Abstract
The Caenorhabditis elegans excretory cell extends tubular processes, called canals, along the basolateral surface of the epidermis. Mutations in the exc-5 gene cause tubulocystic defects in this canal. Ultrastructural analysis suggests that exc-5 is required for the proper placement of cytoskeletal elements at the apical epithelial surface. exc-5 encodes a protein homologous to guanine nucleotide exchange factors and contains motif architecture similar to that of FGD1, which is responsible for faciogenital dysplasia. exc-5 interacts genetically with mig-2, which encodes Rho GTPase. These results suggest that EXC-5 controls the structural organization of the excretory canal by regulating Rho family GTPase activities.
INTRODUCTION
One of the central questions in animal development is how the formation of organ structure during development is controlled. Genetic approaches in model organisms such as the nematode Caenorhabditis elegans have been useful in providing new insights into the molecular mechanisms controlling organ structure formation.
The C. elegans excretory system is believed to regulate osmoregulation and waste elimination, analogous to the renal organs of higher animals. The excretory cell, and its associated gland, duct and pore cells, form the C. elegans renal system (Chitwood and Chitwood, 1974; Nelson et al., 1983; Broeks et al., 1995). The excretory cell is a large ectodermal cell located beneath the posterior pharyngeal bulb, and it extends tube-like canals dorsally on both sides during embryogenesis (Hedgecock et al., 1987). Upon reaching the lateral epidermis, these canals bifurcate and grow anteriorly and posteriorly for nearly the length of the animal, creating an H-shaped canal system (Figure 1A). The longer, posterior canal reaches epidermoblast V3 by the time of hatching, and V6 by the end of the first larval stage (L1), thus completing its active outgrowth. Active outgrowth at the tip of the canal accounts for less than one-third of the adult canal length. The remaining growth occurs along the length of the canal, as the hypodermal cells, to which the canal is connected by gap junctions, expand. The C. elegans excretory cell provides a unique system to study the formation of organ structure in a unicellular epithelium.
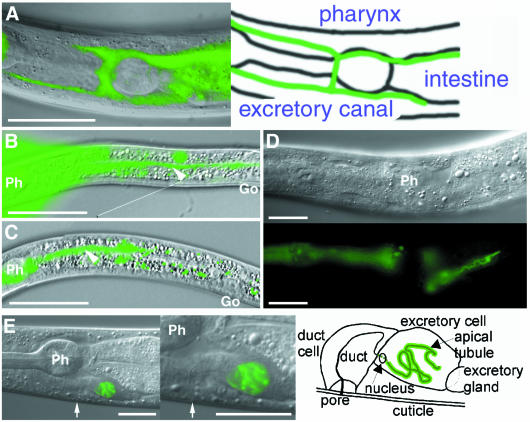
Fig. 1. The exc-5 gene is required for formation of excretory canals. (A) Diagram showing the position of excretory canals. The apical surface is shown in pink, the basolateral in blue. (B–E) The excretory canal (arrowheads) is visualized using sek-1::GFP expression. The photos show posterior excretory canals. Cysts are shown by arrows. Vu, vulva. (B) unc-119(e2498) with sek-1:GFP and pDP#MM016B (unc-119) (Maduro and Pilgrim, 1995). (C) exc-5(km508) with sek-1::GFP and pRF4 (rol-6d). (D) exc-5(rh232) with sek-1::GFP and pRF4 (rol-6d). (E) exc-5(rh232); mig-2(gm38) with sek-1::GFP and pRF4. The right panel shows the excretory canal at high magnification. The cyst (arrow) is much smaller than that in exc-5(rh232) single mutants. Scale bars, 20 µm.
In this study, we show that the exc-5 gene is required for the formation of the excretory canal structure. exc-5 encodes a protein homologous to guanine nucleotide exchange factors (GEFs) and contains motif architecture similar to that of FGD1, which is responsible for faciogenital dysplasia or Aarskog–Scott syndrome (Pasteris et al., 1994). Our results suggest that EXC-5 controls the structural organization of the excretory canal by regulating activity of Rho family GTPases.
