Abstract
Iron starvation of Bordetella avium induced expression of five outer membrane proteins with apparent molecular masses of 95, 92, 91.5, 84, and 51 kDa. Iron-responsive outer membrane proteins (FeRPs) of similar sizes were detected in six of six strains of B. avium, suggesting that the five FeRPs are common constituents of the outer membrane of most, if not all, strains of B. avium. Iron-regulated genes of B. avium were targeted for mutagenesis with the transposon TnphoA. Two mutants with iron-responsive alkaline phosphatase activities were isolated from the transposon library. The transposon insertion did not alter the iron-regulated expression of the five FeRPs in mutant Pho-6. The mutant Pho-20 exhibited a loss in expression of the 95-kDa FeRP and the 84-kDa FeRP. Both Pho-6 and Pho-20 were able to use free iron as a nutrient source. However, Pho-20 was severely compromised in its ability to use iron present in turkey serum. The data indicated that the mutation in Pho-20 affected expression of one or more components of an uptake machinery that is involved in acquisition of iron from organic ferricomplexes.
Bordetella avium is the infectious agent for coryza (bordetellosis), a highly infectious disease of turkeys and chickens. While the disease is usually clinically mild, mortalities in birds can reach up to 75% because of complications of secondary infections (44). Infection by B. avium is accompanied by symptoms in birds that resemble those in humans caused by infection with Bordetella pertussis. These include anorexia, exudative conjunctivitis, sneezing, and serous discharge from the nares (26). In later stages of infection, tracheal rales and dyspnea develop. Pathological changes include rhinitis, sinusitis, bronchopneumonia, and, in severe cases, tracheal collapse. The pathogen has a tropism for ciliated epithelium, but the factors responsible for adherence have not been conclusively identified (18).
As with other organisms, bacterial pathogens require Fe for growth (53). In most animals, Fe is sequestered in high-affinity Fe-binding complexes, such as transferrin in the serum or lactoferrin on mucosal surfaces. These and other Fe-binding molecules maintain the concentration of free Fe in the host at levels below that needed to support the growth of bacterial pathogens. In response to this selective pressure, many pathogenic bacteria have evolved efficient uptake systems to “steal” Fe from the host’s sequestering molecules. As a general rule, expression of genes encoding these Fe-binding molecules is strongly dependent upon the local concentration of Fe.
The ability of a bacterial pathogen to “farm” the tissues and fluids of a host for Fe by using receptors that are specific for the host’s ferricomplexes is considered to be an essential property of virulence (53). A variety of bacterial Fe-binding proteins have been documented. During Fe starvation, Haemophilus ducreyi expresses receptors for hemoglobin on its surface (15). Neisseria meningitidis (1, 12) produces an Fe-regulated transferrin-binding protein that uses the host’s transferrin molecules as a source of Fe. Helicobacter pylori has specific receptors for the host’s lactoferrin molecules (14a). The lactoferrin receptors are expressed only when H. pylori is exposed to Fe-limited conditions. Transferrin- and lactoferrin-binding proteins are found in the outer membranes of B. pertussis and Bordetella bronchiseptica (35). In addition to producing surface receptors for organic ferricomplexes, some pathogenic bacteria have evolved an extracellular system for acquiring Fe from the host. Siderophores are small, nonproteinaceous molecules that are secreted by the cell (53). The Fe-binding affinity constants of siderophores are comparable to that of transferrin, which enables the chelators to competitively remove Fe from that complex and from other organic ferricomplexes. Genes for biosynthesis of siderophores have been identified in B. bronchiseptica (20, 21).
To investigate the role of Fe acquisition in the pathogenesis of B. avium and to identify Fe-regulated outer membrane proteins (OMPs), known as FeRPs, that were likely involved in Fe uptake, B. avium was cultured under conditions that mimicked the low-Fe environment that the bacterium would likely encounter during an infection. Five FeRPs were identified that were expressed by B. avium only during Fe starvation. Infection of B. avium with TnphoA and a rapid plate assay for alkaline phosphatase activity were used to isolate two mutants with transposon insertions in Fe-regulated genes. One mutant was dramatically compromised in its ability to use serum as a source of Fe for growth, indicating that B. avium expresses one or more proteins involved in acquiring organic Fe from serum. These data provide strong evidence that TnphoA mutagenesis provides a rapid and efficient technique to identify genes of B. avium involved in Fe uptake.
