Abstract
Introduction
Totum-070 is a combination of five plant extracts enriched in polyphenols to target hypercholesterolemia, one of the main risk factors for cardiovascular diseases. The aim of this study was to investigate the effects of Totum-070 on cholesterol levels in an animal model of diet-induced hypercholesterolemia.
Methods
C57BL/6JOlaHsd male mice were fed a Western diet and received Totum-070, or not, by daily gavage (1g/kg and 3g/kg body weight) for 6 weeks.
Results
The Western diet induced obesity, fat accumulation, hepatic steatosis and increased plasma cholesterol compared with the control group. All these metabolic perturbations were alleviated by Totum-070 supplementation in a dose-dependent manner. Lipid excretion in feces was higher in mice supplemented with Totum-070, suggesting inhibition of intestinal lipid absorption. Totum-070 also increased the fecal concentration of short chain fatty acids, demonstrating a direct effect on intestinal microbiota.
Discussion
The characterization of fecal microbiota by 16S amplicon sequencing showed that Totum-070 supplementation modulated the dysbiosis associated with metabolic disorders. Specifically, Totum-070 increased the relative abundance of Muribaculum (a beneficial bacterium) and reduced that of Lactococcus (a genus positively correlated with increased plasma cholesterol level). Together, these findings indicate that the cholesterol-lowering effect of Totum-070 bioactive molecules could be mediated through multiple actions on the intestine and gut microbiota.
Keywords: cholesterol, triglycerides, lipoproteins, intestine, microbiota
1. Introduction
Hypercholesterolemia is strongly associated with atherosclerosis. This disease leads to myocardial infarction, stroke and peripheral vascular disease. In people with markedly elevated cholesterol levels, treatment with cholesterol-lowering medication is usually prescribed. For individuals with moderately or slightly elevated cholesterol levels, lifestyle interventions, such as physical activity, dietary changes and dietary supplements, are recommended (1). Plant-based supplements are tested as a nutritional solution for disease prevention. Indeed, phytochemicals that lower serum glucose, cholesterol, and triglyceride concentrations are considered to reduce the risk of cardiovascular diseases (2). Many plants have been investigated to reduce blood cholesterol (3–6). Artichoke (Cynara scolymus L) is a plant rich in natural antioxidants. The pharmacological properties of artichoke leaf extracts are well documented and might contribute to lower cholesterol levels in humans (7, 8). The beneficial effects of artichoke leaf extract are largely explained by their polyphenolic compounds, namely, chlorogenic acid, caffeic acid, 3,4-di-O-caffeoylquinic acid and cynarine, and the main flavonoids apigenin and luteolin (9, 10). Moreover, several studies suggest that olive leaf extracts can reduce cardiovascular disease risk factors, notably through their hypocholesterolemic effect (11, 12). The main phenolic compounds present in olive plant (Olea europaea L) are luteolin, oleuropein, tyrosol, and hydroxytyrosol. Olive leaves contain the highest concentrations of these compounds, especially oleuropein, compared with other parts of the plant (13). Goji berries (Lycium barbarum) have been traditionally used as food and medicinal plants in China and other Asian countries (14). The various pharmacological actions of these berries are mainly attributed to the presence of polysaccharides, flavonoids and carotenoids, especially zeaxanthin, quercetin, and rutin (15, 16). Goji berries also contain betaine, an alkaloid that helps to produce choline, a compound that strengthens the liver. Goji berries significantly reduce total cholesterol and total triglyceride concentrations in older adults (17). Black pepper (Piper nigrum L.) is commonly used as a spice in various food types. The main bioactive ingredient of black pepper fruits (i.e., piperine) has a wide range of pharmacological effects, including anti-oxidant and cholesterol-lowering properties (18). Piperine can enhance the absorption of vitamins and polyphenols (19). Chrysanthellum (Chrysanthellum indicum subsp. afroamericanum B.L. Turner) is a plant that is widespread in the tropics and that reduces cholelithiasis and acts as a hypolipidemic agent (20).
Totum-070 is a polyphenol-rich blend of these five plant extracts (olive leaves, artichoke leaves, chrysanthellum, goji fruits and black pepper) selected because of their abundance in bioactive molecules with the aim of lowering cholesterol in individuals with mild to moderate hypercholesterolemia (French Priority Patent Number: FR1559965). Recently, Wauquier et al. demonstrated that administration of 3 g/kg Totum-070 prevents the development of dyslipidemia in hamsters fed a high-fat diet (HFD) for 12 weeks (21). Moreover, using an explorative clinical approach in healthy volunteers who received a single dose of Totum-070 (5 g), the authors detected the presence of about twenty circulating metabolites, including chlorogenic acid, cynarine, luteolin glucuronides isomers, oleuropein glucuronides isomers, hydroxytyrosol sulfate isomers, and hydroxytyrosol glucuronides isomers. In addition, the ex vivo incubation of palmitate-exposed hepatocytes with human sera enriched in these bioactive molecules following Totum-070 ingestion limited the accumulation of intracellular lipids (21).
Plasma cholesterol concentration is influenced by several factors, including the absorption of dietary cholesterol, endogenous cholesterol synthesis, cholesterol removal from the circulation, and the conversion of hepatic cholesterol into bile acids followed by excretion in the feces (22). Accumulating data from human and animal studies have demonstrate that intestinal microbes can affect the host cholesterol metabolism through multiple direct and indirect biological mechanisms (23, 24). Interestingly, many plant extracts can mitigate dyslipidemia and obesity by regulating the gut microbiota relative abundance and composition (25). There is evidence from animal studies that certain doses of selected polyphenols and polyphenol-rich plant extracts modify the gut microbial composition. For example, supplementation with water extract of Goji fruits ameliorated weight gain and insulin resistance in HFD-fed mice (26). Goji extracts elevated hepatic bile acid biosynthesis and excretion of bile acids in association with decreased relative abundance of Clostridium_XIVa. In rabbits, Goji berry supplementation modulates gastrointestinal microbiota composition and cecal fermentation, with the families Ruminococcaceae and Lachnospiraceae relatively more abundant with Goji than in control group (27). In HFD-fed mice, administration of Olive leaf extract was able to counteract the altered composition in the gut microbiota associated to obesity and restored the main bacteria phyla to the normal values observed in standard diet-fed mice (28). Supplementation with chlorogenic acid in HFD-fed mice protected against metabolic disorders and endotoxemia. Chlorogenic acid significantly changed the composition of the gut microbiota and increased the relative abundance of short chain fatty acid (SFCA)-producers (29). In another study in mice, chlorogenic acid also reversed the HFD-induced gut microbiota dysbiosis, including significantly inhibiting the growth of Desulfovibrionaceae, Ruminococcaceae, Lachnospiraceae, Erysipelotrichaceae, and raising the growth of Bacteroidaceae and Lactobacillaceae (30).
The aim of the present study was to investigate the effects of Totum-070 supplementation in a murine model of hypercholesterolemia and to explore hypotheses of Totum-070 mechanism of action.
2. Materials and methods
2.1. Totum-070 (lipidrive ®) characterization
The batch used in this study was chemically characterized using the Folin–Ciocalteu method (total polyphenol estimation), the Dubois method (total sugar estimation), the Phospho-Vanillin method (total lipid estimation) (31, 32), the O-phthalaldehyde method (total protein estimation) (33), and HPLC-UV/visible/MS (Agilent Technologies Infinity series 1,200 and 1,260, Santa Clara, CA, USA) with a C18 column (250 × 4.6 mm, 5 μm, Phenomenex, USA) and a HILIC Silica column (150 × 4.6 mm, 5 μm, Waters, The Netherlands) (quantification of potential compounds of interest). The results of this analysis are presented in Table 1.
Table 1.
Chemical characterization of Totum-070.
| Compound type (sorted by families) | Extract content (g/100 g dry weight) |
|---|---|
| Total sugars | 41.35 |
| Total lipids | 7.30 |
| Total Proteins | 2.05 |
| Betaine | 0.23 |
| Total phenolic compounds | 15.42 |
| Monocaffeoylquinic acids | |
| Chlorogenic acid | 0.81 |
| Cryptochlorogenic acid | 0.01 |
| Other monocaffeoylquinic acids | 0.25 |
| Dicaffeoylquinic acids | |
| Cynarin | 0.16 |
| 3–5 dicaffeoylquinic acid | 0.18 |
| 4–5 dicaffeoylquinic acid | 0.14 |
| Other dicaffeoylquinic acids | 0.11 |
| Caffeic acid | 0.01 |
| Oleuropein | 4.6 |
| Oleuropein isomers | 0.47 |
| Hydroxytyrosol | 0.08 |
| Luteolin | 0.01 |
| Luteolin-7-O-glucoside | 0.27 |
| Luteolin-7-O-glucoside isomer | 0.02 |
| Luteolin-4-O-glucoside | 0.06 |
| Luteolin-7-O-glucuronide | 0.29 |
| Apigenin-7-O-glucoside | 0.03 |
| Apigenin-7-O-glucuronide | 0.17 |
| Apigenin-6-C-glucoside-8-C-arabinoside (Shaftoside) | 0.01 |
| Apigenin-6,8-C-diglucoside (Vicenin 2) | 0.02 |
| Apigenin-7-O-rutinoside | 0.01 |
| Eriodictyol-7-O-glucoside | 0.04 |
| Flavanomarein | 0.14 |
| Marein | 0.05 |
| Maritimein | 0.05 |
| Rutin | 0.02 |
| Verbascoside | 0.08 |
| Terpenes and terpenoids | |
| Oleanolic acid | 1.64 |
| Saponins | |
| Chrysanthellin A | 0.28 |
| Chrysanthellin B | 0.27 |
| Iridoids | |
| Oleoside | 0.01 |
| Cynaropicrin | 0.07 |
| Alkaloids | |
| Piperine | 0.06 |
2.2. Animals
All animal procedures were approved by the local ethics committee (C2E2A, Auvergne, France) and comply with the ARRIVE guidelines. Male C57BL/6JOlaHsd mice were purchased from Envigo (Gannat, France). All mice were housed at Valbiotis animal facility (Valbiotis R&D Center, Riom, France) at 22°C under standard 12 h light-12 h dark cycle. Upon arrival, 56 mice were fed a normal diet (ND) (D14042701, Research Diets, New Brunswick, NJ, USA) for 2 weeks of acclimatization until the age of 8 weeks. Then, animals were randomly assigned to four groups (n = 14 mice per group) with similar average body weight and fat mass: (i) ND, (ii) WD (41 Kcal % fat, 30 Kcal % sucrose and 0.21% w/w cholesterol; D12079B, Research Diets, New Brunswick, NJ, USA), (iii) WD supplemented with 1 g/kg of Totum-070 (WD-T070 1 g/kg), and (iv) WD supplemented with 3 g/kg of Totum-070 (WD-T070 3 g/kg). Totum-070 solution was prepared in vehicle (1% Tween-20 in tap water) at 100 g/L or 300 g/L. Totum-070 was administered to WD-fed mice by gavage daily. Control groups (WD and ND) received only vehicle by gavage. Food and water were supplied ad libitum. Supplementation was for 6 weeks. Body weight was recorded weekly, and food intake was measured 3 times per week. The ND and WD energy densities were 3.9 kcal·g−1 and 4.7 kcal·g−1, respectively. Plasma lipid parameters were monitored before the study start (week 0), after 3 weeks of supplementation (week 3), and at the study end (week 6). Mice were fasted for 6 h and blood was collected from the tail vein into EDTA-treated capillaries. Blood was centrifuged at 2,000 g at 4°C for 10 min. Plasma was harvested and stored at −80°C until analysis. Body composition was assessed with an echo MRI system (Zynsser Analytic, Frankfurt, Germany). At the end of the 6 weeks of supplementation, animals were fasted for 6 h, anaesthetized with isoflurane, and euthanized by cervical dislocation.
2.3. Plasma lipid parameters
Cholesterol levels in the plasma were quantified enzymatically using the CHOD-PAP colorimetric assay kit (Biolabo SAS, Maizy, France). Plasma triglyceride concentration was measured using a colorimetric assay kit (Cayman chemical, Ann Arbor, MI, USA).
2.4. Liver triglycerides
Liver triglyceride level was determined with the triglyceride assay kit by Cayman Chemical. Mouse livers (100 mg) were homogenized in 1 ml of a solution containing 5% NP40 in water followed by centrifugation (10,000 g at 4°C for 10 min) to recover the supernatant. Triglyceride concentration was normalized to the liver sample weight and expressed as mg/g liver weight.
2.5. Histology
At the study end, liver tissue was fixed in paraformaldehyde at 4°C for 48 h and kept in 70% ethanol for 2 to 3 h before automated tissue processing. Samples were embedded in paraffin and 4μm sections were cut with a microtome. Sections were dewaxed and rehydrated in successive xylene and ethanol baths. Then, they were stained in Gill II hematoxylin for 4 min, and after removal of excess stain in an alcohol acid bath, they were transferred to a 0.25% eosin Y bath for 45 s. Finally, sections were dehydrated with ethanol and xylene, and slides were mounted with the Eukitt medium. Other liver tissue samples were frozen in OCT medium, and 10 μm tissue sections were cut with a cryostat and fixed in cold 10% formalin for 5 min. Slides were stained with Oil Red O at 60°C in an oven for 8 min. Oil Red O areas were quantified using the ImageJ software. Stained areas of at least 10 μm² and with a circularity between 0.1 and 1 were selected for quantifying the Oil Red O surface as percentage of the total surface. At least three images per liver were analyzed.