RESULTS AND DISCUSSION
To understand how the formation of the canal structure is determined, mutants with abnormal excretory canals were isolated using the Tc1 mutator strain NL917. One isolate, km508, exhibits extremely large septate cysts, formed predominantly at the termini of shortened canals (Figure 1C). We mapped the km508 allele to the right arm of linkage group IV using STS mapping (Williams et al., 1992). Among the genes in this region lies exc-5, previously isolated in a screen for mutations with tubulocystic defects in the excretory canal (Buechner et al., 1999). The similarity in phenotype and map position suggested that the km508 mutation might be located in the exc-5 gene. Consistent with this observation, km508 failed to complement exc-5(rh232).
Electron microscopy analysis revealed that the canals in wild-type animals are embedded in the hypodermis and are spread extensively along the basement membrane that separates the hypodermis from the pseudocoelomic space (Figure 2A). The central lumen is surrounded by electron-dense material, presumably actin-based cytoskeleton (Figure 2B) (Nelson et al., 1983). In a large exc-5 cyst, the electron-dense cytoskeletal material appears disrupted, and no longer evenly surrounds the lumen. In addition, the lumen swells immensely (Figure 2C). These defects are consistent with a defect of the cytoskeleton at the apical membrane required for organizing cell shape, and raise the possibility that the EXC-5 protein is required for the proper placement of cytoskeletal elements at the apical epithelial surface.
Fig. 2. Electron micrographs of excretory canals. (A) Schematic section showing an excretory canal. (B) Cross-section of the posterior canal in wild-type animals. A band of electron-dense material (arrowhead) decorates the cytoplasmic face of the membrane surrounding the lumen (lu; arrow) scale bar, 1 µm. (C) Cross-section of a large canal cyst in exc-5(rh232) animals. Apical electron-dense material (arrowheads) is irregularly distributed, and no longer evenly surrounds the lumen. The lumen (lu) swells immensely scale bar, 1 µm. (D) Cross-section of short canal in wild-type animals overexpressing exc-5::GFP. Three lumena (lu; arrows) are visible and they are evenly surrounded by electron-dense materials (arrowheads). The canaliculi are much longer than normal. All panels show transverse views, with lateral side at the top. Scale bar, 0.5 µm.
We cloned the exc-5 gene by transposon tagging. This gene corresponds to the predicted C33D9.1 gene identified by the C. elegans sequencing consortium. A 13.7-kb genomic DNA fragment that contains both regulatory and coding sequences of C33D9.1 was found to rescue the exc-5(km508) mutant phenotype. Analysis of the exc-5 cDNA indicated that the exc-5 gene has 15 exons (Figure 3A) and encodes a single protein of 826 amino acids (Figure 3B). To confirm that we had cloned the correct gene, we sequenced the genomic DNA of three exc-5 mutant alleles: km508, rh232 and n2672 (Figure 3). km508 has a Tc1 transposon insertion at Cys758. The rh232 mutation is a deletion that removes at least the first 12 exons of exc-5 and thus is presumably a null allele. n2672 is a nonsense mutation occurring at Trp604.
Fig. 3. The exc-5 gene and protein. (A) The exon–intron structure of exc-5. This structure is deduced by comparing genomic and cDNA sequences. Boxes represent exons, lines represent introns, and black boxes represent the coding region. The positions and type of the three exc-5 mutations are indicated. (B) Deduced EXC-5 protein sequence. Asterisks indicate the positions of two exc-5 mutations: stop codon at Trp-604(TGG-TGA) in n2672; Tc1 insertion at Cys758 in km508. The region that is deleted in rh232 is underlined. The DDBJ/EMBL/GenBank accession No. for the EXC-5 sequence is AB060647. (C) Comparison of the domain organization among FGD1, EXC-5 and Frabin. The percentage identity compared with EXC-5 is indicated in each of the conserved domains. Asterisks indicate the positions of two exc-5 mutations, n2672 and km508. PRD, proline-rich domain; ABD, actin-binding domain.