MATERIALS AND METHODS
Bacterial strains and antibiotics.
The pathogenic strain B. avium 4169 was a generous gift of R. B. Rimler (39). B. avium 838 and 75 were obtained from L. H. Arp. Strains W, 950, and 955 were obtained from Mark Blakely. Oxidase assays, the ability to grow on MacConkey agar, and reactivity with serum prepared against B. avium (supplied by L. Arp) were used to confirm that the bacteria used in the experiments were B. avium. Escherichia coli DH5αF′tet (Life Technologies, Inc., Gaithersburg, Md.) and DH5αmcr (Life Technologies, Inc.) were used as hosts for recombinant plasmids. E. coli SM10 was used as a donor strain in conjugations (45). B. avium strains were maintained on brain heart infusion agar (BHI) (Difco Laboratories, Detroit, Mich.), while E. coli cultures were maintained on Luria-Bertani agar. Unless otherwise noted, ampicillin was used at 150 μg/ml, streptomycin was used at 300 μg/ml, kanamycin was used at 50 μg/ml, chloramphenicol was used at 10 μg/ml, and rifampin was used at 50 μg/ml. All antibiotics were purchased from Sigma Biochemicals (St. Louis, Mo.).
Culture media for iron starvation of B. avium.
Chemical reagents were obtained from Sigma Biochemicals, Life Technologies, Inc., and Fisher Scientific (Springfield, N.J.). All media and medium components were prepared in sterile polypropylene tubes (Falcon; Becton Dickinson Labware, Franklin Lakes, N.J.) or in Pyrex glass containers that had been washed sequentially with 3 M nitric acid and deionized water to remove surface-bound Fe. Deionized water for all solutions and for washing of glassware was obtained from a Millipore Milli-Q Plus PF deionizer and had a minimum resistance of 18 MΩ.
A formulation for a chemically defined medium (CDM) for growth of B. avium was obtained from Vince Collins and is similar to the defined medium of Stainer and Scholte (47) (Table 1). All chemicals used in the preparation of CDM were of the highest purity. To reduce the concentration of Fe in CDM, the Fe-binding activity of the metal chelator Chelex (Bio-Rad, Hercules, Calif.) was employed. Briefly, 10 g of Chelex beads was washed for 90 min at room temperature in an acid-washed flask containing 400 ml of deionized water to remove potential contaminants. After 90 min, the water was decanted from the beads and 1 liter of CDM was added to the flask. Chelex-containing CDM was stirred with a magnetic bar at room temperature for 90 min. The mixture was filtered through a Stericup-GV ultrafiltration device (Millipore Corp., Marlborough, Mass.) to remove the Chelex beads and to sterilize the Fe-depleted medium. Chelex-treated CDM could be stored at 4°C in the dark for up to 2 weeks.
TABLE 1.
Formula for the CDM for growth of B. aviuma
| Step | Ingredient or procedure | Amt |
|---|---|---|
| Base solution | KH2PO4 | 0.5 g |
| Sodium glutamate | 10.72 g | |
| NaCl | 2.5 g | |
| KCl | 0.2 g | |
| MgCl2 · 6H2O | 0.1 g | |
| Tris | 1.525 g | |
| α-Ketoglutamic acid | 2.0 g | |
| Sodium pyruvate | 2.0 g | |
| CaCl2 | 20.0 mg | |
| Deionized H2O | To 909.0 ml | |
| Adjust to pH 7.8 with NaOH, autoclave, and store at 4°C | ||
| CDM supplement | Ascorbic acid | 0.2 g |
| Glutathione, reduced | 1.0 g | |
| Deionized H2O | To 100.0 ml | |
| Store at −20°C | ||
| l-Cystine supplement | Deionized H2O | 3.0 ml |
| HCl (11.6 M) | 1.0 ml | |
| l-Cystine | 0.4 g | |
| Deionized H2O | To 100.0 ml | |
| Store at −20°C | ||
| l-Proline supplement | l-Proline | 3.0 g |
| Deionized H2O | To 200.0 ml | |
| Autoclave and store at −20°C | ||
| l-Phenylalanine supplement | l-Phenylalanine | 4.0 g |
| Deionized H2O | To 400.0 ml | |
| Autoclave and store at 4°C | ||
| Vitamins | p-Aminobenzoic acid | 10 mg |
| Biotin | 1 mg | |
| Folic acid | 10 mg | |
| Pyridoxyl HCl | 80 mg | |
| Riboflavin | 40 mg | |
| Thiamine HCl | 40 mg | |
| Calcium pantothenate | 80 mg | |
| Nicotinamide | 200 mg | |
| Deionized H2O | 800 ml | |
| Add 2.5 N NaOH until solution clears | ||
| Add deionized H2O to a final volume of 1,000 ml | ||
| Store at −20°C | ||
| Prepare final medium | Base medium | 909 ml |
| l-Cystine stock | 10 ml | |
| l-Proline stock | 16 ml | |
| CDM supplement | 10 ml | |
| l-Phenylalanine stock | 40 ml | |
| Vitamins | 10 ml | |
| Sterilize by ultrafiltration (0.2-μm pore size) |
CDM may be stored for up to 2 weeks at 4°C. Stock solutions may be stored for up to 3 months.