2.6. Quantitative reverse transcription PCR (Rt-qPCR)
Total mRNA was extracted from snap-frozen tissues using TRIzol® (Invitrogen, Life Technologies). cDNA was synthesized from 2 μg RNA with the High-Capacity cDNA transcription kit (Applied Biosystems, Life Technologies). PCR amplification was carried out using the CFX system (Bio-Rad, Marnes-la-Coquette, France) with Taqman probes (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) and the ΔΔCt method was used to quantify mRNA levels. Gene expression was normalized to the housekeeping genes Gapdh (liver samples) and Polr2a (intestine samples).
2.7. Protein extraction and western blot analysis
Liver samples were homogenized in 20 µl/mg RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) on ice using a glass potter. Just before use, protease (P8340, Sigma, Saint-Louis, MO, USA) and phosphatase (88,667, Thermo Fisher Scientific, Whaltham, MA, USA) inhibitors were added to the buffer. Homogenized samples were centrifuged at 14,000 g at 4°C for 10 min, and supernatant was collected. Protein content was determined using a commercial Lowry-based DC protein assay (Bio-Rad, Marnes-la-Coquette, France). All samples were adjusted to a standard concentration (15 µg) and then diluted with 2× Laemmli buffer. Western blotting was performed as described by Chavanelle et al. (34) with the following specificities: membranes were incubated overnight at 4°C with primary antibodies against LDL Receptor (Biovision, Milpitas, CA, USA) diluted to 1:1,000. After overnight incubation, membranes were washed with Tris-buffered saline/0.5% Tween-20 and incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibodies (1:2,000) at room temperature for 1 h. Then, membranes were washed three times in Tris-buffered saline/0.5%Tween-20 before addition of an enhanced chemiluminescent solution (Clarity Western ECL; Bio-Rad) for 1 min. Images were acquired with the Bio-Rad ChemiDoc system, and band density was determined using Image Lab V6.0 (Bio-Rad). Stain-free blot images were used as total protein loading control for data normalization (35).
2.8. Fecal lipid content
For each cage (1 animal per cage), feces were collected over a week at the study end. Feces were dried at 60°C for 72 h. Fecal lipid quantification was performed using 0.5 g of dried sample per cage by chloroform/methanol lipid extraction as described previously (36). The lipid phase was placed in a glass recipient and solvent was evaporated. The total lipid mass per g of feces was calculated by subtracting the empty glass recipient weight.
2.9. Quantification of fecal short chain fatty acids (SCFAs)
Lyophilized feces (± 50 mg) were mixed in 750 µl of 0.04 M sulfuric acid using an Ultra-Turrax® (Ika, Staufen, Germany). Samples were centrifuged (16,000 g, 4°C, 15 min) and the supernatants were filtered (0.22 μm). The three major SCFAs (acetate, propionate and butyrate) were quantified by high-performance liquid chromatography (HPLC, Elite LaChrom, Merck HITACHI, USA) coupled to a UV detector (λ = 205 nm). Elution was done using an ICE-COREGEL 87H3 9 µm 150 × 7.8 mm column and its pre-column (Concise Separations, San Jose, CA, USA) at 50°C with 0.04 M sulfuric acid as mobile phase (0.6 ml/min). Data were analyzed with the EZChrom Elite software. SCFA concentrations were calculated from standard curves established with a known equimolar concentration solution of acetate, propionate, and butyrate (0, 10, 25, and 40 mM).
2.10. Lipase inhibition assay
Lipase (type II, from porcine pancreas, Sigma, Saint-Louis, MO, USA) hydrolytic activity was assessed by monitoring the conversion of 4-methylumbelliferyl oleate (4-MUO, Sigma, Saint-Louis, MO, USA) into 4-methylumbelliferone (4-MU). The enzymatic activity was determined by measuring 4-MU (Sigma, Saint-Louis, MO, USA) fluorescence signal at 460 nm (after excitation at 355 nm) over time. A commercial reference inhibitor, orlistat (Sigma, Saint-Louis, MO, USA), was used as positive control, and citrate-phosphate buffer (0.1 M, pH 7.4) as reaction buffer. A series of 4-MU dilutions in buffer was used to generate the standard curve to convert the absorbance values into 4-MU concentrations. A 4 g/L solution of lipase in buffer was centrifuged at 5 000 g, 10°C for 10 min, then the supernatant was aliquoted. The final in-well concentration used in the assay was 250 mg/L. In each microplate well, 20 µl of test products diluted to 10:90 (v:v) in a DMSO:water mixture (or 20 µl of this solvent mixture for controls), 20 µl of lipase in buffer (or 20 µl of buffer for the blanks), and 110 µl of buffer were incubated at 37°C in a microplate reader for 10 min. Then, 10 µl of 4-MUO substrate (48 µM in DMSO) was added to each well to initiate the reaction. Fluorescence was measured at 355 nm/460 nm (excitation/emission) using a FLUO Star Omega (BMG LabTech) 96-well microplate reader, maintained at 37°C, every 30 s for 15 min. The relative lipase activity was calculated using Equation 1. The IC50 of each compound corresponded to the lowest concentration at which the lipase activity was halved.
| (1) |
2.11. Lipidomics analysis
Lipidomic analysis was performed at the Center for Proteomics and Metabolomics, Leiden University, with feces collected as described above. Lipids were extracted from 50 mg of dry feces using the methyl-tert-butylether method, and analyzed using Lipidyzer™, a direct infusion-tandem mass spectrometry (DI-MS/MS)-based platform (Sciex, Redwood City, USA), as described previously (37, 38). Lipid concentrations were expressed as pmol/mg of dry feces.
2.12. Microbiota analysis
Fresh feces were collected during week 6 of the study and immediately frozen in liquid nitrogen. DNA was extracted from 50 to 100 mg of sample using the FastDNATM Spin Kit for feces (MP Biomedicals, Illkirch, France) and FastPrep-24 5G (MP Biomedicals, Illkirch, France) following the manufacturer's instructions. DNA was eluted in 50 µl of elution buffer, pre-heated at 55°C, and normalized to 20 ng/µl. Microbial 16S amplicon libraries were prepared at the Plateforme Génome Transcriptome de Bordeaux, Bordeaux, France, by amplification and sequencing of the V3–V4 region of the 16S rRNA amplicon on an Illumina MiSeq machine, using the 2 × 250 base pair Illumina v2 kit. Quality filtering and processing were done as described previously (39) with cutadapt v4.0 (40) and Python v3.9.9, usearch v11.0.667 (41, 42) and mothur v1.48.0 (43).Classification of quality filtered reads was performed by comparison with the ribosomal database project (RDP trainset 14) (44, 45).
2.13. Statistical analysis
Prism V.7.0 and 8.0 (GraphPad Software) were used for statistical tests and figure drawing. The Shapiro–Wilk normality test was used to determine whether data followed a Gaussian distribution. If data were not normally distributed, the Kruskal–Wallis nonparametric test was used followed by the Dunn test for post hoc comparison. When the normal distribution was confirmed, data were compared with the one-way or two-way ANOVA and Tukey's test for multiple comparisons. For measurements repeated over time, differences between groups and time points were evaluated using a repeated-measure two-way ANOVA followed by the Tukey's post hoc test for multiple comparisons. If missing data did not allow using the repeated-measures two-way ANOVA, a mixed-effects analysis was performed. Values are presented as the mean ± SEM, unless specified otherwise. Differences were considered significant at p < 0.05.
For microbiota analyses, relative abundance tables were imported and analyzed in R v4.1. Alpha (Chao and Shannon indices) and beta diversity (Bray-Curtis and Jaccard distances) were estimated using vegan 2.6–2 (46) and compared using the Kruskal-Wallis and MANOVA tests, respectively. Indicator taxa were selected using the KW test and multiple testing correction with False Discovery Rate (FDR) correction. For indicator taxa, an additional linear model was tested to determine the type of observed effect. Correlations between the relative abundances of taxa and arbitrary values were tested using the Pearson correlation coefficient. Arbitrary units were allocated to the experimental groups and six models were tested: (1) Totum-070 induces a return to normal that goes beyond the ND value (ND = 25, WD = 75; WD-T070 3 g/kg = 10), (2) Totum-070 induces a return to normal (ND = 25, WD = 75; WD-T070 3 g/kg = 25), (3) Totum-070 induces a partial return to normal (ND = 25, WD = 75; WD-T070 3 g/kg = 50), (4) Totum-070 does not rescue the changes induced by WD (ND = 25, WD = 75; WD-T070 3 g/kg = 75), (5) Totum-070 accentuates the WD effect (ND = 25, WD = 75; WD-T070 3 g/kg = 100), (6) Totum-070, but not WD, induces a change (ND = 100, WD = 100; WD-T070 3 g/kg = 10). Indicator taxa were identified by selecting taxa significantly associated with the supplementation group in the Kruskal-Wallis test, and for which at least one of the linear models was significant. If multiple linear models were significant, the model with the highest effect size was reported in Table 2. Correlations between indicator taxa and metabolic parameters were assessed using the Pearson correlation with FDR correction for multiple testing.
Table 2.
Fecal microbiota: relative abundance differences at the phylum level in the different groups.
| % of relative abundance | Adjusted p. values | |||||
|---|---|---|---|---|---|---|
| Phylum | ND | WD | WD-T070 3 g/kg | ND vs. WD | ND vs. WD-T070 3 g/kg | WD vs. WD-T070 3 g/kg |
| Actinobacteria | 8.50 ± 5.43 | 0.99 ± 0.40 | 1.22 ± 1.05 | 0.0014 | 0.0085 | 0.9292 |
| Firmicutes | 56.03 ± 5.35 | 59.16 ± 4.86 | 53.07 ± 8.78 | 0.4500 | 0.9342 | 0.8885 |
| Bacteroidetes | 26.14 ± 4.44 | 30.16 ± 6.59 | 32.78 ± 4.11 | 0.4500 | 0.0214 | 0.3490 |
| Verrucomicrobia | 3.42 ± 4.31 | 2.59 ± 4.38 | 4.76 ± 6.62 | 0.4500 | 0.5654 | 0.3490 |
| uncl. Bacteria | 2.40 ± 0.45 | 2.11 ± 0.18 | 2.23 ± 0.48 | 0.1318 | 0.5654 | 0.7574 |
| Deferribacteres Candidatus | 1.88 ± 3.92 | 2.63 ± 4.71 | 1.43 ± 2.38 | 0.4500 | 0.5654 | 0.3490 |
| Saccharibacteria | 0.24 ± 0.19 | 0.29 ± 0.36 | 0.56 ± 0.70 | 0.9438 | 0.5654 | 0.8032 |
| Campilobacterota | 0.00 ± 0.00 | 0.00 ± 0.01 | 0.00 ± 0.00 | 0.9438 | 0.5654 | 0.6680 |
| Tenericutes | 0.06 ± 0.09 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.0405 | 0.0408 | |
| Proteobacteria | 1.33 ± 1.04 | 2.07 ± 1.29 | 3.93 ± 1.38 | 0.1318 | 0.0085 | 0.0227 |
| F/B ratio | 2.22 ± 0.57 | 2.06 ± 0.51 | 1.67 ± 0.45 | 0.5262 | 0.0390 | 0.1309 |
Relative abundances are presented as the mean ± SD, and p values were adjusted for multiple testing (n = 11 ND, n = 10 WD, n = 8 WD- T070 3 g/kg). F/B, firmicutes/bacteroidetes.
3. Results
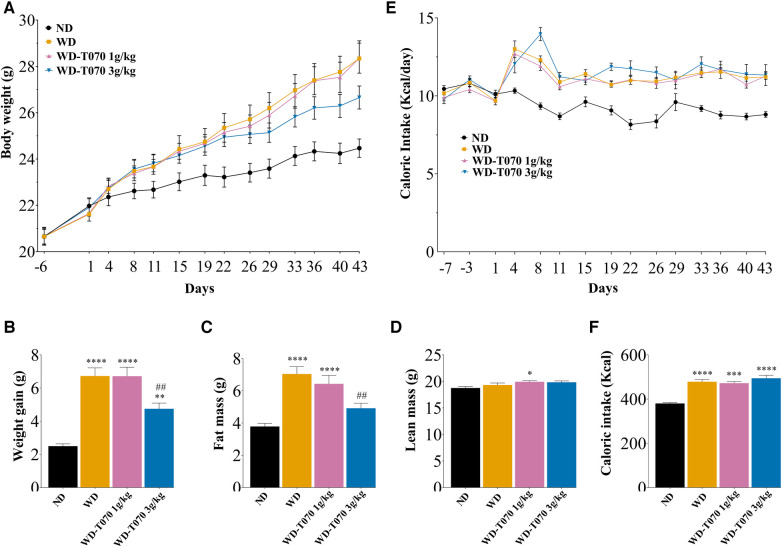
3.1. Totum-070 supplementation limits body weight gain in WD-fed mice
To establish a model of lipid metabolic disorder, mice were fed a WD (n = 42) or a normal diet (ND; n = 14) for 6 weeks. Animals fed a WD were subdivided in three groups: WD alone (WD; n = 14), WD supplemented with 1 g/kg of Totum-070 (WD-T070 1 g/kg; n = 14), and WD supplemented with 3 g/kg of Totum-070 (WD-T070 3 g/kg; n = 14). Totum-070 (or vehicle alone in the WD and ND groups) was administered by gavage daily. The final body weight was significantly higher in the WD groups than ND group (Figure 1A). Body weight was similar between the WD and WD-T070 1 g/kg groups throughout the study (Figures 1A,B). Conversely, after 4 weeks of supplementation, body weight increase was reduced in the WD-T070 3 g/kg group compared with WD mice (Figure 1A). This resulted in a 30% lower (p < 0.01) body weight gain (Figure 1B) and fat mass gain (Figure 1C) compared with the WD group at the supplementation end. No change was observed in lean mass between the WD group and mice supplemented with Totum-070 (both concentrations) (Figure 1D). During the study, the daily caloric intake was higher in the WD groups compared with the ND group (Figure 1E), and was similar among the WD groups (WD and WD-T070) (Figures 1E,F). Therefore, Totum-070 gavage did not affect energy intake in WD mice.