To determine the expression pattern of EXC-5, we generated a transgene, called exc-5 5′::GFP, carrying a predicted exc-5 promoter fused to the coding region for green fluorescent protein (GFP). exc-5 5′::GFP drives GFP expression in the excretory canal (see Figure 4B). We also constructed a gene fusion containing the exc-5 upstream regulatory sequence and its entire coding region fused to the coding region for GFP. This transgene, exc-5::GFP, rescued the exc-5 phenotype, suggesting that it was functional and expressed in all cells that require exc-5 activity. GFP fluorescence was observed in the excretory canal (Figure 4A), consistent with the role this protein plays in the formation of canal structure. In addition, exc-5 expression was also seen in parts of pharyngeal muscles, rectal epithelial cells, and several head and tail neurons.
Fig. 4. Effect of exc-5 overexpression on structure of the excretory canal. (A) Expression of exc-5 is visualized using exc-5::GFP in exc-5(km508). The photo shows the ventral view. The dotted fluorescence results from the autofluorescence of gut granules. (B) L1 unc-119(e2498) larva with exc-5 5′::GFP and pDP#MM016B (unc-119). The excretory canal is visualized using exc-5 5′::GFP expression. High fluorescence in pharynx (Ph) is overexposed to show expression extending throughout the narrow canal (arrowhead). Go, gonad. (C–E) The excretory canal is visualized using exc-5::GFP expression in unc-119(e2498) with exc-5::GFP and pDP#MM016B. (C) The photo shows a truncated posterior excretory canal (arrowhead) from a left lateral aspect in L1 larva. (D) The photos show a shortened posterior excretory canal in L4 larva. Upper and lower panels show Nomarski and epifluorescence images, respectively. The animal has a convoluted tubule in the excretory canal. exc-5::GFP is predominantly expressed at the lumenal (apical) surface of the cell. (E) The photos show a large non-cystic cell body in a mosaic L4 larval animal expressing exc-5::GFP in the excretory canal. The middle panel shows the cell body at high magnification. The arrow indicates a pore connecting the excretory canals to the surface of the animal. GFP expression is seen at the apical surface of the cell. The right panel shows a diagram of the middle panel. Scale bars, 20 µm.
The EXC-5 protein contains, in order, a Dbl-pleckstrin homology (DH-PH) domain, a cysteine-rich evolutionarily conserved zinc-finger motif termed a FYVE domain, and a second PH domain (Figure 3C). This type of tandem DH-PH domain is conserved among GEFs that act on members of the Rho GTPase family (Cerione and Zheng, 1996). The structural organization of EXC-5 is strikingly similar to that of mammalian FGD1 and Frabin (Figure 3C). FGD1 was determined to be the locus responsible for faciogenital dysplasia or Aarskog–Scott syndrome, a multi-systemic developmental disease affecting skeletal and urogenital systems (Pasteris et al., 1994). Frabin was identified as an F-actin-binding protein (Obaishi et al., 1998). FGD1 and Frabin act as GEFs for the Rho family GTPase Cdc42 (Olson et al., 1996; Zheng et al., 1996; Nagata et al., 1998; Obaishi et al., 1998; Umikawa et al., 1999). Taken together, these results suggest that EXC-5 may function as a GEF for the C. elegans Rho family GTPases.