For experiments with BHI, broth cultures were supplemented with ethylenediamine di(o-hydroxy-phenylacetic acid) (EDDA) (Sigma Biochemicals) or a,a′-dipyridyl (a,a′-Dip) (Fisher Scientific Co., Fairlawn, N.J.) to a final concentration of 100 or 200 μM, respectively.
Growth curves.
Acid-washed 250-ml Nephelo flasks (Bellco Glass, Inc., Vineland, N.J.) containing 25 ml of medium were inoculated with 250 μl of B. avium cells that had been cultured at 37°C to stationary phase in the appropriate medium. Flask cultures were incubated at 37°C with vigorous agitation in a New Brunswick Series 25 Floor Shaker incubator (New Brunswick Scientific Co., Inc., Edison, N.J.). Optical densities (ODs) of the cultures at various time points were measured with a Klett-Summerson Photoelectric Colorimeter (320 Klett units = 1.64 OD at 600 nm [OD600] units = 5.0 × 109 CFU/ml).
Preparation of outer membranes.
Outer membranes were prepared by a modification of the protocol of Leyh and Griffith (31). Briefly, 500-ml broth cultures of B. avium were cultured to early stationary phase at 37°C. Cells were pelleted by centrifugation at 4°C at 3,000 × g for 20 min. Cells were resuspended in 15 ml of ice-cold HEPES buffer (10 mM HEPES [pH 7.4], 0.1 mM phenylmethylsulfonyl fluoride). The resuspended cells were frozen at −70°C, thawed in a room temperature water bath, and immediately stored on ice. Cells were placed in an ice bath and sonicated five times for 1 min each with a microtip and a Branson Sonifier 450 ultrasonicator (Branson Ultrasonics Corp., Danbury, Conn.) at a setting of 7 and a 50% cycle. Sonicated cells were centrifuged at 3,000 × g for 20 min at 4°C to pellet cellular debris. Total membranes were obtained by centrifugation of the supernatant fraction at 100,000 × g for 60 min at 4°C. Total membranes were resuspended in a small volume of extraction buffer (1% N-lauroyl-sarcosine in HEPES buffer). After incubation at room temperature for 60 min, the extracted membranes were centrifuged at 100,000 × g for 60 min at 4°C. The pelleted, insoluble outer membranes were resuspended in extraction buffer and after an additional 60-min incubation at room temperature were pelleted by centrifugation, as described above. The outer membranes were resuspended in deionized water and stored at −70°C. The total protein in the outer membrane samples was determined by using the Bio-Rad Protein Assay (Bio-Rad Laboratories) with bovine serum albumin as the standard.
Gel electrophoresis of OMPs.
OMPs were resolved by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (29). One microgram of total OMPs was resuspended in loading buffer (31 mM Tris [pH 6.8], 2% SDS, 2.5% 2-mercaptoethanol, 10% glycerol), and the proteins in the sample were solubilized by boiling the solution for 10 min. Proteins were resolved by electrophoresis in 10% polyacrylamide gels. Proteins in the gels were visualized by staining with Coomassie brilliant blue (38) or silver (37).
Immunoblotting with anti-PhoA antibodies.
Details for immunoblotting with monoclonal antibodies have been described previously (9). Cells grown in BHI and in BHI-EDDA were resuspended in loading buffer and solubilized by boiling for 10 min. Proteins in the samples were resolved by SDS-PAGE with 10% polyacrylamide gels. After transfer to nitrocellulose, proteins on the filters were detected by immunoblotting with a mouse monoclonal anti-PhoA antibody (Caltag Laboratories, Inc., Burlingame, Calif.).
