Abstract
The regulation of the tou operon of Pseudomonas stutzeri OX1, for degradation of toluene and o-xylene via phenolic intermediates, has been faithfully reconstructed in vitro with purified proteins. The set-up included the prokaryotic enhancer-binding protein TouR, the σ54-dependent PToMO promoter and the σ54-containing RNA polymerase. With this system we prove that direct binding of 2-methylphenol (o-cresol) to TouR is the only regulatory step for activation of PToMO in response to aromatic effectors, thereby ruling out the involvement of other factors or a need for protein processing. In addition, we found that while TouR failed entirely to activate PToMO in the absence of inducers, the protein had per se a very significant ATPase activity, which was only moderately increased by o-cresol addition. The results presented here support the view that TouR-like proteins are particularly suitable as evolutionary assets to endow recently evolved pathways for the degradation of environmental pollutants with an optimal degree of transcriptional regulation.
INTRODUCTION
Bacterial strains of the genus Pseudomonas that colonize sites polluted with recalcitrant chemicals frequently evolve novel metabolic pathways for the biodegradation of otherwise toxic compounds (van der Meer et al., 1992). The regulation of such pathways is a key asset for the evolutionary success of any new catabolic operon (Díaz and Prieto, 2000). Hence, the elements involved in their transcriptional control reveal how bacteria acquire the ability to respond to recent and unusual carbon sources (Cases and de Lorenzo, 2001). Pseudomonas stutzeri OX1 is an isolate able to grow on toluene and o-xylene as the only carbon donor (Bertoni et al., 1998). This peculiar property is largely based on the activity of the so-called tou operon (touABCDEF), which encodes a multicomponent toluene and o-xylene mono-oxygenase (Bertoni et al., 1998). This enzyme converts o-xylene into the corresponding methyl-catechols, which are eventually channelled towards the Krebs cycle through a second metabolic pathway that involves the meta-cleavage of the catechol ring (Bertoni et al., 1998). The TouR protein is the regulator of the promoter PToMO of the tou operon in response to methylphenols (Arenghi et al., 1999). TouR belongs to the class of regulators generically known as the NtrC family of prokaryotic enhancer-binding proteins (EBPs). These factors activate at a distance promoters dependent on the alternative sigma factor σ54 (Gralla, 2000), to which type PToMO belongs. According to its sequence (Arenghi et al., 1999), TouR pertains to the subclass of EBPs whose N-terminal portion (A domain; Figure 1) interacts directly with the chemical inducer. This is followed by the relief of an intramolecular repression caused by the N-terminal module on the central, activating domain of the protein (Shingler, 1996). This group includes two well-studied activators (XylR and DmpR) and a growing list of structural variants found in γ-proteobacteria (for a recent inventory see Jaspers et al., 2000). Their central domain (C module) is the most conserved region amongst the protein family members. It is involved in the binding and hydrolysis of ATP that is at the basis of the activation of σ54 promoters (Weiss et al., 1991; Austin and Dixon, 1992; Morett and Segovia, 1993; Porter et al., 1993; Mettke et al., 1995; Shingler and Pavel, 1995; Wedel and Kustu, 1995).
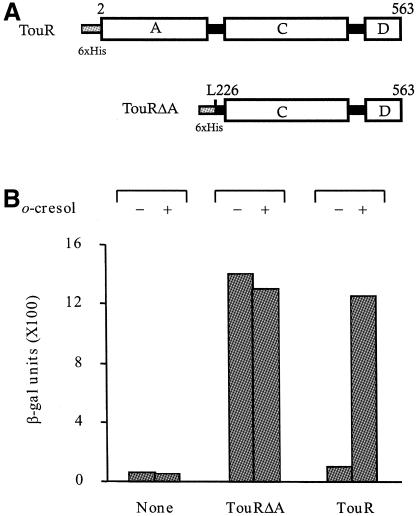
Fig. 1. In vivo properties of the TouR protein. (A) Functional domains of TouR (563 amino acids) and its A-domain-deleted derivative TouRΔA. The signal reception N-terminal module (called the A domain) is connected to the central C activating module, which bears an NTP binding motif. The A domain is deleted in the TouRΔA protein at amino acid L226, as indicated. The D module at the carboxyl end of the regulator includes a helix–turn–helix fold for DNA binding. To facilitate protein purification, a His6 tail was engineered at the N-terminus of either TouR variant. (B) Transcriptional activation in vivo of the σ54-dependent Pu promoter by TouRΔA and TouR. The reporter strain E. coli CC118Pu-lacZ (bearing a chromosomally inserted lacZ fusion to the Pu promoter of the TOL plasmid of P. putida mt-2) was transformed with either pHNΔAR (touRΔA+) or pTRR6N (touR+). Each of the transformants was grown at 37°C in LB medium with ampicillin (200 µg/ml) until cultures reached an OD600 of 1.4. o-cresol (1 mM) was then added and the incubation continued for 2 h. Accumulation of β-galactosidase in each strain, measured in Miller’s units (Miller, 1972), is shown.