A number of Rho family members have been identified in C. elegans, including MIG-2, CED-10/RAC-1, RAC-2, CDC-42 and RHO-1 (Chen et al., 1996a,b; Zipkin et al., 1997; Reddien and Horvitz, 2000). Among them, activated and null mutations in the mig-2 gene have been identified (Zipkin et al., 1997). To test whether EXC-5 could act as a GEF for GTPases, we examined the genetic interaction between exc-5 and mig-2. Animals with the activated gm38 allele of mig-2 exhibit normal canal structure (data not shown). We constructed exc-5(rh232); mig-2(gm38) double mutants. Activated MIG-2 partially suppressed the Exc phenotype; these animals have longer posterior canals and much smaller cysts than do exc-5(rh232) single mutants (Figure 1D and E). This supports the possibility that EXC-5 functions as a GEF for Rho family GTPases in the formation of canal structure. However, unlike exc-5 loss-of-function mutants, the mig-2(mu28) null mutant animals have normal excretory canals (data not shown), suggesting that there are additional GTPase targets for EXC-5 activity that control the structural organization of the excretory canal.
To examine the effect of EXC-5 overexpression on canal structure formation, we generated transgenic animals carrying a high copy array of the exc-5 5′::GFP or exc-5::GFP genes. Whereas wild-type animals carrying exc-5 5′::GFP have longer posterior canals that reach a point in V6 at the L1 stage (Figure 4B), the canal in animals overexpressing exc-5::GFP stops at a point between V1 and V2 (Figure 4C). Even at the L4 stage, animals overexpressing exc-5::GFP have drastically shortened canal extension, which contains a convoluted tubule. Furthermore, exc-5::GFP was found to be expressed at the apical surface of the cell (Figure 4D). Mosaic animals expressing exc-5::GFP only in the excretory cells show a shortened canal phenotype also seen in mosaic animals deficient in canal expression of the basement membrane-binding β1 integrin (data not shown). GFP fluorescence was visible only in the center of the cell body as streaks representing convolutions of the lumen (Figure 4E). Mosaic animals that express EXC-5::GFP in cells other than the excretory canal show normal canal structure. This suggests that overexpression of EXC-5 acts in a cell-autonomous manner in the formation of abnormal canal structure. Electron microscopy analysis revealed that in animals overexpressing exc-5::GFP, the excretory cell body is much larger than normal and exhibits multiple lumena due to convolutions of the lumenal surface within the canal (Figure 2D). However, the lumena have an apparently normal structure and are evenly surrounded by electron-dense materials. In addition, in EXC-5-overexpressing canals, the canals have lost some of their attachment to the basement membrane. These results suggest that EXC-5 overexpression causes the apical surface to form normally; tubule convolution results from the failure of canal extension, known to require basolateral protein function (Hedgecock et al., 1987; Gettner et al., 1995). In contrast to exc-5 overexpression, the activated mig-2 mutation does not affect the canal structure. This suggests that EXC-5 may have another activity in addition to the activation of GTPases and that both activities may be required for the phenotypes induced by exc-5 overexpression. In fact, Frabin possesses Cdc42-activating and F-actin-binding activities, both of which are necessary for microspike formation (Umikawa et al., 1999).
In a developing multicellular organism, dynamic reorganization of the cytoskeleton is essential for many cellular functions, including changes in cell shape, adhesion and motility. Rho family GTPases regulate cytoskeletal reorganization (Hall, 1998). Our results suggest that the EXC-5 protein is involved in the regulation of Rho family GTPase activity. exc-5 loss-of-function mutations induce the formation of cysts in the excretory canal, whereas overexpression of exc-5 causes convolutions of the apical surface within the canal. These results raise the possibility that EXC-5 facilitates the formation of the canal structure by regulating the polarized organization of the cytoskeleton on the apical surfaces through activation of Rho family GTPases. Therefore, elucidating the role of exc-5 in C. elegans development should lead to a better understanding of the role GEF-GTPases play more generally in the formation of organ structure, and may shed light on the function of the FGD1 family of proteins involved in signal transduction.
METHODS
Mutant isolation and genetic mapping. exc-5(km508) was isolated as a mutant with abnormal excretory canals by visual screening of ∼12 000 individuals of the mut-7(pk204) strain NL917. km508 mapped between stP44 and stP4 on linkage group IV by PCR STS mapping (Williams et al., 1992).