TnphoA mutagenesis.
A method similar to that used by Manoil and Beckwith (34) was utilized to mobilize TnphoA into the mobilizable cosmid vector pCOS5 (10). pCOS5 was transformed into CC118, a sup0 strain of E. coli, and a transformant with the plasmid was transfected with bacteriophage λTnphoA (34). Transfected CC118(pCOS5) cells were plated on Luria-Bertani agar containing 300 μg of kanamycin per ml to favor growth of cells having TnphoA transpositions into pCOS5. Kanamycin-resistant survivors of the transfection were pooled, and plasmid DNA isolated from the pooled colonies was used to transform DH5αmcr. Transformants were selected on Luria-Bertani agar containing chloramphenicol and kanamycin. Colonies displaying resistance to both antibiotics were then counterscreened on ampicillin to identify a clone having a transposition of TnphoA into the ampicillin resistance gene of pCOS5. The plasmid isolated from one of the Amps Chlr Kanr colonies was denoted pCOS5::TnphoA.
An isogenic derivative of B. avium 4169 expressing a spontaneous resistance to rifampin was used as the recipient in the conjugations. Strain 4169 encodes a natural resistance to streptomycin. Matings between 4169 and SM10(pCOS5::TnphoA) were accomplished on the surface of BHI by mixing equal aliquots of log-phase cultures. After overnight incubation at 30°C, cells were scraped from the agar surface and transconjugants were isolated by plating the cells on BHI containing rifampin, streptomycin, and kanamycin. Ten random transconjugants were screened for resistance to chloramphenicol to verify loss of pCOS5::TnphoA. All 10 clones were sensitive to chloramphenicol. Approximately 19,500 transposon mutants of strain 4169 were obtained.
BHI culture dishes supplemented with 200 μM a,a′-Dip were inoculated with 400 to 500 CFU of the TnphoA mutant library. After overnight incubation at 37°C, colonies were transferred by blotting to 82-mm-diameter Optitran BA-S 85 nitrocellulose membranes (0.45-μm pore size) (Schleicher & Schuell, Keene, N.H.). Colony-bound membranes were placed colony side up on the surface of circular 3MM (Schleicher & Schuell) paper disks saturated with 150 mM NaCl. After 5 min, the membranes were transferred to the surface of 3MM paper disks that were saturated with a solution of 430 μM bromo-chloro-indolyl phosphate (BCIP) (Sigma) in 150 mM Tris (pH 8.0). Membranes were incubated at 37°C, and the colonies on the membrane were monitored periodically for 24 h for development of a blue coloration. Blue colonies were streaked onto BHI containing 36 μM ferrous sulfate and were rescreened in the BCIP assay for repression of the transposon-encoded alkaline phosphatase activity.
NPP assay for measuring PhoA activity.
Alkaline phosphatase activities of TnphoA mutants were determined with p-nitrophenyl phosphate (NPP) (41). Thirty microliters of bacteria cultured under Fe-limiting conditions was used to inoculate 3 ml of BHI broth. For experiments to measure alkaline phosphatase activity of bacteria grown under conditions of Fe limitation, the BHI broth was supplemented with 100 μM EDDA. For measurements of alkaline phosphatase activity of bacteria cultured under Fe-replete conditions, the BHI broth was supplemented with 36 μM ferrous sulfate. Cultures were incubated at 37°C until attaining the stationary phase. Bacteria in 2 ml of the stationary-phase culture were pelleted by centrifugation at 3,000 × g for 5 min. The pelleted cells were resuspended to an OD600 of 0.3 to 0.7 in ice-cold 1 M Tris (pH 8.0) and permeabilized by sonication with a microtip and a Branson Sonifier 450 (2 min of sonication at a setting of 6, 50% cycle). One hundred microliters of 0.4% (wt/vol) NPP in water was added to 1 ml of the sonicated cells, and the OD420/550 was determined at intervals of 41 s over a period of 6 min. Alkaline phosphatase activity was calculated by the formula
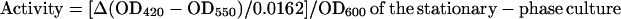 |
Growth of B. avium in turkey serum.