One intriguing feature of this type of effector-inducible, single-component EBP is how the binding of the aromatic effector gives rise to a protein that activates cognate promoters to the same extent as the protein entirely deleted of the A domain (Fernández et al., 1995; Pérez-Martín and de Lorenzo, 1996b; Shingler, 1996). In other words, what is the effect of inducer binding that is at the basis of the regulatory quality of the protein? The current model proposes that inducer binding suffices to hinder the inter-A–C domain repression, that such a hindrance triggers an ATPase activity, and that this is instantly translated into transcription initiation (Delgado and Ramos, 1994; Shingler and Moore, 1994; Pérez-Martín and de Lorenzo, 1995, 1996a,b; Shingler and Pavel, 1995; Neuhard and Nygaard, 1987; O’Neill et al., 1998, 1999; Skarfstad et al., 2000). However, this could never be tested directly because of the lack of suitable assay systems where transcription and ATP hydrolysis could be followed in parallel. In this context, we found the TouR protein to be particularly attractive since its properties in vivo are virtually identical to those of DmpR or XylR, while it can be purified in large amounts in a soluble and fully active form (see below).
In this report, we describe for the first time the faithful reproduction in vitro of the entire effector-responsive transcription activation of a σ54 promoter by a xenobiotic-responsive EBP. The data shown below rule out the involvement of additional factors in the transcriptional activation of the PToMO promoter in vivo. In addition, the results suggest that the key regulatory effect of inducer binding to TouR may be unrelated to the increase in ATPase activity. Taken together, the findings reported here reveal the adequacy of this type of EBP for regulating biodegradative pathways that evolve rapidly in response to recalcitrant chemicals in the environment.
RESULTS AND DISCUSSION
TouR is an archetype of effector-inducible EBPs
Since the nucleotide sequence of TouR (Arenghi et al., 1999) indicates that it is a regulator of the NtrC family, homologous to XylR and DmpR, a reasonable guess is that its mechanism of activation by methylphenols will also be similar to that of XylR, or DmpR (Shingler, 1996). The main attribute of this class of regulators is the acquisition of a fully constitutive phenotype when such a domain A is deleted. To confirm this, we produced expression plasmids pTRR6N and pHNΔAR. These encoded, respectively, the predicted full-length TouR and its ΔA derivative truncated at position L266 of the amino acid sequence (Figure 1A). To examine whether the response of the tou genes to 2-methylphenol (o-cresol) observed in P. stutzeri OX1 could be unequivocally traced to TouR, we transformed Escherichia coli strain CC118Pu-lacZ with pTRR6N (touR+). This strain bears a chromosomal insertion of a reporter fusion to the σ54 Pu promoter of the TOL plasmid. The upstream activating sequence (UAS) of these promoters can be bound by a variety of XylR/DmpR regulators (Inouye et al., 1988; Shingler et al., 1993; Fernández et al., 1994; Arenghi et al., 1999). Any β-galactosidase accumulated by such a strain can thus be unequivocally traced to the activity of the regulator encoded by the plasmid. Figure 1B shows the result of growing E. coli strain CC118Pu-lacZ in the presence or absence of o-cresol. Levels of β-galactosidase became significant only when the cultures were added with the aromatic effector, thus meaning that TouR suffices to cause such an induction of the reporter system. In a different experiment, E. coli CC118Pu-lacZ was transformed with pHNΔAR (touRΔA+) and examined in induction experiments with o-cresol as before. The data of Figure 1B show that regardless of the presence of the aromatic compound, the Pu promoter was activated to the same level as it was by the full-length protein with o-cresol. These experiments proved that TouR grossly behaved in vivo as XylR and DmpR, and can thus be considered a suitable model to examine general features of the activation mechanism for this type of σ54-dependent regulator, which responds to environmental pollutants.