Figure 1.
Effect of supplementation with Totum-070 (1 g/kg and 3 g/kg) in Western diet-fed mice on body weight, body composition and caloric intake during the study period. (A) Starting at day 1, male C57BL/6JOlaHsd mice were fed a normal diet (ND), a Western diet (WD) or a WD plus daily gavage with Totum-070 (WD-T070, 1 g/kg or 3 g/kg) (n = 14 per group). Body weight was recorded during the study. (B) Body weight gain (difference between the last body weight at day 43 and the body weight at the study start, day 1). (C) Fat mass and (D) lean mass measured by EchoMRI at the study end. (E) Caloric intake during the study period. (F) Cumulative caloric intake during the study. * p < 0.05, ** p < 0.01, and *** p < 0.001 vs. ND. ## p < 0.01 Totum-070 groups vs. WD. Data are the mean ± SEM.
3.2. Totum-070 supplementation prevents cholesterol elevation in WD-fed mice
At the study start (week 0), plasma cholesterol concentrations were identical in all groups (Figure 2A). After 3 weeks, plasma cholesterol was significantly increased (+38%) in the WD group, and this level was maintained at the study end (+36%) (Figure 2A). Totum-070 (1 g/kg and 3 g/kg) dose-dependently reduced plasma cholesterol concentration after 3 weeks (−8.7%, p < 0.05 and −12.7%, p < 0.01 vs. WD group, respectively), and after 6 weeks of supplementation (−7.3%, p = 0.08 and −11%, p < 0.01 vs. WD group, respectively) (Figure 2A). Circulating triglyceride concentration was similar in all groups throughout the study (Figure 2B).
Figure 2.
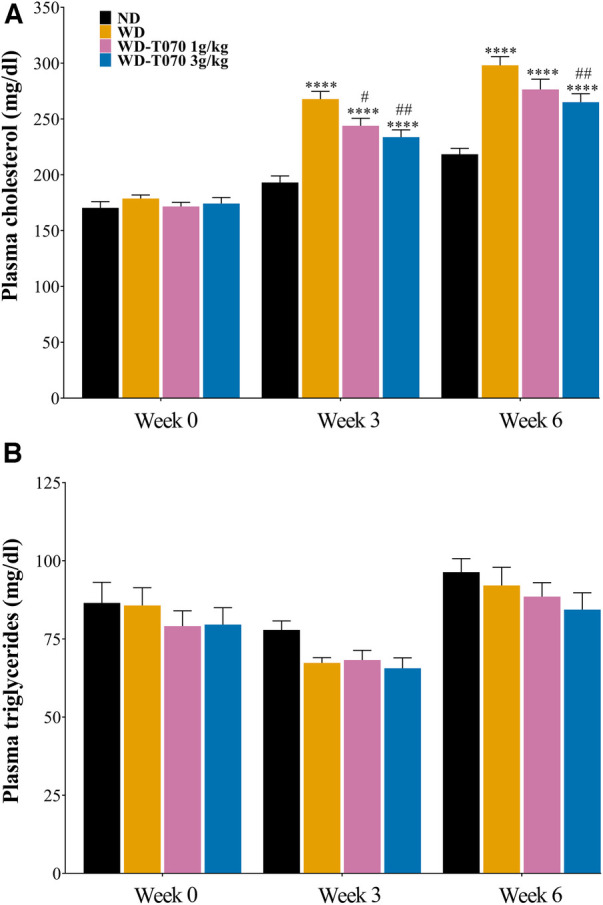
Effect of Totum-070 supplementation on plasma lipid concentration. (A) Plasma concentration of cholesterol (n = 14 per group) and (B) of triglycerides (n = 14 per group) at week 0 (before starting the WD), during the study (week 3), and at the study end (week 6). **** p < 0.0001 vs. ND. # p < 0.05 and ## p < 0.01 Totum-070 (T070) groups vs. WD. Data are the mean ± SEM.
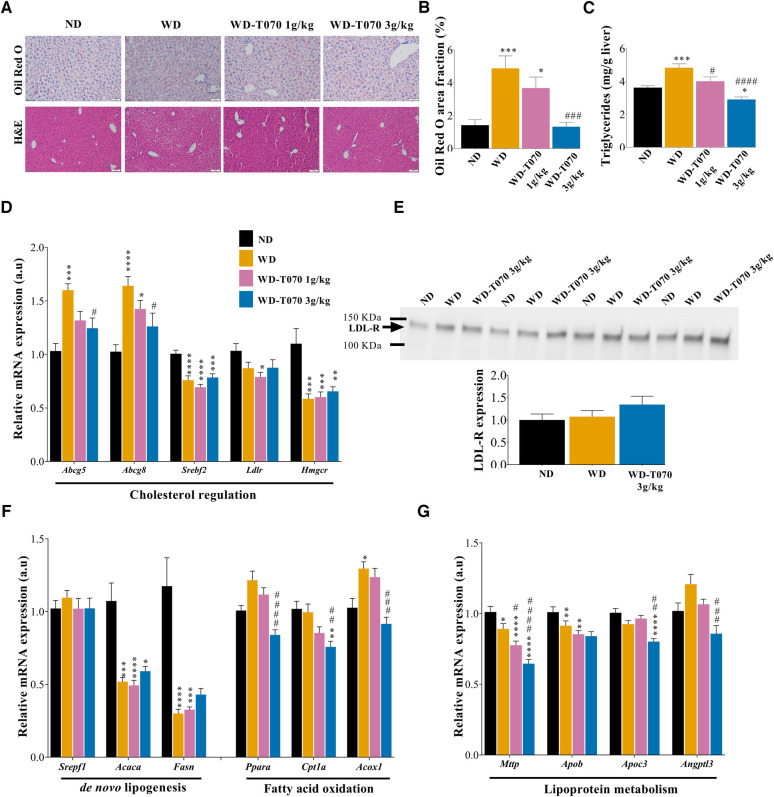
3.3. Totum-070 supplementation improves hepatic lipid metabolism
Histological analysis of liver samples revealed abnormal accumulation of lipid droplets induced by WD that was reduced by Totum-070 supplementation (Figure 3A). In agreement, quantification of Oil Red O staining in liver sections confirmed in the WD-T070 3 g/kg group, the reduction of neutral lipid accumulation to the level observed in the ND group (Figure 3B). Moreover, hepatic triglyceride content was reduced by 17% (p < 0.05) in the WD-T070 1 g/kg group and by 40% (p < 0.0001) in the WD-T070 3 g/kg group (Figure 3C) compared with the WD group. Analysis of the expression of genes implicated in liver lipid and lipoprotein metabolism showed that Totum-070 supplementation influenced the expression of some genes involved in cholesterol metabolism (Figure 3D). Specifically, Totum-070 supplementation limited the upregulation of the cholesterol transporters Abcg5 and Abcg8 induced by the WD. Conversely, it did not have any effect on the expression levels of Srebf2 and the downstream genes Ldlr and Hmgcr that were decreased in all WD groups. WD and Totum-070 supplementation did not have any effect on LDL receptor (LDL-R) protein expression in liver (Figure 3E). Totum-070 supplementation did not have any effect on the expression of genes implicated in de novo lipogenesis, whereas it decreased (vs. WD) the expression of genes involved in fatty acid oxidation (Ppara, Cpt1a, Acox1), but only when used at the concentration of 3 g/kg (Figure 3F). Totum-070 further decreased the expression of genes involved in lipoprotein metabolism, such as Mttp (both doses compared with the WD and ND groups) and also Apoc3 and Angptl3, but only at the higher dose (Figure 3G).
Figure 3.
Effect of Totum-070 on liver lipid metabolism. (A) Frozen mouse liver tissue sections stained with Oil Red O and counterstained with hematoxylin (upper panels). Scale bars, 50 µm. Paraffin-embedded sections of mouse livers stained with hematoxylin and eosin (H&E) (lower panels). Scale bar, 100 µm. (B) Quantification of Oil Red O in liver sections (at least three images per liver, n = 14 per group) shows reduction of neutral lipid accumulation in mice supplemented with Totum-070. (C) Quantification of hepatic triglyceride content (n = 14 per group). (D) Relative expression in liver of selected genes encoding proteins implicated in cholesterol metabolism (n = 14 per group). (E) Immunoblot to monitor LDL receptor (LDL-R) expression in liver samples from the indicated groups and quantification. (F) Relative expression in mouse liver samples from the indicated groups of selected genes encoding proteins implicated in lipid metabolism. (G) Relative expression in mouse liver samples from the indicated groups of selected genes encoding proteins implicated in lipoprotein metabolism. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 vs. ND. #p < 0.05, ##p < 0.01, ###p < 0.001 and ####p < 0.001 Totum-070 (T070) groups vs. WD. Data are the mean ± SEM.
3.4. Totum-070 modulates the intestinal lipid metabolism
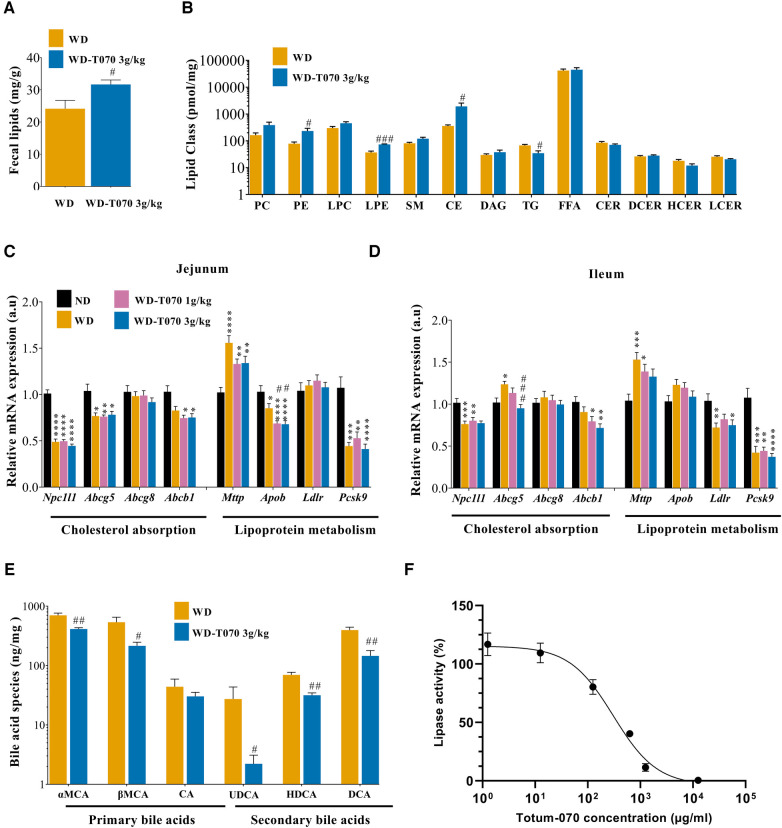
To better understand the regulation of lipid homeostasis by Totum-070, total lipid content was quantified in feces from mice in the WD and WD-T070 3 g/kg groups (Figure 4A). This showed that fecal lipid content was increased by 31% in the supplemented group (p < 0.05). Lipidomic analyses in stool samples highlighted the increase of specific lipid classes in the WD-T070 3 g/kg group compared with the WD group: cholesterol ester (CE) (+437%, p < 0.05), lysophosphatidylethanolamine (LPE) (+98%, p < 0.001) and phosphatidylethanolamine (PE) (+197%, p < 0.05). Conversely, triglycerides (TG) were decreased by 49% (p < 0.05) in feces from the WD-T070 3 g/kg group compared with the WD group (Figure 4B). Genes implicated in intestinal cholesterol absorption were assessed for differential expression between groups in the duodenum (Supplementary Figure S1A), jejunum (Figure 4C) and ileum (Figure 4D). Most investigated genes were not influenced by Totum-070 in the three intestinal segments. Apob mRNA level was repressed specifically in the jejunum (Figure 4C). The transcript for Abcg5 was downregulated by Totum-070 in the ileum only (Figure 4D).
Figure 4.
Effect of Totum-070 on intestinal lipid metabolism. (A) Total lipid content in feces (n = 5 per group). (B) Quantification of different lipid classes in feces. CE, cholesterol ester; CER, ceramides; DAG, diacylglycerides; DCER, dihydroceramides; FFA, free fatty acids; HCER, hexosylceramides; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; and TG, triacylglycerides. (C,D) Relative expression of selected genes implicated in cholesterol and lipoprotein metabolism in jejunum (C), and ileum samples (D) (n = 14 samples per tissue and per group). (E) Quantification of different bile acid classes in feces (n = 5 per group). αMCA, α-muricholic acid; βMCA, β-muricholic acid; CA, cholic acid; UDCA, ursodeoxycholic acid; HDCA, hyodeoxycholic acid; DCA, deoxycholic acid. (F) Inhibition of lipase activity by Totum-070, n = 4–11 replicates. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 vs. ND. #p < 0.05, ##p < 0.01, ###p < 0.001 and ####p < 0.0001 Totum-070 (T070) groups vs. WD. Data are the mean ± SEM.