Turkey serum from an egg-laying hen (Crane Laboratories Inc., Syracuse, N.Y.) was inactivated by incubation at 56°C for 30 min. A 3-ml broth culture of CDM in 15-ml polypropylene tubes (Falcon) was inoculated with 30 μl of an overnight Chelex-treated CDM culture of B. avium. The culture medium was supplemented with one or more of the following reagents: ferrous sulfate to a final concentration of 36 or 86 μM, heat-inactivated turkey serum to 1.7%, and EDDA to 50 μM. Streptomycin was added to all cultures to inhibit growth of contaminants. As a measure of cell growth, the OD600 of stationary-phase cultures was determined with a Beckman DU-640B spectrophotometer.
Biochemical and genetic detection of siderophores.
Chrome azurol S-iron(III)-hexadecyltrimethylammonium bromide (CAS) (43) was used for the detection of siderophores in culture supernatants of B. avium. Control reactions demonstrated that undiluted Chelex-treated CDM caused a small degree of interference in the CAS assay. Interference was undetectable when Chelex-treated CDM was diluted 1:10 with deionized water. B. avium 4169 was grown in Chelex-treated CDM and in Chelex-treated CDM supplemented with 36 μM FeSO4. Culture supernatants which had been separated from cells by centrifugation were diluted 1:10 in deionized water, and the diluted samples were analyzed for Fe-binding activities with the CAS assay. The A630 of the reactions was measured with a Beckman DU-640 spectrophotometer. Desferal (CIBA Pharmaceutical Co., Summit, N.J.), a siderophore produced by Streptomyces pilosus, was used as a positive control.
To prepare chromosomal DNA, cells were resuspended in a solution of 10 mM Tris (pH 8.0), 1 mM EDDA, 0.5% SDS, and 100 μM proteinase K (Sigma Biochemicals). After incubation at 56°C for 60 min, NaCl was added to 714 mM, and hexadecyl trimethyl ammonium bromide (CTAB) was added to 1%. The solution was incubated at 37°C for 10 min, and the precipitated proteins and polysaccharides were removed by repeated extraction with a 1:1 solution of phenol-chloroform. Isopropanol was used to precipitate the chromosomal DNA.
Southern hybridizations were done as described previously (33). A 4.7-kbp EcoRI fragment containing the siderophore biosynthesis genes of B. bronchiseptica was obtained from restriction digestion of pDLA5 (20, 21). The EcoRI fragment was radiolabelled with [32P]dCTP (DuPont/New England Nuclear, Boston, Mass.) with a Random Priming Kit (Life Technologies). Filters hybridized with the radiolabelled fragment were washed twice for 60 min each at 45°C in a solution of 0.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5 mM sodium pyrophosphate, and 0.1% SDS. Filters were exposed to Kodak X-Omat AR film (Rochester, N.Y.) to detect radiolabelled signals.
RESULTS
Growth of B. avium under Fe-limiting conditions.
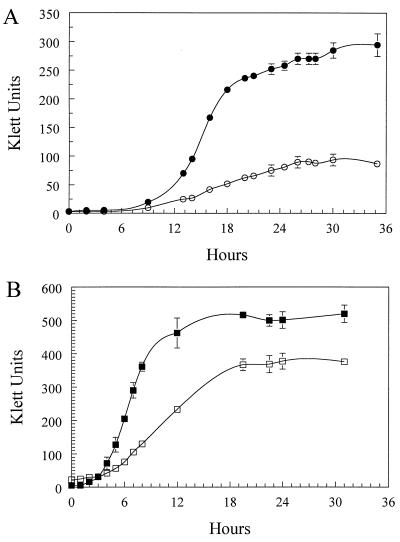
To investigate the effects of Fe starvation on B. avium, strain 4169 was cultured in a CDM that contained all essential nutrients for growth of the bacterium with the exception of Fe. Although ultrapure reagents had been used to prepare CDM, it was likely that a small, but biologically significant amount of Fe was present as a contaminant. To further reduce the concentration of Fe and produce conditions of Fe stress, CDM was pretreated in a batchwise manner with Chelex, a high-affinity Fe chelator. When 4169 was cultured in Chelex-treated CDM, stationary-phase culture densities were 66% lower than culture densities of 4169 cultured in Fe-supplemented Chelex-treated CDM (Fig. 1A). Growth rates during the log phase were also inhibited in the Fe-deficient medium. 4169 had a log-phase rate of growth of 4.97 Klett units/h when grown in Chelex-treated CDM. Supplementation of Chelex-treated CDM with FeSO4 increased the growth rate to 29.2 Klett units/h.