Cloning of exc-5. The genomic sequence corresponding to exc-5 was identified by use of a transposon display method. The mutant-specific Tc1 insertion was found in the C33D9.1 gene. The mixture of two cosmids, C33D9 and W07F6, was found to rescue exc-5(rh232), as did a subclone containing only the C33D9.1 gene. The 5′ terminus of the exc-5 mRNA was analyzed by use of the 5′ RACE System for Rapid Amplification of cDNA Ends (Life Technologies). Molecular lesions were identified by direct sequencing of PCR products spanning the exc-5 gene.
Analysis of canal defects. Caenorhabditis elegans hermaphrodites were observed using a Zeiss Axioplan microscope equipped with a Plan 63 objective and differential interference contrast optics. Images were captured with a Hamamatsu Photonics C5810 Color Chilled 3-CCD camera connected to a Macintosh G3 computer. To visualize the excretory canal, a construct expressing GFP under control of the sek-1 promoter was coinjected with pRF4, which contains the rol-6d gene (Mello et al., 1991). The sek-1 gene encodes MAP kinase kinase (Kawasaki et al., 1999) and is expressed in excretory canals (Tanaka-Hino and Matsumoto, unpublished result).
Electron microscopy. L4 larvae and young adults were cut through the midbody and fixed immediately in buffered (199 mM HEPES pH 7.5) 3% glutaraldehyde, followed by post-fixation in buffered 1% OsO4. After encasement in 1% agar, samples were dehydrated and embedded in Polybed 812 resin (Polysciences). Serial sections (∼70 nm) were post-stained in uranyl acetate followed by lead citrate.
Transgene experiments. To construct a plasmid that contains only the C33D9.1 gene, the SalI–PstI fragment was cloned into the SalI–PstI site of the pBSII KS(–) vector. This construct contained 12.5 kb of the upstream sequence, coding regions, and all of the introns of the exc-5 gene, as well as 1.1 kb of its downstream sequence. The plasmid containing only the C33D9.1 gene was co-injected into exc-5(rh232) with a marker plasmid pEF1a, which contains the EF1a promoter fused to GFP (Kawasaki et al., 1999). To construct the exc-5::GFP protein fusion gene, the 12.6 kb fragment from the cosmid clones W07F6 and C33D9 was fused to a SalI–BamHI DNA fragment containing GFP plus the unc-54 3′ UTR from pPD95.75 (from A. Fire) using appropriate synthetic oligonucleotides. This construct contained 7.2 kb of the exc-5 upstream sequence and 5.3 kb of the exc-5 coding region and all introns. The plasmid containing the exc-5::GFP fusion gene was coinjected with pRF4 into exc-5(km508).
Acknowledgments
ACKNOWLEDGEMENTS
We thank G. Stephney for expert assistance in electron microscopy; C.I. Bargmann, A. Coulson, A. Fire, C.J. Kenyon, Y. Kohara, H. Qadota and the Caenorhabditis Genetics Center for materials; and M. Lamphier for critical reading of the manuscript. Supported by special grants for CREST and Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan (N.H. and K.M.); Daiko Foundation (K.M.); Uehara Memorial Foundation (K.M.); NIH DK55526, DK99-012 and RR12596 (M.B. and D.H.H.); and Polycystic Kidney Research Foundation (M.B.).