To complete the characterization of pathways linked to cholesterol metabolism, the concentration of the major bile acid species was measured in feces. With the exception of cholic acid (CA), all bile acid classes were significantly reduced in feces from mice in the WD-T070 3 g/kg group compared with the WD group: α-muricholic acid (αMCA) −41%; β-muricholic acid (βMCA) −60%; ursodeoxycholic acid (UDCA) −92%; hyodeoxycholic acid (HDCA) −54.5%; and deoxycholic acid (DCA) −63% (Figure 4E). Based on these data, the expression of genes associated with bile acid regulation was assessed in liver and ileum (Supplementary Figure S1B,C). In liver, Cyp8b1 and Cyp27a1 were significantly downregulated in the WD-T070 3 g/kg group compared with the WD group as well as Slc10a1 and Slco1a5, which encode transporters implicated in the regulation of bile acid uptake by hepatocytes. In ileum, none of the genes investigated was affected by Totum-070 administration.
Lastly, an in vitro pancreatic lipase activity inhibition test was used to directly assess Totum-070 effect on intestinal lipid digestion. Comparison of Totum-070 and orlistat (the reference lipase inhibitor drug) effects on lipase activity (Figure 4F and Supplementary Figure S1D) showed that Totum-070 inhibited lipase activity (IC50 = 305.6 µg/ml), but not as strongly as orlistat (IC50 = 0.0084 µg/ml.
3.5. Effect of Totum-070 supplementation on fecal microbiota
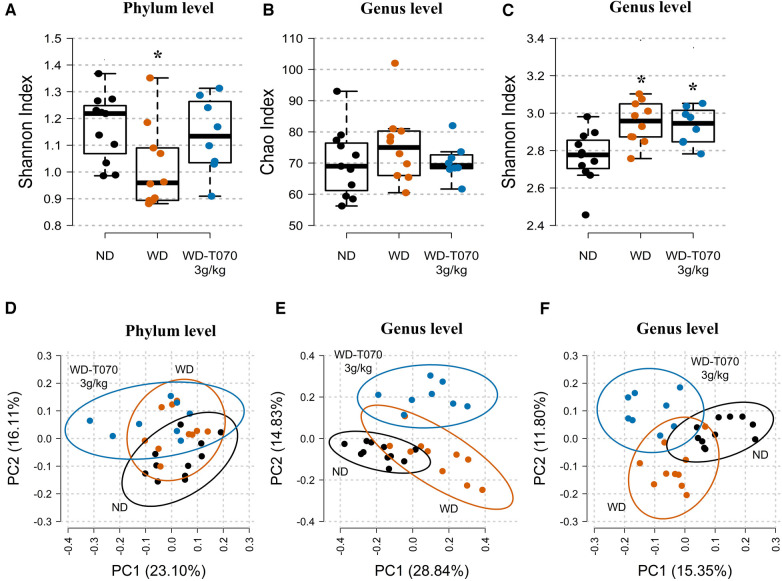
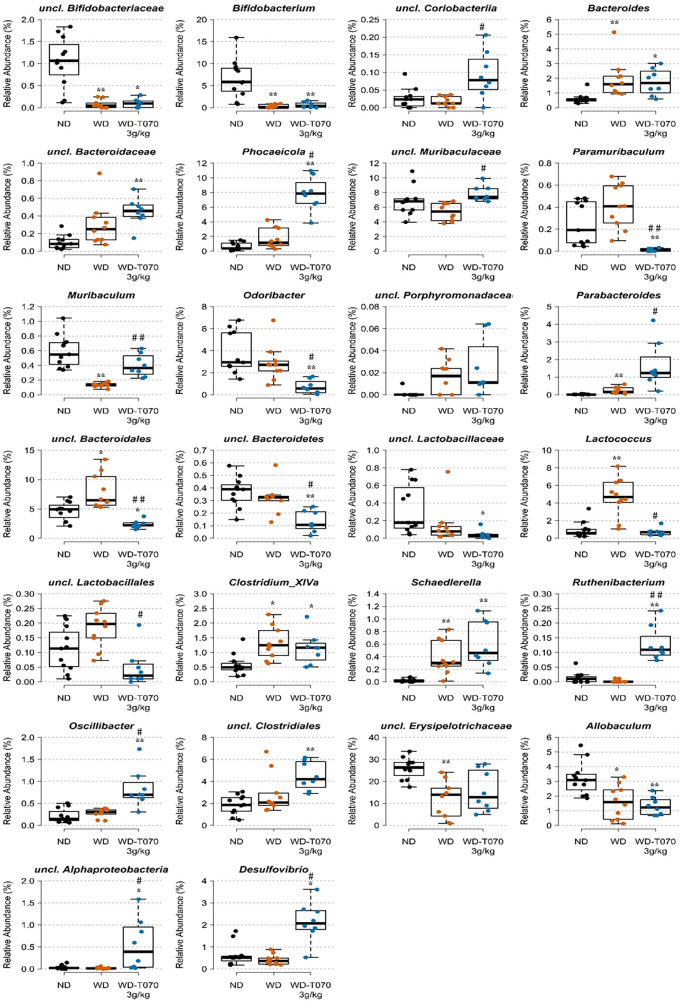
Next, 16S rRNA amplicon sequencing was used to identify the microbiota structure in feces collected from mice in the ND, WD and WD-T070 3 g/kg groups at the study end. At the phylum level, evenness (Shannon index) was significantly decreased in the WD group (p < 0.05), compared with the ND group, but not in the WD-T070 3 g/kg group (Figure 5A). At the genus level, species richness (Chao index) was comparable among groups, whereas evenness (Shannon index) was increased in both WD and WD-T070 3 g/kg groups compared with the ND group (Figure 5B,C). Principal coordinate analysis of the relative abundance of intestinal microbial communities at the phylum level using the Bray-Curtis dissimilarity index followed by PERMANOVA showed that microbial composition differed significantly in the three groups (Bray-Curtis, adonis: p < 0.05 RNDvsWD = 016, p < 0.01 RNDvsWD−T070 3 g/kg = 0.24, p < 0.1 RWDvsWD−T070 3 g/kg = 0.12) (Figure 5D). The same analysis at the genus level confirmed that the WD strongly affected microbial composition compared with the ND (Bray-Curtis, adonis: p < 0.0001, RNDvsWD = 0.32; Figure 5E and Jaccard, adonis: p < 0.0001, RNDvsWD = 0.20; Figure 5F). The microbial composition also differed between the WD-070 group and the ND group (Bray-Curtis, adonis: p < 0.0001 RWDvsWD−T070 3 g/kg = 0.39; Figure 5E and Jaccard, adonis: p < 0.0001, RWDvsWD−T070 3 g/kg = 0.21; Figure 5F) and also the WD group (Bray-Curtis, adonis: p < 0.001, RWDvsWD−T070 3 g/kg = 0.26; Figure 5E and Jaccard, adonis: p < 0.0001, RWDvsWD−T070 3 g/kg = 0.20; Figure 5F). Given the supplementation effect on the alpha and beta diversity measures and the visible differences in the average composition at the phylum and genus levels, an indicator species analysis was performed to identify the taxa that were specifically affected in the different conditions. Data are shown at the phylum level (Table 2) and genus level (Figure 6 and Table 3) as results of the two extremes of the taxonomic levels.
Figure 5.
Effect of Totum-070 on richness, diversity and composition of fecal microbiota. (A) Shannon diversity index at the phylum level. (B) Rarefied Chao index (richness index) at the genus level. (C) Shannon diversity index at the genus level. (D) and (E) Principal coordinate analysis based on the Bray–Curtis dissimilarity index at the phylum and genus levels, with 95% confidence standard deviation ellipses. (F) Principal coordinate analysis based on the Jaccard dissimilarity index at the genus level, with 95% confidence standard deviation ellipses. Boxplots show the median, 25th and 75th percentile, and adjacent values. n = 11 ND, n = 10 WD, n = 8 WD-T070 3 g/kg. * p < 0.05 vs. ND.
Figure 6.
Effect of Totum-070 on composition of fecal microbiota at the genus level. Figures represent relative abundance values of genera that differed significantly between two groups or more. Boxplots show the median, 25th and 75th percentile, and adjacent values. n = 11 ND, n = 10 WD, n = 8 WD-T070 3 g/kg. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 vs. ND. #p < 0.05, ##p < 0.01, ###p < 0.001 and ####p < 0.0001 Totum-070 (T070) group vs. WD.
Table 3.
Fecal microbiota: relative abundance differences at the genus level.
| Genus | % of relative abundance | Adjusted p. values | Plasma cholesterol | |||||
|---|---|---|---|---|---|---|---|---|
| ND | WD | WD-T070 3 g/kg | ND vs. WD | ND vs. WD-T070 3 g/kg | WD vs. WD-T070 3 g/kg | Person | Adjusted p. values | |
| uncl. Bifidobacteriaceae | 1.05 ± 0.58 | 0.07 ± 0.10 | 0.10 ± 0.10 | 0.0067 | 0.0133 | 0.5270 | −59.4% | 0.0058 |
| Bifidobacterium | 6.63 ± 4.43 | 0.30 ± 0.38 | 0.57 ± 0.62 | 0.0065 | 0.0099 | 0.4050 | −55.4% | 0.0119 |
| uncl. Coriobacteriia | 0.03 ± 0.03 | 0.02 ± 0.01 | 0.09 ± 0.07 | 0.7760 | 0.0652 | 0.0360 | 4.6% | 0.8128 |
| Bacteroides | 0.61 ± 0.35 | 1.93 ± 1.26 | 1.74 ± 0.89 | 0.0067 | 0.0133 | 1.0000 | 52.9% | 0.0137 |
| uncl. Bacteroidaceae | 0.10 ± 0.08 | 0.30 ± 0.24 | 0.45 ± 0.16 | 0.0598 | 0.0065 | 0.1645 | 46.4% | 0.0322 |
| Phocaeicola | 0.67 ± 0.52 | 1.71 ± 1.36 | 7.79 ± 2.29 | 0.1621 | 0.0058 | 0.0111 | 32.1% | 0.1549 |
| uncl. Muribaculaceae | 6.86 ± 1.95 | 5.39 ± 1.20 | 7.85 ± 1.06 | 0.1636 | 0.2233 | 0.0162 | −29.0% | 0.1998 |
| Paramuribaculum | 0.25 ± 0.20 | 0.41 ± 0.20 | 0.01 ± 0.01 | 0.2441 | 0.0058 | 0.0096 | 18.3% | 0.4222 |
| Muribaculum | 0.60 ± 0.22 | 0.13 ± 0.03 | 0.40 ± 0.15 | 0.0065 | 0.2086 | 0.0096 | 65.9% | 0.0026 |
| Odoribacter | 3.78 ± 1.89 | 2.85 ± 1.61 | 0.70 ± 0.62 | 0.2910 | 0.0065 | 0.0181 | −21.8% | 0.3337 |
| uncl. Porphyromonadaceae | 0.00 ± 0.00 | 0.02 ± 0.02 | 0.02 ± 0.03 | 0.0590 | 0.0084 | 0.6573 | 47.5% | 0.0322 |
| Parabacteroides | 0.02 ± 0.02 | 0.24 ± 0.18 | 1.64 ± 1.29 | 0.0065 | 0.0058 | 0.0162 | 23.5% | 0.2998 |
| uncl. Bacteroidales | 4.65 ± 1.58 | 7.94 ± 2.92 | 2.38 ± 0.66 | 0.0463 | 0.0133 | 0.0096 | 35.8% | 0.1074 |
| uncl. Bacteroidetes | 0.37 ± 0.12 | 0.32 ± 0.12 | 0.13 ± 0.08 | 0.3998 | 0.0089 | 0.0181 | −11.9% | 0.6376 |
| uncl. Lactobacillaceae | 0.34 ± 0.28 | 0.15 ± 0.22 | 0.04 ± 0.05 | 0.1636 | 0.0133 | 0.1983 | −38.9% | 0.0800 |
| Lactococcus | 0.98 ± 0.92 | 4.66 ± 2.19 | 0.73 ± 0.43 | 0.0074 | 0.7202 | 0.0116 | 46.6% | 0.0322 |
| uncl. Lactobacillales | 0.11 ± 0.08 | 0.19 ± 0.07 | 0.05 ± 0.06 | 0.1452 | 0.1298 | 0.0193 | 9.7% | 0.6663 |
| Clostridium_XlVa | 0.58 ± 0.36 | 1.32 ± 0.54 | 1.14 ± 0.54 | 0.0246 | 0.0442 | 0.6573 | 45.3% | 0.0354 |
| Schaedlerella | 0.02 ± 0.03 | 0.38 ± 0.26 | 0.60 ± 0.36 | 0.0067 | 0.0058 | 0.3201 | 35.6% | 0.1074 |
| Ruthenibacterium | 0.01 ± 0.02 | 0.00 ± 0.00 | 0.13 ± 0.06 | 0.1921 | 0.0058 | 0.0096 | 10.3% | 0.6663 |
| Oscillibacter | 0.22 ± 0.16 | 0.28 ± 0.10 | 0.83 ± 0.43 | 0.4392 | 0.0080 | 0.0181 | 26.6% | 0.2361 |
| uncl. Clostridiales | 1.87 ± 0.87 | 2.84 ± 1.78 | 4.49 ± 1.28 | 0.4392 | 0.0058 | 0.0974 | 44.6% | 0.0361 |
| uncl. Erysipelotrichaceae | 25.87 ± 4.88 | 11.79 ± 8.28 | 15.50 ± 9.24 | 0.0067 | 0.1998 | 0.4050 | −62.9% | 0.0034 |
| Allobaculum | 3.18 ± 1.13 | 1.58 ± 1.14 | 1.31 ± 0.62 | 0.0416 | 0.0099 | 0.7845 | −53.4% | 0.0137 |
| uncl. Alphaproteobacteria | 0.03 ± 0.05 | 0.02 ± 0.02 | 0.55 ± 0.58 | 0.7760 | 0.0302 | 0.0199 | 28.7% | 0.1998 |
| Desulfovibrio | 0.64 ± 0.50 | 0.42 ± 0.23 | 2.14 ± 0.89 | 0.4392 | 0.0133 | 0.0116 | 6.2% | 0.7791 |
Indicator taxa were identified as those with relative abundance values that differed significantly between two groups or more. Relative abundances are presented as mean ± SD and p values were adjusted for multiple testing. Correlations between indicator taxa and plasma total cholesterol was assessed using the Pearson correlation with FDR correction for multiple testing (n = 11 ND, n = 10 WD, n = 8 WD-T070 3 g/kg).