FIG. 1.
Growth curves demonstrating Fe starvation of 4169. (A) Growth curve of 4169 cultured in Chelex-treated CDM (○) and in Chelex-treated CDM–36 μM FeSO4 (•). (B) Growth curve of 4169 cultured in BHI (■) and in BHI–100 μM EDDA (□). Each point represents the mean culture density of three independent cultures. Error bars represent 1 standard deviation of the mean.
To establish whether the inhibitory effect on growth was limited to CDM, 4169 was cultured in BHI alone and in BHI that had been supplemented with the high-affinity Fe chelator EDDA (Fig. 1B). Growth of 4169 in BHI-EDDA reduced the stationary-phase density of the culture to 71% of the stationary-phase density of cells grown in BHI. The rate of growth was reduced from 77.5 Klett units/h in BHI to 25.0 Klett units/h in BHI-EDDA. The effect of EDDA was reversed by addition of FeSO4 to the medium (data not shown), indicating that the inhibition of growth was due solely to the loss of free Fe in the medium and not to sequestration of other nutrients by the chelator. To extend these results, a second series of growth experiments was done in which EDDA was substituted with the high-affinity Fe chelator a,a′-Dip. Addition of a,a′-Dip to BHI resulted in a similar reduction in growth of 4169 (data not shown). These data indicate that addition of high-affinity Fe chelators to BHI is sufficient to produce Fe-limiting conditions for growth of B. avium.
Biochemical and genetic assays for siderophore production.
To determine if B. avium produced siderophores, supernatant from cultures of 4169 grown in Chelex-treated defined medium was analyzed for Fe-binding activity by using the CAS assay (43). No reactivities above background were detected (data not shown), indicating that siderophores had not been secreted into the culture supernatant.
To determine whether B. avium carried genes for siderophore biosynthesis, chromosomal DNA from 4169 was hybridized with a 4.7-kbp EcoRI fragment that carried the genes for production of alcaligin, the siderophore of B. bronchiseptica (20, 21). There was no detectable hybridization of the 4169 chromosome to the B. bronchiseptica biosynthetic genes when either high- or low-stringency conditions were used (data not shown). These data indicated that siderophore biosynthesis genes similar to the genes for alcaligin production in B. bronchiseptica were not present in 4169.
FeRPs.
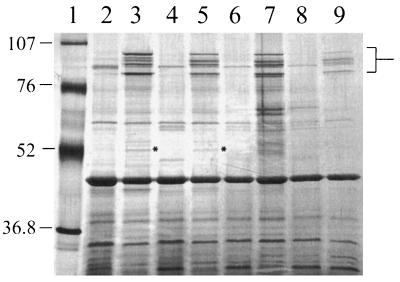
Growth curve experiments established that Fe-limited conditions could be obtained by pretreatment of CDM with Chelex. Fe acquisition systems in a variety of bacteria involve expression of one or more FeRPs. To determine whether culture of 4169 under conditions of Fe starvation induced expression of FeRPs, outer membranes were prepared from cells cultured to early stationary phase in Chelex-treated CDM and in Chelex-treated CDM supplemented with 36 μM FeSO4. SDS-PAGE analysis of the outer membranes revealed that growth of 4169 under Fe limitation induced expression of five FeRPs of between 84 and 97 kDa (Fig. 2). An FeRP of 51 kDa which was expressed at low levels was also evident when larger amounts of total protein were analyzed by SDS-PAGE. Comparison of the mobilities of the FeRPs to the mobilities of standard proteins established the apparent molecular masses of the FeRPs as 95, 92, 91.5, 84, and 51 kDa. Several minor polypeptides that appeared to be expressed only under Fe-limited growth were detected in some outer membrane preparations but were absent in other preparations.
FIG. 2.
Induction of FeRPs by Fe starvation of B. avium. Lanes: 1, molecular mass standards; 2, 4169 in Chelex-treated CDM–36 μM FeSO4; 3, 4169 in Chelex-treated CDM; 4, 4169 in BHI; 5, 4169 in BHI–100 μM EDDA; 6, 838 in BHI; 7, 838 in BHI–100 μM EDDA; 8, 75 in BHI; 9, 75 in BHI–100 μM EDDA. The location of the 84-, 91.5-, 92-, and 95-kDa FeRPs is marked in brackets. The 51-kDa FeRP, which is evident only in lanes 3 and 5, is marked with asterisks. Lanes 2 to 9 each contain 1 μg of total protein. Molecular masses are in kilodaltons. The gel was stained with silver.