TouR has an intrinsic ATPase activity that is stimulated by o-cresol
One of the pillars of the current model to account for the mechanism of transcriptional activation by EPBs of the XylR/DmpR type is that effector addition triggers the ATPase activity, which is then coupled to the formation of an open complex (Pérez-Martín and de Lorenzo, 1995; Shingler, 1996). This is largely based on the observation that purified DmpR bound to a solid matrix shows an increased ATPase activity when exposed to o-cresol (Shingler and Pavel, 1995). This fits the separate observation that ATP binding to the C-domain of XylR (i) triggers the multimerization of the protein at the promoter UAS (Pérez-Martín and de Lorenzo, 1996a), (ii) increases the ATPase activity (Mettke et al., 1995; Farez-Vidal et al., 1996; Pérez-Martín and de Lorenzo, 1996a) and (iii) sustains the regulator multimer that activates σ54 RNA polymerase (RNAP) (Weiss et al., 1991; Porter et al., 1993; Pérez-Martín and de Lorenzo, 1996a). To re-examine these assumptions of the model, we performed a series of assays to monitor the effects of o-cresol on the ability of purified TouR and TouRΔA to hydrolyse ATP, by themselves or in combination with UAS DNA.
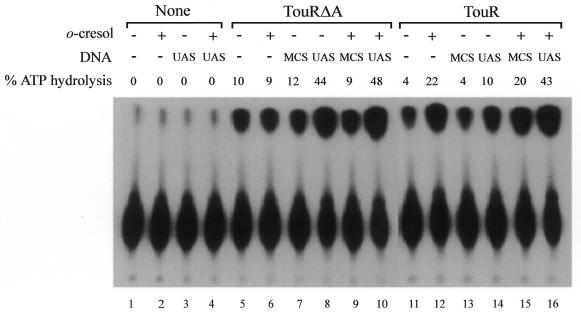
The results of the ATP hydrolysis caused by each protein and condition are presented in Figure 2. The controls with TouRΔA showed that (similarly to XylRΔA; Pérez-Martín and de Lorenzo, 1996b) the presence of UAS DNA borne by plasmid pTE128 specifically increased by 4-fold the already notable levels of ATP hydrolysis (Figure 2, lanes 7 and 8), which were not affected by o-cresol. The behaviour of wild-type TouR in this type of assay was, however, unexpected. First, the protein had per se a significant ATPase activity, which was stimulated by UAS DNA even in the absence of any inducer. o-cresol separately stimulated the ATPase activity of TouR as well, but levels of hydrolysed ATP were only twice as high (lane 12 versus 14). The combination of the two (o-cresol and UAS) increased the ATP hydrolysis caused by TouR up to the maximum level registered for TouRΔA. That the sole addition of UAS to TouR (without effector, lane 14) induces as much as a quarter of the ATP hydrolysis of the fully active TouRΔA (lane 9) would argue against a direct correlation between ATPase activity and transcriptional activation. A side consequence of these experiments is the relatively minor role of the DNA in the stimulation of the ATPase activity of TouR, at least under the conditions of the experiment. o-cresol addition increased 4–5 times the level of ATP hydrolysed by TouR in the absence of DNA (Figure 2, lanes 11 and 12). Effector-dependent derepression of TouR may thus occur prior to multimerization of the EBP at the UAS of the PToMO promoter.