REFERENCES
- Broeks A., Janssen, H.W., Calafat, J. and Plasterk, R.K. (1995) A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J., 14, 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechner M., Hall, D.H., Bhatt, H. and Hedgecock, E.M. (1999) Cystic canal mutants in Caenorhabditis elegans are defective in the apical membrane domain of the renal (excretory) cell. Dev. Biol., 214, 227–241. [DOI] [PubMed] [Google Scholar]
- Cerione R.A. and Zheng, Y. (1996) The Dbl family of oncogenes. Curr. Opin. Cell Biol., 8, 216–222. [DOI] [PubMed] [Google Scholar]
- Chen W.N., Yap, S.F. and Lim, L. (1996a) Isolation of a gene coding for the Caenorhabditis elegans Rac2 homologue, a Ras-related small GTP-binding protein. Gene, 180, 217–219. [DOI] [PubMed] [Google Scholar]
- Chen W.N., Chen, S., Yap, S.F. and Lim, L. (1996b) The Caenorhabditis elegans p21-activating kinase (CePAK) colocalizes with CeRac1 and CDC42 at hypodermal cell boundaries during embryo elongation. J. Biol. Chem., 271, 26362–26368. [DOI] [PubMed] [Google Scholar]
- Chitwood M.B. and Chitwood, B.G. (1974) The excretory system. In Introduction to Nematology. University Park Press, Baltimore, MD, pp. 126–135.
- Gettner S.N., Kenyon, C. and Reichardt, L.F. (1995) Characterization of βPAT-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol., 129, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. (1998) Rho GTPases and the actin cytoskeleton. Science, 279, 509–514. [DOI] [PubMed] [Google Scholar]
- Hedgecock E.M., Culotti, J.G., Hall, D.H. and Stern, B.D. (1987) Genetics of cell and axon migrations in Caenorhabditis elegans. Development, 100, 365–382. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Hisamoto, N., Iino, Y., Yamamoto, M., Ninomiya-Tsuji, J. and Matsumoto, K. (1999) A Caenorhabditis elegans JNK signal transduction pathway regulates coordinated movement via type-D GABAergic motor neurons. EMBO J., 18, 3604–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M. and Pilgrim, D. (1995) Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics, 141, 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer, J.M., Stinchconb, D. and Ambros, V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Driessens, M., Lamarche, N., Gorski, J.L. and Hall, A. (1998) Activation of G1 progression, JNK mitogen-activated protein kinase, and actin filament assembly by the exchange factor FGD1. J. Biol. Chem., 273, 15453–15457. [DOI] [PubMed] [Google Scholar]
- Nelson F.K., Albert, T.S. and Riddle, D.L. (1983) Fine structure of the Caenorhabditis elegans secretory-excretory system. J. Ultrastruct. Res., 82, 156–171. [DOI] [PubMed] [Google Scholar]
- Obaishi H. et al. (1998) Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J. Biol. Chem., 273, 18697–18700. [DOI] [PubMed] [Google Scholar]
- Olson M.F., Pasteris, N.G., Gorski, J.L. and Hall, A. (1996) Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr. Biol., 6, 1628–1633. [DOI] [PubMed] [Google Scholar]
- Pasteris N.G., Cadle, A., Logie, L.J., Porteous, M.E., Schwartz, C.E., Stevenson, R.E., Glover, T.W., Wilroy, R.S. and Gorski, J.L. (1994) Isolation and characterization of the faciogenital dysplasia (Aarskog–Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell, 79, 669–678. [DOI] [PubMed] [Google Scholar]
- Reddien P.W. and Horvitz, R. (2000) CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nature Cell Biol., 2, 131–136. [DOI] [PubMed] [Google Scholar]
- Umikawa M., Obaishi, H., Nakanishi, H., Satoh-Horikawa, K., Takahashi, K., Hotta, I., Matsuura, Y. and Takai, Y. (1999) Association of Frabin with the actin cytoskeleton is essential for microspike formation through activation of Cdc42 small G protein. J. Biol. Chem., 274, 25197–25200. [DOI] [PubMed] [Google Scholar]
- Williams B.D., Schrank, C., Huynh, R., Shownkeen, R. and Waterston, R.H. (1992) A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics, 131, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Fischer, D.J., Santos, M.F., Tigyi, G., Pasteris, N.G., Gorski, J.L. and Xu, Y. (1996) The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J. Biol. Chem., 271, 33169–33172. [DOI] [PubMed] [Google Scholar]
- Zipkin I.D., Kindt, R.M. and Kenyon, C.J. (1997) Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell, 90, 883–894. [DOI] [PubMed] [Google Scholar]