To determine if expression of the five FeRPs was limited to cells grown in Chelex-treated CDM, outer membranes were prepared from 4169 cells cultured in BHI broth and in BHI broth supplemented with EDDA. SDS-PAGE analysis of the outer membranes demonstrated that addition of EDDA to BHI broth was sufficient to induce expression of all five FeRPs (Fig. 2, lane 5). Substitution of EDDA with the chelator a,a′-Dip in BHI broth also produced conditions that induced expression of the five FeRPs (data not shown).
To demonstrate that induction of FeRPs under these conditions was not limited to strain 4169, five additional strains of B. avium were cultured under Fe-limiting and Fe-replete conditions and their outer membranes were analyzed by SDS-PAGE. Strains 838 and 75, which were previously used in animal challenge experiments (16, 25), expressed five FeRPs with apparent molecular masses similar to those of the FeRPs produced by 4169 (Fig. 2, lanes 7 and 9). FeRPs of comparable apparent molecular masses were detected in the outer membranes of strains W, 950, and 955 (44) (data not shown).
TnphoA mutagenesis of 4169.
TnphoA is a derivative of Tn5 that encodes an engineered phoA gene that is lacking a promoter and a translation initiation codon (34). Insertion of the transposon within the structural sequences of a gene in the correct reading frame and orientation produces a hybrid phoA gene whose expression is controlled by the exogenous promoter. PhoA is inactive unless it is transported to an extracytoplasmic location. Therefore, alkaline phosphatase activity of the PhoA hybrid protein is evident only when the transposon insertion occurs within a gene encoding an extracytoplasmic protein. Differences in expression of the inserted gene under different growth conditions were determined by measuring alkaline phosphatase activity.
To isolate mutants of B. avium with insertions in Fe-regulated genes, TnphoA was introduced into a rifampin-resistant derivative of 4169 by conjugation with pCOS5::TnphoA, a mobilizable plasmid that will not replicate in B. avium. Over 19,000 kanamycin-resistant, ampicillin-sensitive transconjugants were obtained from the mating. To detect mutants in the transposon library that expressed active PhoA hybrid proteins only under Fe-limiting conditions, a dual-plating screen was devised. Transconjugants were plated first onto BHI–a,a′-Dip agar and surveyed for alkaline phosphatase activity by using the chromogenic indicator BCIP. BCIP-positive mutants obtained from the Fe-limited agar plates were replica plated onto BHI that had been supplemented with FeSO4. With this dual-plating protocol, two mutants were isolated that displayed Fe-regulated alkaline phosphatase activities. Mutants Pho-6 and Pho-20 expressed BCIP reactivity only when grown under Fe-limited conditions. When Pho-6 or Pho-20 was cultured under Fe-replete conditions, no BCIP reactivities were detectable (data not shown). The parental strain had little or no endogenous alkaline phosphatase activity when cultured under Fe-replete (BHI-FeSO4) or Fe-limited (BHI–a,a′-Dip) conditions.
To precisely determine the amount of alkaline phosphatase produced by Pho-6 and Pho-20 under different conditions of Fe stress, cells grown under Fe-limiting and Fe-replete conditions were permeabilized, and the extracts were measured spectrophotometrically for the ability to hydrolyze NPP, a chromogenic indicator of alkaline phosphatase activity. The results of the NPP assays mirrored those obtained from the semiquantitative BCIP agar assays (Table 2). When Pho-6 was cultured in medium that had been supplemented with FeSO4, alkaline phosphatase activity was not significantly different from the activity determined for 4169 in the same medium (P = 0.13). In contrast, when cultured under conditions in which Fe was a limiting resource, Pho-6 responded by expressing 9.2 times the amount of alkaline phosphatase activity expressed by 4169 grown under the same conditions (P = 0.003). The response of Pho-20 to Fe stress was similar to that of Pho-6. 4169 and Pho-20 had similar levels of alkaline phosphatase activity when cultured in Fe-rich medium (P = 0.28), but Pho-20 produced over 15.5 times more alkaline phosphatase activity than 4169 when the strains were cultured in Fe-poor medium (P = 0.0003).
TABLE 2.