Fig. 2. Monitoring ATPase activity of TouR and TouRΔA. Mixtures of ATP (0.02 µCi/µl [γ-32P]ATP, 3.0 mM ATP), TouRΔA or TouR (500 nM), the 208 bp EcoRI–HindIII fragment of pTE218 (UAS DNA) or a 146 bp fragment containing the multicloning site of pUC18 (MCS; 250 nM) were incubated for 30 min at 30°C and spotted onto TLC slides to separate Pi from non-hydrolysed ATP. Where indicated, samples were amended with 100 µM of o-cresol. The figures for ATPase activities are expressed as the percentage of labelled Pi released from the total [γ-32P]ATP added to the reaction and corrected with the background value of the controls (lanes 1–4). A value of 50% corresponds to a specific activity of ∼210 nmol of hydrolysed ATP/min/nmol of dimeric TouR or TouRΔA.
Different effectors cause a distinct response in the ATPase activity of TouR
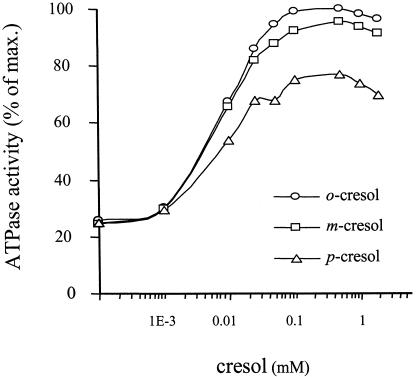
The PToMO promoter is activated in vivo to various degrees in response to different mono-methylated and di-methylated phenols (Arenghi et al., 1999). The non-equal responses to each of the inducers could be attributed to (i) variations in inducer permeability, (ii) different affinities for TouR and (iii) distinct capacity of each effector to derepress TouR. To investigate this issue, we produced dose–response curves for the ATPase activity of TouR with increasing concentrations of each of the three cresols. As shown in Figure 3, each isomer elicited a given maximum level of stimulation. The effector dose required to reach 50% of the maximum ATP hydrolysis caused by the protein was approximatively the same (∼10 µM) for the three cresol isomers, thereby suggesting a similar apparent affinity for the regulator. At higher concentrations of the effectors, all curves in Figure 3 showed a clear inhibition, probably due to the damage caused by phenolic compounds to the protein in the assay. Through the lower effector concentration range, it did appear, however, that each compound caused a distinct degree of stimulation of the ATPase activity of the protein. Hence, it seems that, following binding, every inducer enters a different signal in the protein, which translates into a certain specific activity. When other weaker inducers were tested [i.e. 2,3 dimethylphenol (DMP), 3,4 DMP and 2,4 DMP], the stimulation of the ATPase activity in vitro had a reflection in the induction of PToMO in vivo (not shown). This result in vivo was somehow contradictory with the data of Figure 2, in which effector addition did not parallel ATPase activity in vitro. To settle this discrepancy, we set out to assemble a complete, coupled, in vitro transcription assay in which effector addition could be directly correlated with the production of mRNA.
Fig. 3. Dose-dependent stimulation of the ATPase activity of TouR by o-cresol, m-cresol and p-cresol. TouR (500 nM) was mixed with ATP (0.02 µCi/µl [γ-32P]ATP, 3 mM ATP) and UAS DNA (250 nM) in the presence of increasing concentrations of each of the cresols. After incubation for 30 min at 30°C, samples were spotted onto TLC plates as in Figure 2 to monitor ATP hydrolysis. Activities are expressed as the percentage of the hydrolysis of ATP achieved by TouR with 100 µM o-cresol added.