At the phylum level, supplementation with Totum-070 (3 g/kg) led to an increase in the relative abundance of Proteobacteria compared with the WD group (+89%, p < 0.05) and the ND group (+195%, p < 0.01). Moreover, the Firmicutes to Bacteroidetes ratio remained identical between WD and ND (Table 2) but was 20.5% lower in the WD-T070 3 g/kg group compared with the ND group (p < 0.05).
At the genus level (Figure 6 and Table 3), the proportions of uncl. Coriobacteriia (p = 0.036), Phocaeicola (p = 0.0111), uncl. Muribaculaceae (p = 0.0162), Muribaculum (p = 0.0096), Parabacteroides (p = 0.0162), Ruthenibacterium (p = 0.0096), Oscillibacter (p = 0.0181), uncl. Alphaproteobacteria (p = 0.0199) and Desulfovibrio (p = 0.0116) were significantly increased in the WD-T070 3 g/kg group compared with the WD group. Conversely, Paramuribaculum (p = 0.0096), Odoribacter (p = 0.0181), uncl. Bacteroidales (p = 0.0096), uncl. Bacteroidetes (p = 0.0181), Lactococcus (p = 0.0116) and uncl. Lactobacillales (p = 0.0193) were significantly decreased in the WDT070 3 g/kg group compared with the WD group. Noteworthy, some WD-linked alterations were reversed by Totum-070 supplementation. Specifically, Muribaculum richness was significantly reduced in WD-fed mice (−79%, p < 0.01, WD vs. ND), but was increased by Totum-070 (+207%, p < 0.01, WD-T070 3 g/kg vs. WD). Moreover, the Pearson's correlation analysis showed a significant negative association between Muribaculum richness and plasma cholesterol (p = 0.0026). Conversely, Lactococcus richness was significantly increased by WD (+375%, p < 0.01, WD vs. ND, respectively) and reduced by Totum-070 supplementation (−84%, p < 0.05, WD-T070 3 g/kg vs. WD). The relative abundance of Lactococcus was positively correlated with plasma cholesterol (p = 0.0322) (Table 3).
3.6. The hypocholesterolemic effect of totum-070 is associated with increased microbiota-derived SFCAs
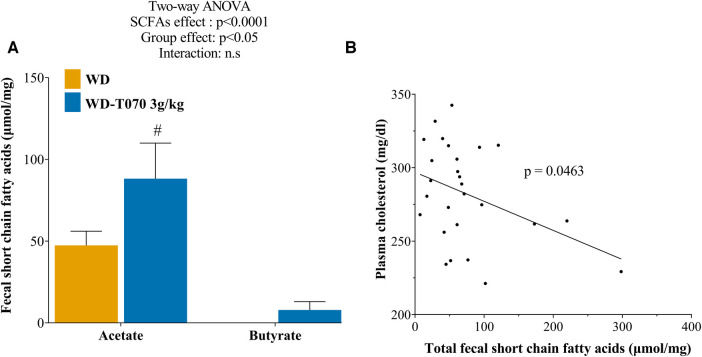
Our analysis showed that the cholesterol-lowering effect of Totum-070 was related to gut microbiota modifications. As fibers and some herb-derived polysaccharides are fermented into SCFAs (mainly acetate, propionate, and butyrate) by intestinal bacteria (47) with potential beneficial effects on lipid disorders, Totum-070 effect on SCFA production was investigated at the study end. Quantification of SCFA species in the feces revealed a significant global increase of the total fecal SCFA amount in the WD-T070 3 g/kg group compared with the WD group (+102.8%, p < 0.05). This effect was mainly driven by fecal acetate production (Figure 7A). Post-hoc analysis indicated a significant increase of acetate in feces from WD-T070 3 g/kg mice compared with WD mice (+86%, p < 0.05). Propionate was not detected in any sample and butyrate only in samples from WD-T070 3 g/kg mice. The total SCFA amount in feces was negatively correlated with plasma cholesterol levels in the WD and WD-T070 3 g/kg groups (Figure 7B, p < 0.05).
Figure 7.
Effect of Totum-070 on fecal short chain fatty acid (SCFA) composition. (A) Quantification of SCFA species content in feces (n = 14 per group) at the study end. (B) Relationship between plasma cholesterol levels and fecal SCFA content in the WD and WD-T070 3 g/kg groups. #p < 0.05, Totum-070 (T070) group vs. WD. Data are the mean ± SEM.
4. Discussion
Atherosclerotic cardiovascular disease is the main cause of mortality worldwide (48, 49). Increased plasma low density lipoprotein cholesterol (LDL-C) level is the primary risk factor for the development of coronary heart disease and atherosclerosis (50, 51). Totum-070 combines five plant extracts involved in the regulation of cholesterol homeostasis: artichoke leaves (52–55), olive leaves (56–59), chrysanthellum (20), goji berries (60, 61) and black pepper (62, 63). Its chemical characterization revealed a high polyphenol content (>15% dry weight, Table 1). Plant-derived polyphenols are considered promising strategies to prevent metabolic disorders such as obesity, type 2 diabetes, and dyslipidemia (64, 65). Here, we evaluated the effects of Totum-070 on cholesterol levels in WD-fed hypercholesterolemic mice. After 3 weeks of supplementation, Totum-070 limited cholesterol elevation in WD-fed mice in a dose-dependent manner. These results are consistent with a recent study in dyslipidemic hamsters showing decreased total cholesterol following daily administration of 3 g/kg Totum-070 by gavage for 12 weeks (21). We also found that 3 g/kg of Totum-070 inhibited body weight and fat mass gain, whereas upon supplementation with only 1 g/kg, cholesterol reduction was not fully related to reduced weight gain. Some of the plants in Totum-070 formulation have already shown similar effects on cholesterol reduction without affecting body weight gain (56–59). In our study, total lipid content in feces from Totum-070 supplemented mice was significantly increased by 31% compared with WD-fed animals (Figure 4A). This result suggests impaired intestinal lipid absorption following Totum-070 administration that could explain, at least in part, its cholesterol-lowering effect. This hypothesis is supported by several in vivo and in vitro studies demonstrating that artichoke leaves (66) and piperine (67–69) and luteolin (70) (two active molecules present in Totum-070) inhibit intestinal cholesterol absorption. Then, we characterized the lipid-related gene response in the intestine after 6 weeks of supplementation (Figures 4C,D), but we could not establish a clear Totum-070-related pattern. Indeed, the supplementation effect on the expression of the selected genes was limited and often not conserved in the three gut segments. A chronic action of Totum-070 on intestinal gene regulation cannot be excluded. Nevertheless, phenolic compounds display very potent inhibitory effects on digestive enzymes that might explain the increased lipid fecal excretion (71). Antilipemic drugs, including orlistat, act as lipase inhibitors and are used in clinic to prevent obesity and hyperlipidemia by decreasing the dietary fat digestion and absorption in blood and by increasing fat excretion in the feces (72). Here, we found that Totum-070 inhibited pancreatic lipase activity (IC50 = 305.6 µg/ml, Figure 4F). This Totum-070 inhibitory effect was comparable to previous results obtained by testing different plant extracts (IC50 > 100 µg/ml), but was lower than the effect observed with orlistat (73–75). Literature data indicate that the artichoke and olive leaf extracts used in Totum-070 have inhibitory effects on digestive enzymes (76, 77). Several polyphenols found in Totum-070 also display inhibitory effects on pancreatic lipase, such as chlorogenic acid, oleanolic acid, di-caffeoylquinic acids, luteolin-7-O-glucoside, luteolin and apigenin (78–81). Although, it is difficult to identify the compounds responsible for a given effect, we can speculate that the inhibition of digestive enzymes by Totum-070 molecules may play a role in the hypocholesterolemic effect and in the weight gain reduction observed at the highest dose (3 g/kg body weight).
Given the importance of liver in lipid and lipoprotein regulation, we quantified the expression of selected genes to determine Totum-070 effects on liver metabolic pathways (Figure 3). Totum-070 did not have any effect on the SREBP2 signaling pathway (Hmgcr and Ldlr expression) and also on hepatic LDL-R protein expression. These results suggest that Totum-070 cholesterol lowering effect does not occur through the liver SREBP2 pathway and LDL-R-mediated LDL-C clearance. This is in contrast with the study by Wauquier et al. who found decreased Hmgcr expression and HMGCR enzyme activity in human HepG2 hepatocytes incubated with human sera enriched in Totum-070 metabolites (21). This discrepancy could be explained by the fact that Hmgcr downregulation by Totum-070 metabolites occurred in a palmitate-induced Hmgcr expression background in cell cultures. Conversely, our mice were fed a WD enriched in cholesterol that inhibited SREPB2 signaling, as attested by Hmgcr and Ldlr downregulation in livers from WD animals compared with the ND group. Cholesterol is eliminated from the body via its conversion into bile acids in the liver through a classic and an alternative pathway (82). In the intestine, 95% of bile acids are reabsorbed by ileum and return to liver. Therefore, bile acid sequestration to prevent their intestinal re-absorption will stimulate bile acid production from cholesterol in liver, and consequently reduce blood cholesterol level. To test whether Totum-070 cholesterol lowering effect was mediated by increased bile acid excretion, bile acid concentrations were quantified in fecal samples. Bile acid concentration was significantly reduced in feces from WD-T070 mice compared with WD mice (Figure 4E). This is in contrast with literature data showing that some Totum-070 components enhance fecal excretion of bile acids. In golden Syrian hamsters fed a HFD associated with 0.24 g/kg artichoke leaf extract (6 weeks of supplementation), plasma cholesterol concentration reduction was associated with a 53% increase in fecal bile acid concentration (66). In Wistar rats fed a HFD for 10 weeks, daily gavage of 40 mg/kg piperine from black pepper increased bile acid excretion in the feces and reduced plasma cholesterol level (67). We cannot explain the difference between our observation and these previous works; however, study design differences (species, intervention, dose, method and duration) may have contributed to these discrepancies. Quantification of the expression of genes encoding enzymes involved in bile acid formation indicated reduced level of Cyp8b1and Cyp27a1 in the WD-T070 groups (Supplementary Figure S1). This suggests that rather than stimulating cholesterol elimination through enhanced bile acid synthesis and fecal excretion, Totum-070 cholesterol-lowering effect is associated with the opposite effect in WD-fed mice. A similar phenotype was described in patients treated with orlistat in whom fecal bile acid amount was decreased (83), while orlistat is known to reduce blood cholesterol level in hypercholesterolemic patients (84). As described earlier, Totum-070 cholesterol-lowering effect might be mediated by inhibition of intestinal cholesterol absorption. Thus, reduced cholesterol elimination though the hepatic bile acid pathway could be compensated by lower cholesterol supply, maintaining Totum-070 cholesterol-lowering global net action.
Recently, the gut microbiota has emerged as a crucial factor that influences cholesterol metabolism. Gut flora modulation by some probiotic and prebiotic agents has cholesterol-lowering property (85). The intestinal microbiota contributes to determine the circulating cholesterol level and may represent a novel therapeutic target in the management of dyslipidemia and cardiovascular diseases (86). Many studies have already linked the microbial community imbalance or maladaptation, defined as “dysbiosis”, to lipid metabolism and metabolic disorders in the host (87, 88). To determine whether Totum-070 hypocholesterolemic effect was associated with the gut microbiota, the association between plasma cholesterol concentration and altered intestinal bacteria at the genus level was tested by Pearson's correlation analysis. The relative abundance of Muribaculum was decreased in the WD group but restored by Totum-070 supplementation (Table 3), whereas the relative abondance of Lactococcus was increased by WD and reduced by Totum-070 supplementation. Moreover, the relative abundances of Muribaculum and of Lactococcus were negatively and positively, respectively, correlated with plasma cholesterol. This suggest that Lactococcus might have adverse effects on lipid metabolism homeostasis, and Muribaculum beneficial effects on plasma cholesterol. This is consistent with previous studies showing that Muribaculum relative abundance is lower in WD-fed mice (89) and obese mice fed a HFD (90, 91) than in ND mice, and that Lactococcus richness is increased in animals fed a HFD (92, 93). Thus, the present results suggest that Muribaculum and Lactococcus are bacteria associated with Totum-070 beneficial effect on lipid disorders caused by WD.
