM mRNA. Green fluorescent protein [GFP cycle 3 mutant (Crameri et al., 1996)] mRNA was synthesized in vitro with T7 RNA polymerase (Gurevich et al., 1991) from pMGFP plasmid linearized with EcoRI. The plasmid was constructed by the subcloning of the GFP cycle 3 gene from pBAD-GFP (Maxygene, Redwood City, CA) into pTU#58 (Chalfie et al., 1994), in place of the wild-type GFP gene.
Cell-free translation of poly(U)
Translation of poly(U) was performed as described in the previous section except that the concentration of S100 extract was 1 mg/ml, ribosomal subunits were added to a final concentration of 0.22 µM each, 0.2 mg/ml poly(U) was used instead of GFP mRNA, 16 µM [14C]phenylalanine and no other amino acids were present, and buffer A contained 95 mM NH4Cl instead of 85 mM NH4Cl.
Binding of aminoacyl-tRNA to the 70S ribosome
1.2 µM 70S ribosomes prepared as described previously (Agafonov et al., 1999) were first incubated at 37°C for 10 min with 2.4 µM pY in buffer A containing 100 mM NH4Cl instead of 85 mM NH4Cl. In order to form the aminoacyl-tRNA–EF-Tu–GTP ternary complex, EF-Tu–GDP was converted into EF-Tu–GTP by incubation of 17 µM EF-Tu–GDP with 5 mM phosphoenolpyruvate, 2 mM GTP and 0.2 µg/µl pyruvate kinase in buffer containing 10 mM MgCl2, 50 mM NH4Cl, 50 mM KCl, 50 mM Tris–HCl pH 7.6 and 10 mM β-mercaptoetanol (Ribeiro et al., 1995), and then [14C]phenylalanyl-tRNAPhe was added to a final concentration of 10 µM. The [14C]phenylalanyl-tRNAPhe–EF-Tu–GTP complex was added to the ribosomes preincubated with pY (see above) in the presence of 0.2 mg/ml poly(U). After another 10 min incubation at 37°C the mixture was filtered through a 0.45 µm nitrocellulose membrane and the filter-retained radioactivity measured. The non-enzymatic [14C]phenylalanyl-tRNAPhe binding tests were similarly conducted but EF-Tu was omitted from the reaction mixture.
Tetracycline binding assay.
1 µM 70S ribosomes were incubated at 37°C for 10 min with 50 µM [3H]tetracycline (NEN, 850 d.p.m./pmol) in the presence of 1 µM or 3 µM pY in buffer containing 10 mM MgCl2, 100 mM NH4Cl, 20 mM Tris–HCl pH 7.6, 0.1 mM EDTA and 1 mM DTT. After incubation the mixture was filtered through a 0.45 µm nitrocellulose membrane and the filter-retained radioactivity measured.
Supplementary data
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank K.S. Vassilenko for critical reading of the munuscript, A.V. Golubtsov, V.A. Shirokov and A. Kommer for helpful suggestions and comments, M.A. Posedko, I.R. Zakeeva and D.N. Ermolenko for participation in some experiments and A.P. Alimov for GFP mRNA preparation. This work was supported by grants 99-04-48087 and 00-15-97903 from the Russian Foundation for Basic Research and by the Russian Academy of Sciences.
References
- Agafonov D.E., Kolb, V.A., Nazimov, I.V. and Spirin, A.S. (1999) A protein residing at the subunit interface of bacterial ribosome. Proc. Natl Acad. Sci. USA, 96, 12345–12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Yusupov, M.M., Yusupova G.Zh., Earnest, T.N. and Noller, H.F. (1999) X-ray crystal structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu, Y., Euskirchen, G., Ward, W.W. and Prasher, D.C. (1994) Green fluorescent protein as a marker for gene expression. Science, 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Crameri A., Whitehorn, E.A., Tate, E. and Stemmer, W.P.C. (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nature Biotechnol., 14, 315–319. [DOI] [PubMed] [Google Scholar]
- Dammel C.S. and Noller, H.F. (1995) Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev., 9, 626–637. [DOI] [PubMed] [Google Scholar]
- Das H.K. and Goldstein, A. (1968) Limited capacity for protein synthesis at zero degrees centigrade in Escherichia coli. J. Mol. Biol., 31, 209–226. [DOI] [PubMed] [Google Scholar]
- Flessel C.P., Ralph, P. and Rich, A. (1967) Polyribosomes of growing bacteria. Science, 158, 658–660. [DOI] [PubMed] [Google Scholar]
- Friedman H., Lu, P. and Rich, A. (1969) Ribosomal subunits produced by cold sensitive initiation of protein synthesis. Nature, 223, 909–913. [DOI] [PubMed] [Google Scholar]
- Gavrilova L.P., Kostiashkina, O.E., Koteliansky, V.E., Rutkevitch, N.M. and Spirin, A.S. (1976) Factor-free (‘non-enzymic’) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol., 101, 537–552. [DOI] [PubMed] [Google Scholar]
- Gold L.M. and Schweiger, M. (1971) Synthesis of bacteriophage-specific enzymes directed by DNA in vitro. Methods Enzymol., 20, 537–542. [Google Scholar]
- Goldstein J., Pollitt, N.S. and Inouye, M. (1990) Major cold shock protein of Escherichia coli. Proc. Natl Acad. Sci. USA, 87, 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P. and Marahiel, M.A. (1996) Some like it cold: response of microorganisms to cold shock. Arch. Microbiol., 166, 293–300. [DOI] [PubMed] [Google Scholar]
- Gurevich V.V., Pokrovskaya, I.D., Obukhova, T.A. and Zozulya, S.A. (1991) Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal. Biochem., 195, 207–213. [DOI] [PubMed] [Google Scholar]
- Jiang W., Hou, Y. and Inouye, M. (1997) CspA, the major cold shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem., 272, 196–202. [DOI] [PubMed] [Google Scholar]
- Jones P.G. and Inouye, M. (1994) The cold shock response—a hot topic. Mol. Microbiol., 11, 811–818. [DOI] [PubMed] [Google Scholar]
- Jones P.G. and Inouye, M. (1996) RbfA, a 30S ribosomal binding factor, is a cold shock protein whose absence triggers the cold shock response. Mol. Microbiol., 21, 1207–1218. [DOI] [PubMed] [Google Scholar]
- Jones P.G., Mitta, M., Kim, Y., Jiang, W. and Inouye, M. (1996) Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S., Nock, S. and Sprinzl, M. (1995) Purification of aminoacyl-tRNA by affinity chromatography on immobilized Thermus termophilus EF-Tu–GTP. Anal. Biochem., 228, 330–335. [DOI] [PubMed] [Google Scholar]
- VanBogelen R.A. and Neidhard, F.C. (1990) Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl Acad. Sci. USA, 87, 5589–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.