o-cresol enables wild-type TouR to activate the PToMO promoter in vitro
The ability of TouR and TouRΔA proteins to activate the PToMO promoter was examined with a reaction mixture containing supercoiled plasmid pTE128, σ54 and core RNAP. The test plasmid contains the PToMO promoter region cloned in front of a T7 terminator. Figure 4 shows the results of the experiment. As a control, lanes 3 and 4 show the maximum accumulation of transcripts brought about by the constitutive TouRΔA protein, the activity of which was unaffected by addition of o-cresol. On the contrary, lanes 4 and 5 of Figure 4 show that the wild-type TouR was unable to activate per PToMO in the absence of o-cresol. No band corresponding to the transcript under scrutiny could be detected in lane 5, even after prolonged exposure of the gel, thus suggesting that TouR is basically inert for transcription in the absence of effectors. On the contrary, addition of o-cresol revived TouR activity to produce PToMO transcripts at the levels generated by the constitutive TouRΔA. The TouR-mediated control of transcription in response to o-cresol occurred in vitro at physiological concentrations of nucleoside triphosphate (2.5–3.0 mM; Neuhard and Nygaard, 1987). Until now, such an effector-dependent transcriptional activation of a promoter of this type had not been fully reconstructed in vitro, since so far only the ATPase activity of DmpR (and TouR, see above) could be shown to be inducer dependent (Shingler and Pavel, 1995). This is, thus, the first demonstration of effector-responsive transcription activation in vitro by a member of the aromatic-responsive subgroup of σ54 regulators. The otherwise simple experiment of Figure 4 has three important consequences. First, it shows unequivocally that direct effector binding to TouR is the only regulatory step required for activation of PToMO in response to aromatic effectors, thereby ruling out the involvement of other factors or a need for protein processing in vivo. Secondly, it proves that TouR is not a semi-constitutive protein, as could be predicted both from its significant basal level of ATPase activity (Figure 2) and the logic of the regulatory circuit of the touABCDEF operon of P. stutzeri OX1 (Arenghi et al., 1999). Thirdly, it affirms that under conditions of substantial ATP hydrolysis (close to a quarter of the full potential of TouR; Figure 2), the PToMO promoter remains completely silent in the absence of o-cresol.
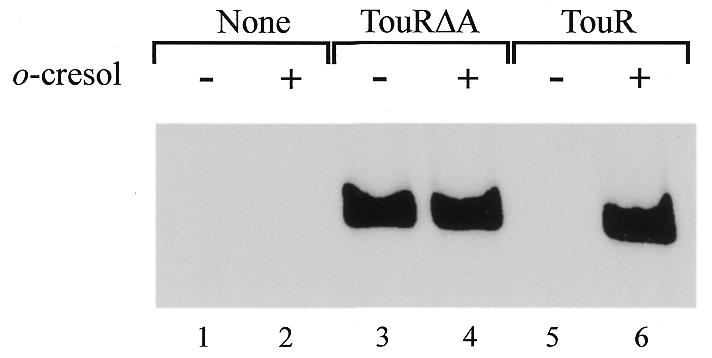
Fig. 4. In vitro activation of the σ54-dependent PToMO promoter by TouRΔA or TouR. Supercoiled plasmid pTE218 bearing the PToMO sequence was mixed with 0.5 U of core RNAP of E. coli, 150 nM purified σ54 factor from P. putida, 2.5 mM ATP and 250 nM TouRΔA or TouR and subjected to a multiple-round transcription reaction as explained in Methods. Where indicated, the inducer o-cresol was added to a final concentration of 100 µM. Transcription of the PToMO promoter in the template employed generates an mRNA of 311 nucleotides, which becomes apparent in the autoradiograph of the gel shown.
Conclusion
The results of this work reveal some interesting facets of the existing model for xenobiotic-responsive regulators that belong to the σ54-dependent class. On one hand, our data support the notion that the main regulatory event brought about by inducer binding to TouR is the coupling of an ongoing ATPase activity to the transcriptional machinery. In the wild-type protein, such a coupling is probably inhibited by the A domain, which could keep the protein in a form unable to oligomerize, yet still able to bind and hydrolyse ATP. A second facet of this work is that TouR and related factors (e.g. DmpR, XylR) turn out to be archetypes of built-in protein multifunctionality in the prokaryotic world. Just one transcriptional factor (TouR) is able to bind target DNA in the UAS of the cognate promoter, recognize the substrate of the biodegradative pathway (or one of its intermediate metabolites), and elicit a whole collection of intramolecular events that end up in activation of the σ54-RNAP. As shown in Figure 4, all this occurs without any extra factor or signal transduction pathway. We believe that such a property makes this type of protein optimal for rapidly acquiring a responsiveness to novel effectors, as is obligatory for the evolution of recent catabolic operons (Cases and de Lorenzo, 2000).