It has been reported that members of the Muribaculum genus can ferment complex polysaccharides (94), and animal studies demonstrated that increased SCFA production is associated with Muribaculum abondance (95, 96). The analysis of the microbiota modifications induced by Totum-070 supplementation highlighted increased abondance of several other genera implicated in SFCA production, such as Desulfovibrio (97), Phocaeicola (98, 99) and Parabacteroides (100–102) for acetate, and Oscillibacter (103, 104) and Ruthenibacterium (105) for butyrate. Moreover, many in vitro studies tested the fermentability of artichoke leaf or olive leaf extracts (included in Totum-070 mix) and demonstrated increased formation of total SCFAs, mainly driven by acetate (106–108). This correlation between increased acetate production upon plant ingredient supplementation and reduction of circulating cholesterol was previously reported. In a clinical trial in patients with hypercholesteremia, oat bran consumption for 3 weeks reduced LDL-C by 12.1% and serum acetate peak was significantly higher than in controls (109). In individuals with normal weight, intake of resistant starch for 4 weeks significantly decreased LDL-C by 5.8% in association with higher acetate concentration (110). In a HFD-fed mouse model, Astragalus polysaccharide supplementation improved hepatic steatosis, obesity and dyslipidemia. This effect was dependent on the presence of acetate-producing bacteria (97). Thus, a significant increase in acetate production in humans and animal models contributes to the hypocholesterolemic effects of plant extracts. Although acetate is a lipogenesis substrate in many cells, evidences from animal models suggest that direct administration of acetate improves the lipid profile. In rats fed a cholesterol-rich diet, supplementation with 0.3% acetic acid for 19 days resulted in a significant total cholesterol reduction by 18.7% and a tendency to triglyceride concentration reduction in liver (111). Oral administration of 5.2 mg/kg of acetate for 6 months in obesity-linked type 2 diabetic Otsuka Long-Evans Tokushima Fatty rats protected against fat accumulation in liver and decreased hypercholesterolemia by 35.9% (112). Lastly, meta-analyses of data from clinical trials indicate that dietary acetic acid supplementation significantly reduces triglyceride levels in individuals with overweight and obesity and in patients with type 2 diabetes (113).
In accordance with the effect of Totum-070, consumption of oleanolic acid in mice increased the relative abundance Oscillibacter (114). In mice, both tyrosol (115) and hydroxytyrosol (116) increased the relative abundance of family Muribaculaceae. Interestingly, previous report showed that intake of Goji juice in DSS-induced ulcerative colitis mice increased relative abundance of Oscillibacter, Desulfovibrio, Parabacteroides and uncl.Muribaculaceae (117), which is consistent with the effects of Totum-070 supplementation. However, the administration of polysaccharides from Goji fruits in streptozotocin-induced mice modified composition of intestinal flora, including increased relative abundance of Bacteroides and decreased relative abundance of Allobaculum (118), while these last two genera were not significantly regulated by Totum-070. Similar discrepancy was observed with administration of Goji extracts which decreased relative abundance of Clostridium_XIVa in HFD-fed mice (26). Contrary to our results, the administration of chlorogenic acid in HFD-fed mice decreased the relative abundance of Desulfovibrio and Bifidobacterium, while Bacteroides was increased (30). These observations demonstrate the complexity behind the various effects generated by the combination of each extract and biomolecule included in Totum-070 mix to produce this specific regulation of intestinal microbiota in WD-fed mice.
Collectively, the present findings indicate that the improved plasma cholesterol level following Totum-070 supplementation, could be, at least partly, mediated by modulation of the gut microbiota and the related acetate production. However, this study has potential limitations. The use of germ-free mice or treatment with broad-spectrum antibiotics to deplete the gut microbiota (119) would give insight in Totum-070 beneficial effects; and also provide knowledge on the importance of the production of SFCAs as a mechanistic part of the Totum-070 hypocholesterolemic effect. Secondly, in order to decipher the molecular mechanism of actions of Totum-070, genetically modified mouse models would be of great interest. For example, mice deficient for the cholesterol transporter NPC1L1 (120) or knock-out for the LDL-receptor (121) would help to understand the cholesterol-lowering effect of Totum-070 in the intestinal cholesterol absorption and through regulation of hepatic LDL-C clearance, respectively. Finally, future good quality clinical studies will have to confirm the hypocholesterolemic potential of this formulation in humans.
In conclusion, we showed that supplementation with Totum-070 prevents the WD-induced plasma cholesterol increase and concomitantly increases fecal lipid excretion and SCFA production. Furthermore, Totum-070 modulated beneficially the gut microbiota composition, by increasing the relative abundance of Muribaculum and reducing the relative abundance of Lactococcus. These results demonstrates that Totum-070 could be used as an effective dietary supplement to prevent blood cholesterol increase, a major risk factor for cardiovascular disease.
Acknowledgments
We thank Elisabetta Andermarcher for reviewing and correcting the manuscript. Part of the experiments (16S rRNA gene sequencing) were performed at the Genome Transcriptome Facility of Bordeaux (Grants from In-vestissements d'avenir, Convention attributive d'aide EquipEx Xyloforest ANR-10-EQPX-16-01).
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This work was supported by a FEDER (Fonds Européen de Développement Régional) grant number 2015RPC-BAFE-114.
Data availability statement
The datasets presented in this study can be found in online repositories. The 16S rRNA gene sequencing data from mouse fecal microbiota presented in this study has been deposited in the Sequence Read Archive under BioProject PRJNA952522 with the following accession numbers: SRR24071998 to SRR24072026 (BioSamples: SAMN34074972 to SAMN34075000).
Ethics statement
The animal study was approved by local Ethics Committee C2E2A, Auvergne, France. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CL: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. MV: Conceptualization, Formal Analysis, Methodology, Software, Validation, Writing – review & editing. AB: Conceptualization, Formal Analysis, Methodology, Validation, Writing – review & editing. YO: Conceptualization, Validation, Writing – review & editing. MM: Conceptualization, Formal Analysis, Methodology, Writing – review & editing. FL: Conceptualization, Formal Analysis, Methodology, Writing – review & editing. NB: Conceptualization, Formal Analysis, Methodology, Writing – review & editing. MG: Conceptualization, Formal Analysis, Methodology, Writing – review & editing. BG: Conceptualization, Methodology, Writing – review & editing. TM: Conceptualization, Methodology, Writing – review & editing. BC: Methodology, Writing – review & editing. SP: Supervision, Writing – review & editing. SB: Methodology, Writing – review & editing. JB: Supervision, Writing – review & editing. PS: Supervision, Writing – review & editing.
Conflict of interest
CL, MV, YO, MM, FL and PS are employees of Valbiotis. SP is CEO of Valbiotis. JB is member of the scientific committee and stock shareholder of Valbiotis. BG is member of the scientific committee of Valbiotis. BC reports honorarium and consulting fees from Nestlé, Procter and Nobles and Qiagen.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2024.1342388/full#supplementary-material
References
- 1.Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. (2020) 41(1):111–88. 10.1093/eurheartj/ehz455 [DOI] [PubMed] [Google Scholar]
- 2.Howard BV, Kritchevsky D. Phytochemicals and cardiovascular disease. A statement for healthcare professionals from the American heart association. Circulation. (1997) 95(11):2591–3. 10.1161/01.CIR.95.11.2591 [DOI] [PubMed] [Google Scholar]
- 3.Zanotti I, Dall'Asta M, Mena P, Mele L, Bruni R, Ray S, et al. Atheroprotective effects of (poly)phenols: a focus on cell cholesterol metabolism. Food Funct. (2015) 6(1):13–31. 10.1039/C4FO00670D [DOI] [PubMed] [Google Scholar]
- 4.Morze J, Osadnik T, Osadnik K, Lejawa M, Jakubiak G, Pawlas N, et al. Comparative effect of nutraceuticals on lipid profile: a protocol for systematic review and network meta-analysis. BMJ Open. (2020) 10(8):e032755. 10.1136/bmjopen-2019-032755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osadnik T, Golawski M, Lewandowski P, Morze J, Osadnik K, Pawlas N, et al. A network meta-analysis on the comparative effect of nutraceuticals on lipid profile in adults. Pharmacol Res. (2022) 183:106402. 10.1016/j.phrs.2022.106402 [DOI] [PubMed] [Google Scholar]
- 6.Cicero AFG, Colletti A, Bajraktari G, Descamps O, Djuric DM, Ezhov M, et al. Lipid-lowering nutraceuticals in clinical practice: position paper from an international lipid expert panel. Nutr Rev. (2017) 75(9):731–67. 10.1093/nutrit/nux047 [DOI] [PubMed] [Google Scholar]
- 7.Wider B, Pittler MH, Thompson-Coon J, Ernst E. Artichoke leaf extract for treating hypercholesterolaemia. Cochrane Database Syst Rev. (2013) 28(3):CD003335. 10.1002/14651858.CD003335.pub3 [DOI] [PubMed] [Google Scholar]
- 8.Sahebkar A, Pirro M, Banach M, Mikhailidis DP, Atkin SL, Cicero AFG. Lipid-lowering activity of artichoke extracts: a systematic review and meta-analysis. Crit Rev Food Sci Nutr. (2018) 58(15):2549–56. 10.1080/10408398.2017.1332572 [DOI] [PubMed] [Google Scholar]
- 9.Pandino G, Courts FL, Lombardo S, Mauromicale G, Williamson G. Caffeoylquinic acids and flavonoids in the immature inflorescence of globe artichoke, wild cardoon, and cultivated cardoon. J Agric Food Chem. (2010) 58(2):1026–31. 10.1021/jf903311j [DOI] [PubMed] [Google Scholar]
- 10.Ben Salem M, Affes H, Ksouda K, Dhouibi R, Sahnoun Z, Hammami S, et al. Pharmacological studies of artichoke leaf extract and their health benefits. Plant Foods Hum Nutr. (2015) 70(4):441–53. 10.1007/s11130-015-0503-8 [DOI] [PubMed] [Google Scholar]
- 11.Acar-Tek N, Agagunduz D. Olive leaf (Olea europaea L. folium): potential effects on glycemia and lipidemia. Ann Nutr Metab. (2020) 76(1):10–5. 10.1159/000505508 [DOI] [PubMed] [Google Scholar]
- 12.Vogel P, Kasper Machado I, Garavaglia J, Zani VT, de Souza D, Morelo Dal Bosco S. Polyphenols benefits of olive leaf (Olea europaea L) to human health. Nutr Hosp. (2014) 31(3):1427–33. 10.3305/nh.2015.31.3.8400 [DOI] [PubMed] [Google Scholar]
- 13.Lockyer S, Yaqoob P, Spencer JPE, Rowland IR. Olive leaf phenolics and cardiovascular risk reduction: physiological effects and mechanisms of action. Nutr Aging. (2012) 1(2):125–40. 10.3233/NUA-2012-0011 [DOI] [Google Scholar]
- 14.Kulczyński B, Gramza-Michałowska A. Goji berry (Lycium barbarum): composition and health effects—a review. Pol J Food Nutr Sci. (2016) 66(2):67–75. 10.1515/pjfns-2015-0040 [DOI] [Google Scholar]
- 15.Cheng J, Zhou ZW, Sheng HP, He LJ, Fan XW, He ZX, et al. An evidence-based update on the pharmacological activities and possible molecular targets of Lycium barbarum polysaccharides. Drug Des Devel Ther. (2015) 9:33–78. 10.2147/DDDT.S72892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mocan A, Zengin G, Simirgiotis M, Schafberg M, Mollica A, Vodnar DC, et al. Functional constituents of wild and cultivated Goji (L. barbarum L.) leaves: phytochemical characterization, biological profile, and computational studies. J Enzyme Inhib Med Chem. (2017) 32(1):153–68. 10.1080/14756366.2016.1243535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo XF, Li ZH, Cai H, Li D. The effects of Lycium barbarum L. (L. barbarum) on cardiometabolic risk factors: a meta-analysis of randomized controlled trials. Food Funct. (2017) 8(5):1741–8. 10.1039/C7FO00183E [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan K. Black pepper and its pungent principle-piperine: a review of diverse physiological effects. Crit Rev Food Sci Nutr. (2007) 47(8):735–48. 10.1080/10408390601062054 [DOI] [PubMed] [Google Scholar]
- 19.Lambert JD, Hong J, Kim DH, Mishin VM, Yang CS. Piperine enhances the bioavailability of the tea polyphenol (-)-epigallocatechin-3-gallate in mice. J Nutr. (2004) 134(8):1948–52. 10.1093/jn/134.8.1948 [DOI] [PubMed] [Google Scholar]
- 20.Honore-Thorez D. Description, identification and therapeutic use of chrysanthellum “americanum": chrysanthellum indicum DC. Subsp afroamericanum B. L. Turner. J Pharm Belg. (1985) 40(5):323–31. [PubMed] [Google Scholar]
- 21.Wauquier F, Boutin-Wittrant L, Krisa S, Valls J, Langhi C, Otero YF, et al. Circulating human metabolites resulting from TOTUM-070 absorption (a plant-based, polyphenol-rich ingredient) improve lipid metabolism in human hepatocytes: lessons from an original ex vivo clinical trial. Nutrients. (2023) 15(8):1903. 10.3390/nu15081903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. (2020) 21(4):225–45. 10.1038/s41580-019-0190-7 [DOI] [PubMed] [Google Scholar]
- 23.Ghazalpour A, Cespedes I, Bennett BJ, Allayee H. Expanding role of gut microbiota in lipid metabolism. Curr Opin Lipidol. (2016) 27(2):141–7. 10.1097/MOL.0000000000000278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kriaa A, Bourgin M, Potiron A, Mkaouar H, Jablaoui A, Gerard P, et al. Microbial impact on cholesterol and bile acid metabolism: current status and future prospects. J Lipid Res. (2019) 60(2):323–32. 10.1194/jlr.R088989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng G, Duan Y, Zhong Y, Song B, Zheng J, Zhang S, et al. Plant extracts in obesity: a role of gut microbiota. Front Nutr. (2021) 8:727951. 10.3389/fnut.2021.727951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong W, Huang Y, Shu Y, Fan X, Tian X, Yan Y, et al. Water extract of goji berries improves neuroinflammation induced by a high-fat and high-fructose diet based on the bile acid-mediated gut-brain axis pathway. Food Funct. (2023) 14(18):8631–45. 10.1039/D3FO02651E [DOI] [PubMed] [Google Scholar]
- 27.Cremonesi P, Curone G, Biscarini F, Cotozzolo E, Menchetti L, Riva F, et al. Dietary supplementation with Goji berries (Lycium barbarum) modulates the microbiota of digestive tract and caecal metabolites in rabbits. Animals. (2022) 12(1):121. 10.3390/ani12010121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vezza T, Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, Romero M, Sanchez M, et al. The metabolic and vascular protective effects of olive (Olea europaea L.) leaf extract in diet-induced obesity in mice are related to the amelioration of gut microbiota dysbiosis and to its immunomodulatory properties. Pharmacol Res. (2019) 150:104487. 10.1016/j.phrs.2019.104487 [DOI] [PubMed] [Google Scholar]
- 29.Ye X, Liu Y, Hu J, Gao Y, Ma Y, Wen D. Chlorogenic acid-induced gut microbiota improves metabolic endotoxemia. Front Endocrinol. (2021) 12:762691. 10.3389/fendo.2021.762691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Lam KL, Hu J, Ge S, Zhou A, Zheng B, et al. Chlorogenic acid alleviates obesity and modulates gut microbiota in high-fat-fed mice. Food Sci Nutr. (2019) 7(2):579–88. 10.1002/fsn3.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cottet J, Etienne J. Determination of serum lipids by the sulfo-phospho-vanillic method of E. Chabrol and R. Charonnat. Bull Acad Natl Med. (1965) 149(16):331–8. [PubMed] [Google Scholar]
- 32.Cheng YS, Zheng Y, VanderGheynst JS. Rapid quantitative analysis of lipids using a colorimetric method in a microplate format. Lipids. (2011) 46(1):95–103. 10.1007/s11745-010-3494-0 [DOI] [PubMed] [Google Scholar]
- 33.Benson JR, Hare PE. O-phthalaldehyde: fluorogenic detection of primary amines in the picomole range. Comparison with fluorescamine and ninhydrin. Proc Natl Acad Sci U S A. (1975) 72(2):619–22. 10.1073/pnas.72.2.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chavanelle V, Otero YF, Le Joubioux F, Ripoche D, Bargetto M, Vluggens A, et al. Effects of Totum-63 on glucose homeostasis and postprandial glycemia: a translational study. Am J Physiol Endocrinol Metab. (2021) 320(6):E1119–E37. 10.1152/ajpendo.00629.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilda JE, Gomes AV. Western blotting using in-gel protein labeling as a normalization control: stain-free technology. Methods Mol Biol. (2015) 1295:381–91. 10.1007/978-1-4939-2550-6_27 [DOI] [PubMed] [Google Scholar]
- 36.Kraus D, Yang Q, Kahn BB. Lipid extraction from mouse feces. Bio Protoc. (2015) 5(1):e1375. 10.21769/BioProtoc.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghorasaini M, Mohammed Y, Adamski J, Bettcher L, Bowden JA, Cabruja M, et al. Cross-laboratory standardization of preclinical lipidomics using differential mobility spectrometry and multiple reaction monitoring. Anal Chem. (2021) 93(49):16369–78. 10.1021/acs.analchem.1c02826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su B, Bettcher LF, Hsieh WY, Hornburg D, Pearson MJ, Blomberg N, et al. A DMS shotgun lipidomics workflow application to facilitate high-throughput, comprehensive lipidomics. J Am Soc Mass Spectrom. (2021) 32(11):2655–63. 10.1021/jasms.1c00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallier M, Suwandi A, Ehrhardt K, Belheouane M, Berry D, Cepic A, et al. Pathometagenomics reveals susceptibility to intestinal infection by Morganella to be mediated by the blood group-related B4galnt2 gene in wild mice. Gut Microbes. (2023) 15(1):2164448. 10.1080/19490976.2022.2164448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. (2011) 17(1):10–2. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 41.Edgar RC. UNOISE2: improved error-correction for illumina 16S and ITS amplicon sequencing. bioRxiv. (2016):081257. 10.1101/081257 [DOI] [Google Scholar]
- 42.Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. (2015) 31(21):3476–82. 10.1093/bioinformatics/btv401 [DOI] [PubMed] [Google Scholar]
- 43.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75(23):7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. (2014) 42(Database issue):D633–42. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41(Database issue):D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philip D. VEGAN, a package of R functions for community ecology. J Veg Sci. (2003) 14(6):927–30. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- 47.Ilyes T, Silaghi CN, Craciun AM. Diet-related changes of short-chain fatty acids in blood and feces in obesity and metabolic syndrome. Biology. (2022) 11(11):1556. 10.3390/biology11111556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dagenais GR, Leong DP, Rangarajan S, Lanas F, Lopez-Jaramillo P, Gupta R, et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet. (2020) 395(10226):785–94. 10.1016/S0140-6736(19)32007-0 [DOI] [PubMed] [Google Scholar]
- 49.Farzadfar F. Cardiovascular disease risk prediction models: challenges and perspectives. Lancet Glob Health. (2019) 7(10):e1288–e9. 10.1016/S2214-109X(19)30365-1 [DOI] [PubMed] [Google Scholar]
- 50.Boren J, Chapman MJ, Krauss RM, Packard CJ, Bentzon JF, Binder CJ, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: pathophysiological, genetic, and therapeutic insights: a consensus statement from the European atherosclerosis society consensus panel. Eur Heart J. (2020) 41(24):2313–30. 10.1093/eurheartj/ehz962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prospective Studies C, Lewington S, Whitlock G, Clarke R, Sherliker P, Emberson J, et al. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. (2007) 370(9602):1829–39. 10.1016/S0140-6736(07)61778-4 [DOI] [PubMed] [Google Scholar]
- 52.Kusku-Kiraz Z, Mehmetcik G, Dogru-Abbasoglu S, Uysal M. Artichoke leaf extract reduces oxidative stress and lipoprotein dyshomeostasis in rats fed on high cholesterol diet. Phytother Res. (2010) 24(4):565–70. 10.1002/ptr.2985 [DOI] [PubMed] [Google Scholar]
- 53.Kucukgergin C, Aydin AF, Ozdemirler-Erata G, Mehmetcik G, Kocak-Toker N, Uysal M. Effect of artichoke leaf extract on hepatic and cardiac oxidative stress in rats fed on high cholesterol diet. Biol Trace Elem Res. (2010) 135(1-3):264–74. 10.1007/s12011-009-8484-9 [DOI] [PubMed] [Google Scholar]
- 54.Ben Salem M, Affes H, Dhouibi R, Charfi S, Turki M, Hammami S, et al. Effect of artichoke (cynara scolymus) on cardiac markers, lipid profile and antioxidants levels in tissue of HFD-induced obesity. Arch Physiol Biochem. (2022) 128(1):184–94. 10.1080/13813455.2019.1670213 [DOI] [PubMed] [Google Scholar]
- 55.Ben Salem M, Ksouda K, Dhouibi R, Charfi S, Turki M, Hammami S, et al. LC-MS/MS analysis and hepatoprotective activity of artichoke (Cynara scolymus L.) leaves extract against high fat diet-induced obesity in rats. Biomed Res Int. (2019) 2019:4851279. 10.1155/2019/4851279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheurfa M, Abdallah HH, Allem R, Noui A, Picot-Allain CMN, Mahomoodally F. Hypocholesterolaemic and antioxidant properties of Olea europaea L. Leaves from chlef province, algeria using in vitro, in vivo and in silico approaches. Food Chem Toxicol. (2019) 123:98–105. 10.1016/j.fct.2018.10.002 [DOI] [PubMed] [Google Scholar]
- 57.Poudyal H, Campbell F, Brown L. Olive leaf extract attenuates cardiac, hepatic, and metabolic changes in high carbohydrate-, high fat-fed rats. J Nutr. (2010) 140(5):946–53. 10.3945/jn.109.117812 [DOI] [PubMed] [Google Scholar]
- 58.Yoon L, Liu YN, Park H, Kim HS. Olive leaf extract elevates hepatic PPAR alpha mRNA expression and improves Serum lipid profiles in ovariectomized rats. J Med Food. (2015) 18(7):738–44. 10.1089/jmf.2014.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Geng C, Jiang L, Gong D, Liu D, Yoshimura H, et al. The anti-atherosclerotic effect of olive leaf extract is related to suppressed inflammatory response in rabbits with experimental atherosclerosis. Eur J Nutr. (2008) 47(5):235–43. 10.1007/s00394-008-0717-8 [DOI] [PubMed] [Google Scholar]
- 60.Cui B, Liu S, Lin X, Wang J, Li S, Wang Q, et al. Effects of Lycium barbarum aqueous and ethanol extracts on high-fat-diet induced oxidative stress in rat liver tissue. Molecules. (2011) 16(11):9116–28. 10.3390/molecules16119116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo Q, Cai Y, Yan J, Sun M, Corke H. Hypoglycemic and hypolipidemic effects and antioxidant activity of fruit extracts from Lycium barbarum. Life Sci. (2004) 76(2):137–49. 10.1016/j.lfs.2004.04.056 [DOI] [PubMed] [Google Scholar]
- 62.Parim B, Harishankar N, Balaji M, Pothana S, Sajjalaguddam RR. Effects of Piper nigrum extracts: restorative perspectives of high-fat diet-induced changes on lipid profile, body composition, and hormones in sprague-dawley rats. Pharm Biol. (2015) 53(9):1318–28. 10.3109/13880209.2014.980585 [DOI] [PubMed] [Google Scholar]
- 63.Vijayakumar RS, Surya D, Senthilkumar R, Nalini N. Hypolipidemic effect of black pepper (Piper nigrum linn.) in rats fed high fat diet. J Clin Biochem Nutr. (2002) 32:31–42. 10.3164/jcbn.32.31 [DOI] [Google Scholar]
- 64.Guasch-Ferre M, Merino J, Sun Q, Fito M, Salas-Salvado J. Dietary polyphenols, mediterranean diet, prediabetes, and type 2 diabetes: a narrative review of the evidence. Oxid Med Cell Longev. (2017) 2017:6723931. 10.1155/2017/6723931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graf BL, Raskin I, Cefalu WT, Ribnicky DM. Plant-derived therapeutics for the treatment of metabolic syndrome. Curr Opin Investig Drugs. (2010) 11(10):1107–15. [PMC free article] [PubMed] [Google Scholar]
- 66.Qiang Z, Lee SO, Ye Z, Wu X, Hendrich S. Artichoke extract lowered plasma cholesterol and increased fecal bile acids in golden Syrian hamsters. Phytother Res. (2012) 26(7):1048–52. 10.1002/ptr.3698 [DOI] [PubMed] [Google Scholar]
- 67.Vijayakumar RS, Nalini N. Lipid-lowering efficacy of piperine from piper nigrum l. In high-fat diet and antithyroid drug-induced hypercholesterolemic rats. J Food Biochem. (2006) 30(4):405–21. 10.1111/j.1745-4514.2006.00074.x [DOI] [PubMed] [Google Scholar]
- 68.Duangjai A, Ingkaninan K, Praputbut S, Limpeanchob N. Black pepper and piperine reduce cholesterol uptake and enhance translocation of cholesterol transporter proteins. J Nat Med. (2013) 67(2):303–10. 10.1007/s11418-012-0682-7 [DOI] [PubMed] [Google Scholar]
- 69.Zhu H, Chen J, He Z, Hao W, Liu J, Kwek E, et al. Plasma cholesterol-lowering activity of soybean germ phytosterols. Nutrients. (2019) 11(11):2784. 10.3390/nu11112784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogawa M, Yamanashi Y, Takada T, Abe K, Kobayashi S. Effect of luteolin on the expression of intestinal cholesterol transporters. J Funct Foods. (2017) 36:274–9. 10.1016/j.jff.2017.07.008 [DOI] [Google Scholar]
- 71.You Q, Chen F, Wang X, Luo PG, Jiang Y. Inhibitory effects of muscadine anthocyanins on alpha-glucosidase and pancreatic lipase activities. J Agric Food Chem. (2011) 59(17):9506–11. 10.1021/jf201452v [DOI] [PubMed] [Google Scholar]