METHODS
Overproduction and in vivo assays for TouR and TouRΔA. Both TouR and TouRΔA were purified by metalloaffinity chromatography of the proteins, engineered at their N-termini with a His6 label. Such a tag was entered in TouR (Arenghi et al., 1999) by PCR with primers 5′-CGGAATTCATCACCATCACCATCACGCAACAAGCTATAAGCCC-3′ (EcoRI site in bold) and 5′-CCCAAGCTTTCAGGCTTCAGAAAAAATGCC-3′ (HindIII site in bold). The amplified fragment was digested with EcoRI and HindIII, and cloned in expression plasmid pTrc99B (Amann et al., 1988), generating plasmid pTRR6N. Similarly, the TouRΔA protein sequence was His tagged through amplification with primers 5′-CGGGATCCGCTTGAG AAACAGCAGGGCCAG-3′ (BamHI site in bold) and 5′-CCCAAGCTTTCAGGCT TCAGAAAAAATGCC-3′ (HindIII site in bold) of the touR coding region spanning from L226 to the stop codon. The PCR product was digested with BamHI and HindIII, and cloned between the corresponding sites of plasmid pQE10 (Qiagen Inc.), generating plasmid pHNΔAR. This placed codon 226 of TouR in-frame with a tail of the six histidines of the multicloning site (MCS) of the vector. To test the functionality of the His-tagged proteins in vivo, the Pu-lacZ reporter strain E. coli CC118Pu-lacZ was transformed with either pTRR6N (touR+) or pHNΔAR (touRΔA+), along with the compatible Kmr and lacI+ plasmid pREP4. Each of the strains was grown at 37°C in LB medium (Sambrook et al., 1989) with ampicillin (Ap; 200 µg/ml) and kanamycin (Km; 50 µg/ml) where needed, until cultures reached an OD600 of 1.4. o-cresol (1 mM) was then added and the incubation continued for 2 h. Accumulation of β-galactosidase in each strain was measured with the procedure of Miller (1972) on cells permeabilized with SDS and chloroform.
Purification of active TouR and TouRΔA proteins. TouR and TouRΔA were prepared in an active and soluble form by a metalloaffinity purification procedure, as follows. Strains E. coli CC118Pu-lacZ (pTRR6N) and E. coli CC118Pu-lacZ (pHNΔAR, pREP4) were grown at 30°C in LB medium amended with suitable antibiotics up to an OD600 of 0.6, at which point 0.1 mM isopropyl-β-d-thiogalactopyranoside was added. Incubation was continued overnight and the cells of each litre of culture were collected by centrifugation, resuspended in 20 ml of buffer A (20 mM Na phosphate pH 8.0, 0.5 M NaCl, 0.1% v/v Triton X-100, 10% v/v glycerol, 1.0 mM β-mercaptoethanol, 10 mM imidazole) and passed twice at 5°C through a French press. The lysate was cleared for 1 h at 24 000 g for 60 min. The supernatant was then diluted 3-fold with the same buffer and mixed with 2 ml of a slurry of Talon metal affinity resin (Clontech Inc.) in buffer A. Following 2 h at 4°C, the resin was collected at 700 g for 10 min, washed twice with 20 ml of buffer A, and packed in a small chromatographic column. Proteins bound to the resin were eluted with a linear gradient of imidazole (10–100 mM). Most of the His-tagged protein peaks eluted from the column between 50 and 80 mM. Fractions containing TouR or TouRΔA were pooled, dialysed against buffer B (20 mM Na phosphate pH 8.0, 0.5M NaCl, 0.1% v/v Triton X-100, 30% v/v glycerol, 1.0 mM β-mercaptoethanol) and stored in aliquots at –80°C until use at an estimated 95% purity and concentrations of 0.8 mg/ml (TouR) and 2 mg/ml (TouRΔA).