- 72.Heck AM, Yanovski JA, Calis KA. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy. (2000) 20(3):270–9. 10.1592/phco.20.4.270.34882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim YS, Lee Y, Kim J, Sohn E, Kim CS, Lee YM, et al. Inhibitory activities of cudrania tricuspidata leaves on pancreatic lipase in vitro and lipolysis in vivo. Evid Based Complement Alternat Med. (2012) 2012:878365. 10.1155/2012/878365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seyedan A, Alshawsh MA, Alshagga MA, Koosha S, Mohamed Z. Medicinal plants and their inhibitory activities against pancreatic lipase: a review. Evid Based Complement Alternat Med. (2015) 2015:973143. 10.1155/2015/973143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haguet Q, Le Joubioux F, Chavanelle V, Groult H, Schoonjans N, Langhi C, et al. Inhibitory potential of alpha-amylase, alpha-glucosidase, and pancreatic lipase by a formulation of five plant extracts: TOTUM-63. Int J Mol Sci. (2023) 24(4):3652. 10.3390/ijms24043652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ben Salem M, Ben Abdallah Kolsi R, Dhouibi R, Ksouda K, Charfi S, Yaich M, et al. Protective effects of Cynara scolymus leaves extract on metabolic disorders and oxidative stress in alloxan-diabetic rats. BMC Complement Altern Med. (2017) 17(1):328. 10.1186/s12906-017-1835-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Komaki E, Yamaguchi S, Maru I, Kinoshita M, Kakehi K, Ohta Y, et al. Identification of anti-α-amylase components from olive leaf extracts. Food Sci Technol Res. (2003) 9(1):35–9. 10.3136/fstr.9.35 [DOI] [Google Scholar]
- 78.Ahmad I, Syakfanaya AM, Azminah A, Saputri FC, Mun'im A. Optimization of Betaine-Sorbitol Natural Deep Eutectic Solvent-Based Ultrasound-Assisted Extraction and Pancreatic Lipase Inhibitory Activity of Chlorogenic Acid and Caffeine Content from Robusta Green Coffee Beans. Heliyon. (2021) 7:e07702. 10.1016/j.heliyon.2021.e07702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li MM, Chen YT, Ruan JC, Wang WJ, Chen JG, Zhang QF. Structure-activity relationship of dietary flavonoids on pancreatic lipase. Curr Res Food Sci. (2023) 6:100424. 10.1016/j.crfs.2022.100424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Chen L, Liu H, Xie J, Yin W, Xu Z, et al. Characterization of the synergistic inhibitory effect of cyanidin-3-O-glucoside and catechin on pancreatic lipase. Food Chem. (2023) 404(Pt B):134672. 10.1016/j.foodchem.2022.134672 [DOI] [PubMed] [Google Scholar]
- 81.Hu B, Cui F, Yin F, Zeng X, Sun Y, Li Y. Caffeoylquinic acids competitively inhibit pancreatic lipase through binding to the catalytic triad. Int J Biol Macromol. (2015) 80:529–35. 10.1016/j.ijbiomac.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 82.Chiang J. Recent advances in understanding bile acid homeostasis [version 1; peer review: 2 approved]. F1000Res. (2017) 6:2029. 10.12688/f1000research.12449.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahnen DJ, Guerciolini R, Hauptman J, Blotner S, Woods CJ, Wargovich MJ. Effect of orlistat on fecal fat, fecal biliary acids, and colonic cell proliferation in obese subjects. Clin Gastroenterol Hepatol. (2007) 5(11):1291–9. 10.1016/j.cgh.2007.07.009 [DOI] [PubMed] [Google Scholar]
- 84.Muls E, Kolanowski J, Scheen A, Van Gaal L, ObelHyx Study G. The effects of orlistat on weight and on serum lipids in obese patients with hypercholesterolemia: a randomized, double-blind, placebo-controlled, multicentre study. Int J Obes Relat Metab Disord. (2001) 25(11):1713–21. 10.1038/sj.ijo.0801814 [DOI] [PubMed] [Google Scholar]
- 85.Ooi LG, Liong MT. Cholesterol-lowering effects of probiotics and prebiotics: a review of in vivo and in vitro findings. Int J Mol Sci. (2010) 11(6):2499–522. 10.3390/ijms11062499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Roy T, Lecuyer E, Chassaing B, Rhimi M, Lhomme M, Boudebbouze S, et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol. (2019) 17(1):94. 10.1186/s12915-019-0715-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu Y, Raka F, Adeli K. The role of the gut microbiota in lipid and lipoprotein metabolism. J Clin Med. (2019) 8(12):2227. 10.3390/jcm8122227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang X, Gerard P. Diet-gut microbiota interactions on cardiovascular disease. Comput Struct Biotechnol J. (2022) 20:1528–40. 10.1016/j.csbj.2022.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Do MH, Lee HB, Lee E, Park HY. The effects of gelatinized wheat starch and high salt diet on gut microbiota and metabolic disorder. Nutrients. (2020) 12(2):301. 10.3390/nu12020301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu Q, Niu Y, Yang Y, Mao Q, Lu Y, Ran H, et al. Polydextrose alleviates adipose tissue inflammation and modulates the gut microbiota in high-fat diet-fed mice. Front Pharmacol. (2021) 12:795483. 10.3389/fphar.2021.795483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Do MH, Lee E, Oh MJ, Kim Y, Park HY. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients. (2018) 10(6):761. 10.3390/nu10060761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lang JM, Sedgeman LR, Cai L, Layne JD, Wang Z, Pan C, et al. Dietary and pharmacologic manipulations of host lipids and their interaction with the gut microbiome in non-human primates. Front Med. (2021) 8:646710. 10.3389/fmed.2021.646710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang M, Han H, Wan F, Zhong R, Do YJ, Oh SI, et al. Dihydroquercetin supplementation improved hepatic lipid dysmetabolism mediated by gut microbiota in high-fat diet (HFD)-fed mice. Nutrients. (2022) 14(24):5214. 10.3390/nu14245214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. (2016) 4(1):36. 10.1186/s40168-016-0181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamane T, Handa S, Imai M, Harada N, Sakamoto T, Ishida T, et al. Exopolysaccharides from a scandinavian fermented milk viili increase butyric acid and muribaculum members in the mouse gut. Food Chem. (2021) 3:100042. 10.1016/j.fochms.2021.100042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith BJ, Miller RA, Schmidt TM. Muribaculaceae genomes assembled from metagenomes suggest genetic drivers of differential response to acarbose treatment in mice. mSphere. (2021) 6(6):e0085121. 10.1128/msphere.00851-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hong Y, Sheng L, Zhong J, Tao X, Zhu W, Ma J, et al. Desulfovibrio vulgaris, a potent acetic acid-producing bacterium, attenuates nonalcoholic fatty liver disease in mice. Gut Microbes. (2021) 13(1):1–20. 10.1080/19490976.2021.1930874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi JY, Choi SH, Park JE, Kim JS, Lee J, Lee MK, et al. Phocaeicola faecicola sp. Nov., isolated from porcine faeces. Int J Syst Evol Microbiol. (2021) 71(9). 10.1099/ijsem.0.005008 [DOI] [PubMed] [Google Scholar]
- 99.Sun Z, Jiang X, Wang B, Tian F, Zhang H, Yu L. Novel phocaeicola strain ameliorates dextran sulfate sodium-induced colitis in mice. Curr Microbiol. (2022) 79(12):393. 10.1007/s00284-022-03054-6 [DOI] [PubMed] [Google Scholar]
- 100.Wang YJ, Xu XJ, Zhou N, Sun Y, Liu C, Liu SJ, et al. Parabacteroides acidifaciens sp. Nov., isolated from human faeces. Int J Syst Evol Microbiol. (2019) 69(3):761–6. 10.1099/ijsem.0.003230 [DOI] [PubMed] [Google Scholar]
- 101.Cui Y, Zhang L, Wang X, Yi Y, Shan Y, Liu B, et al. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol Lett. (2022) 369(1):fnac072. 10.1093/femsle/fnac072 [DOI] [PubMed] [Google Scholar]
- 102.Lei Y, Tang L, Liu S, Hu S, Wu L, Liu Y, et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome. (2021) 9(1):115. 10.1186/s40168-021-01065-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee GH, Rhee MS, Chang DH, Lee J, Kim S, Yoon MH, et al. Oscillibacter ruminantium sp. Nov., isolated from the rumen of Korean native cattle. Int J Syst Evol Microbiol. (2013) 63(Pt 6):1942–6. 10.1099/ijs.0.041749-0 [DOI] [PubMed] [Google Scholar]
- 104.Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. (2017) 2(6):e00130-17. 10.1128/mSystems.00130-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker A, Schmartz GP, Groger L, Grammes N, Galata V, Philippeit H, et al. Effects of resistant starch on symptoms, fecal markers, and gut microbiota in Parkinson’s disease—the RESISTA-PD trial. Genomics Proteomics Bioinformatics. (2022) 20(2):274–87. 10.1016/j.gpb.2021.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holgado F, Campos-Monfort G, Cdl H, Rupérez P. In vitro fermentability of globe artichoke by-product by Lactobacillus acidophilus and Bifidobacterium bifidum. Bioactive Carbohydrates Dietary Fibre. (2021) 26:100286. 10.1016/j.bcdf.2021.100286 [DOI] [Google Scholar]
- 107.Van den Abbeele P, Ghyselinck J, Marzorati M, Villar A, Zangara A, Smidt CR, et al. In vitro evaluation of prebiotic properties of a commercial artichoke inflorescence extract revealed bifidogenic effects. Nutrients. (2020) 12(6):1552. 10.3390/nu12061552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haddadin MSY. Effect of olive leaf extracts on the growth and metabolism of two probiotic bacteria of intestinal origin. Pak J Nutr. (2010) 9:787–93. 10.3923/pjn.2010.787.793 [DOI] [Google Scholar]
- 109.Bridges SR, Anderson JW, Deakins DA, Dillon DW, Wood CL. Oat bran increases serum acetate of hypercholesterolemic men. Am J Clin Nutr. (1992) 56(2):455–9. 10.1093/ajcn/56.2.455 [DOI] [PubMed] [Google Scholar]
- 110.Zhang L, Ouyang Y, Li H, Shen L, Ni Y, Fang Q, et al. Metabolic phenotypes and the gut microbiota in response to dietary resistant starch type 2 in normal-weight subjects: a randomized crossover trial. Sci Rep. (2019) 9(1):4736. 10.1038/s41598-018-38216-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fushimi T, Suruga K, Oshima Y, Fukiharu M, Tsukamoto Y, Goda T. Dietary acetic acid reduces serum cholesterol and triacylglycerols in rats fed a cholesterol-rich diet. Br J Nutr. (2006) 95(5):916–24. 10.1079/BJN20061740 [DOI] [PubMed] [Google Scholar]
- 112.Yamashita H, Fujisawa K, Ito E, Idei S, Kawaguchi N, Kimoto M, et al. Improvement of obesity and glucose tolerance by acetate in type 2 diabetic otsuka long-evans tokushima fatty (OLETF) rats. Biosci Biotechnol Biochem. (2007) 71(5):1236–43. 10.1271/bbb.60668 [DOI] [PubMed] [Google Scholar]
- 113.Valdes DS, So D, Gill PA, Kellow NJ. Effect of dietary acetic acid supplementation on plasma glucose, lipid profiles, and body mass Index in human adults: a systematic review and meta-analysis. J Acad Nutr Diet. (2021) 121(5):895–914. 10.1016/j.jand.2020.12.002 [DOI] [PubMed] [Google Scholar]
- 114.Xue C, Lv H, Li Y, Dong N, Wang Y, Zhou J, et al. Oleanolic acid reshapes the gut microbiota and alters immune-related gene expression of intestinal epithelial cells. J Sci Food Agric. (2022) 102(2):764–73. 10.1002/jsfa.11410 [DOI] [PubMed] [Google Scholar]
- 115.Li X, Wei T, Li J, Yuan Y, Wu M, Chen F, et al. Tyrosol ameliorates the symptoms of obesity, promotes adipose thermogenesis, and modulates the composition of gut microbiota in HFD fed mice. Mol Nutr Food Res. (2022) 66(15):e2101015. 10.1002/mnfr.202101015 [DOI] [PubMed] [Google Scholar]
- 116.Wang Q, Wang C, Abdullah Tian W, Qiu Z, Song M, et al. Hydroxytyrosol alleviates dextran sulfate sodium-induced colitis by modulating inflammatory responses, intestinal barrier, and microbiome. J Agric Food Chem. (2022) 70(7):2241–52. 10.1021/acs.jafc.1c07568 [DOI] [PubMed] [Google Scholar]
- 117.Liu Y, Fang H, Liu H, Cheng H, Pan L, Hu M, et al. Goji berry juice fermented by probiotics attenuates dextran sodium sulfate-induced ulcerative colitis in mice. J Funct Foods. (2021) 83:104491. 10.1016/j.jff.2021.104491 [DOI] [Google Scholar]
- 118.Ma Q, Zhai R, Xie X, Chen T, Zhang Z, Liu H, et al. Hypoglycemic effects of Lycium barbarum polysaccharide in type 2 diabetes mellitus mice via modulating gut microbiota. Front Nutr. (2022) 9:916271. 10.3389/fnut.2022.916271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. (2018) 9:1534. 10.3389/fphys.2018.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davies JP, Scott C, Oishi K, Liapis A, Ioannou YA. Inactivation of NPC1L1 causes multiple lipid transport defects and protects against diet-induced hypercholesterolemia. J Biol Chem. (2005) 280(13):12710–20. 10.1074/jbc.M409110200 [DOI] [PubMed] [Google Scholar]
- 121.Kamato D, Ilyas I, Xu S, Little PJ. Non-mouse models of atherosclerosis: approaches to exploring the translational potential of new therapies. Int J Mol Sci. (2022) 23(21):12964. 10.3390/ijms232112964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The 16S rRNA gene sequencing data from mouse fecal microbiota presented in this study has been deposited in the Sequence Read Archive under BioProject PRJNA952522 with the following accession numbers: SRR24071998 to SRR24072026 (BioSamples: SAMN34074972 to SAMN34075000).