In vitro transcription assays. Plasmid pTE218, used as the supercoiled template for the assays, was obtained by cloning a 208 bp EcoRI–HindIII fragment spanning positions –185 to +23 of the PToMO promoter in vector pTE103 (Elliott and Geiduschek, 1984). This places the promoter upstream of a T7 terminator and allows the formation of a 311 nucleotide transcript. Reactions of 25 µl were set up by mixing on ice 5 nM DNA template with 0.5 U of E. coli core RNAP (Epicentre Technologies), 150 nM P. putida σ54 factor, 2.5 mM ATP in 35 mM Tris–acetate pH 8.0, and 250 nM TouR or TouRΔA dimers in a buffer containing 5 mM MgAc2, 70 mM KAc, 20 mM NH4Ac, 1 mM dithiothreitol (DTT). Reactions were run as described in Pérez-Martín and de Lorenzo (1996b). Where specified, 100 µM o-cresol was added as well. These mixtures were incubated for 5 min at room temperature and then at 30°C for 10 min, before initiating the transcription reactions by the addition of [α-32P]UTP (0.5 µCi; 6000 Ci/mmol), 12 µM cold UTP, and 400 µM ATP, CTP and GTP. Ten minutes later, the reactions were stopped by adding an equal volume of stop mix (5 M NH4Ac, 100 mM EDTA, 0.5 mg/ml carrier tRNA) and precipitated with 2 vols of ethanol. The reactions were analysed in denaturing gels with 7 M urea–4% polyacrylamide gel and autoradiographed on X-ray films.
Monitoring ATP hydrolysis. Hydrolysis of ATP was examined as described in Pérez-Martín and de Lorenzo (1996b). To this end, 25 µl samples were set up in a buffer (35 mM Tris–acetate pH 8.0, 5 mM MgAc2, 70 mM KAc, 20 mM NH4Ac, 1.0 mM DTT) with 500 nM TouR or TouRΔA added. Where indicated, the 208 bp EcoRI–HindIII insert of pTE218 containing the PToMO promoter, or a 146 bp DNA fragment including pUC18 MCS, was added at 250 nM. Reactions were also carried out in the presence of aromatic compounds at the concentrations indicated in each case. Following incubation at 30°C for 5 min, the assays were started by adding 2.5 µl of an ATP-Mg mix (0.2 µCi/µl [γ-32P]ATP + 30 mM ATP-Mg) and incubated further for 30 min at 30°C. The reactions were stopped with 25 µl of 1.0 M formic acid. Free Pi was separated from nucleotide mono-, di- and triphosphates by spotting 5 µl of the samples on polyethyleneimine–cellulose TLC plates (Cel 300/PEI/UV254; Macherey and Nagel Inc.) and running them with a buffer containing 0.4 M K2HPO4 and 0.7 M boric acid. Following chromatography, TLC plates were dried and autoradiographed on X-ray film or exposed to a PhosphorImager screen system for quantification.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to J. Pérez-Martin for inspiring discussions and to F. Bartels for the gift of purified σ54 from P. putida. This work was supported by grant CT99.00287.PF49.115.30637 of the Italian Consiglio Nazionale delle Ricerche (Rome) to P.B., Contracts QLK3-CT20000-00170 and QLRT-99-00041 of the EC, and grant BIO98-0808 of the Spanish CICYT to V.d.L.
References
- Amann E., Ochs, B. and Abel, K.J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli.Gene, 69, 301–315. [DOI] [PubMed] [Google Scholar]
- Arenghi F.L., Pinti, M., Galli, E. and Barbieri, P. (1999) Identification of the Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase regulatory gene (touR) and of its cognate promoter. Appl. Environ. Microbiol., 65, 4057–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S. and Dixon, R. (1992) The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J., 11, 2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni G., Martino, M., Galli, E. and Barbieri, P. (1998) Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol., 64, 3626–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I. and de Lorenzo, V. (2001) The black cat/white cat principle of signal integration in bacterial promoters. EMBO J., 20, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A. and Ramos, J.L. (1994) Genetic evidence for activation of the positive transcriptional regulator Xy1R, a member of the NtrC family of regulators, by effector binding. J. Biol. Chem., 269, 8059–8062. [PubMed] [Google Scholar]
- Díaz E. and Prieto, M.A. (2000) Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotech., 11, 467–475. [DOI] [PubMed] [Google Scholar]
- Elliott T. and Geiduschek, E.P. (1984) Defining a bacteriophage T4 late promoter: absence of a ‘–35’ region. Cell, 36, 211–219. [DOI] [PubMed] [Google Scholar]
- Farez-Vidal M.E., Wilson, T.J., Davidson, B.E., Howlett, G.J., Austin, S. and Dixon, R.A. (1996) Effector-induced self-association and conformational changes in the enhancer-binding protein NTRC. Mol. Microbiol., 22, 779–788. [DOI] [PubMed] [Google Scholar]
- Fernández S., Shingler, V. and de Lorenzo, V. (1994) Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J. Bacteriol., 176, 5052–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández S., de Lorenzo, V. and Pérez-Martín, J. (1995) Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol. Microbiol., 16, 205–213. [DOI] [PubMed] [Google Scholar]
- Gralla J. (2000) Signaling through σ. Nature Struct. Biol., 7, 530–532. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa, A. and Nakazawa, T. (1988) Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene, 66, 301–306. [DOI] [PubMed] [Google Scholar]
- Jaspers M.C., Suske, W.A., Schmid, A., Goslings, D.A., Kohler, H.P. and van der Meer, J.R. (2000) HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol., 182, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettke I., Fiedler, U. and Weiss, V. (1995) Mechanism of activation of a response regulator: interaction of NtrC-P dimers induces ATPase activity. J. Bacteriol., 177, 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Morett E. and Segovia, L. (1993) The σ 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol., 175, 6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J. and Nygaard, P. (1987) Purines and pyrimidines. In Neidhardt, F.C., Ingraham, J.L., Low, K.B., Magasanik, B., Schaechter, M. and Umbarger, H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 445–473.
- Ng L.C., O’Neill, E. and Shingler, V. (1996) Genetic evidence for interdomain regulation of the phenol-responsive final σ54-dependent activator DmpR. J. Biol. Chem., 271, 17281–17286. [DOI] [PubMed] [Google Scholar]
- O’Neill E., Ng, L.C., Sze, C.C. and Shingler, V. (1998) Aromatic ligand binding and intramolecular signalling of the phenol-responsive σ54-dependent regulator DmpR. Mol. Microbiol., 28, 131–141. [DOI] [PubMed] [Google Scholar]
- O’Neill E., Sze, C.C. and Shingler, V. (1999) Novel effector control through modulation of a pre-existing binding site of the aromatic-responsive σ54-dependent regulator DmpR. J. Biol. Chem., 274, 32425–32432. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo, V. (1995) The N-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc. Natl Acad. Sci. USA, 92, 9392–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo, V. (1996a) ATP binding to the σ54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell, 86, 331–339. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo, V. (1996b) In vitro activities of an N-terminal truncated form of XylR, a σ54-dependent transcriptional activator of Pseudomonas putida.J. Mol. Biol., 258, 575–587. [DOI] [PubMed] [Google Scholar]
- Porter S.C., North, A.K., Wedel, A.B. and Kustu, S. (1993) Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev., 7, 2258–2273. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shingler V. (1996) Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol. Microbiol., 19, 409–416. [DOI] [PubMed] [Google Scholar]
- Shingler V. and Moore, T. (1994) Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J. Bacteriol., 176, 1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingler V. and Pavel, H. (1995) Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol. Microbiol., 17, 505–513. [DOI] [PubMed] [Google Scholar]
- Shingler V., Bartilson, M. and Moore, T. (1993) Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J. Bacteriol., 175, 1596–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarfstad E., O’Neill, E., Garmendia, J. and Shingler, V. (2000) Identification of an effector specificity subregion within the aromatic-responsive regulators DmpR and XylR by DNA shuffling. J. Bacteriol., 182, 3008–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J.R., de Vos, W.H., Harayama, S. and Zehnder, A. (1992) Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev., 56, 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A. and Kustu, S. (1995) The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev., 9, 2042–2052. [DOI] [PubMed] [Google Scholar]
- Weiss D.S., Batut, J., Klose, K.E., Keener, J. and Kustu, S. (1991) The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell, 67, 155–167. [DOI] [PubMed] [Google Scholar]