Alkaline phosphatase activities of the TnphoA mutants of B. avium
| Strain | Fe availability (sp act)a
|
Fold inductionb | |
|---|---|---|---|
| Fe replete | Fe limited | ||
| 4169 | 0.018 ± 0.001 | 0.014 ± 0.001 | 0.8 |
| Pho-6 | 0.004 ± 0.005 | 0.129 ± 0.042 | 32.3 |
| Pho-20 | 0.033 ± 0.021 | 0.217 ± 0.045 | 6.6 |
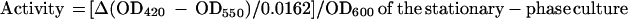 |
Activity during Fe-limited culture/activity during Fe-replete culture.
Induction of alkaline phosphatase activity of Pho-20 was 6.6 times greater in Fe-limited conditions than in Fe-rich conditions. Pho-6, however, produced over 32-fold the amount of alkaline phosphatase activity during Fe starvation compared with the amount produced during culture of the mutant in Fe-replete medium.
OMP profile of Pho-6 and Pho-20.
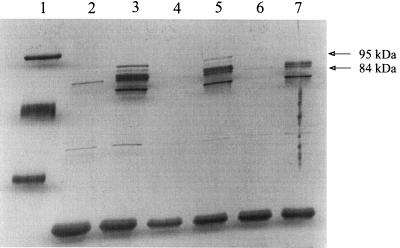
Outer membranes obtained from Fe-stressed Pho-6 and Fe-stressed Pho-20 were analyzed by SDS-PAGE to ascertain if the transposon insertions in Pho-6 and Pho-20 affected expression of the 95-, 92-, 91.5-, 84-, and 51-kDa FeRPs (Fig. 3). Comparison of the OMP profile of Pho-6 to the OMP profile of 4169 demonstrated that the mutant expressed all five FeRPs. Analysis of the outer membranes of Pho-20 established that the transposon insertion in that mutant caused a loss in expression of the 95-kDa FeRP and the 84-kDa FeRP. Expression of the 92-, 91.5-, and 51-kDa FeRPs by Pho-20 was unaffected. Within the limits of detection afforded by silver staining of the gels, the loss of expression of the 95- and 84-kDa FeRPs by Pho-20 was not accompanied by acquisition of a new OMP encoded by the Fe-regulated PhoA hybrid gene.
FIG. 3.
SDS-PAGE analysis of the OMPs of B. avium 4169 and the transposon mutants Pho-6 and Pho-20. Lanes: 1, molecular mass standards; 2, 4169 in BHI; 3, 4169 in BHI-EDDA; 4, Pho-6 in BHI; 5, Pho-6 in BHI-EDDA; 6, Pho-20 in BHI; 7, Pho-20 in BHI-EDDA. Lanes 2 to 7 each contain 1 μg of total protein. Molecular masses are in kilodaltons. The positions of the 95- and 84-kDa FeRPs that are missing in the outer membrane of Pho-20 are denoted by arrows. The gel was stained with silver.
An anti-PhoA monoclonal antibody was used to analyze whole-cell preparations of Pho-6 and Pho-20 for expression of the hybrid proteins. Immunoblotting with the anti-PhoA antibodies did not detect immunoreactive protein in either 4169, Pho-6, or Pho-20 (data not shown).
Growth of Pho-6 and Pho-20 under Fe stress.
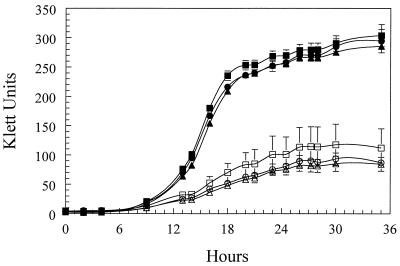
To determine if the transposon-induced mutations affected the ability of the mutants to use FeSO4 as a nutrient source, Pho-6, Pho-20, and 4169 were cultured in Chelex-treated CDM and in Chelex-treated CDM–36 μM FeSO4. The results of the growth curve experiments revealed that there were no detectable differences in the rates of growth of 4169, Pho-6, or Pho-20 in Chelex-treated CDM or in Chelex-treated CDM containing FeSO4 (Fig. 4).
FIG. 4.
Growth of 4169 and the transposon mutants Pho-6 and Pho-20. ○, 4169 in Chelex-treated CDM; •, 4169 in Chelex-treated CDM–36 μM FeSO4; □, Pho-6 in Chelex-treated CDM; ■, Pho-6 in Chelex-treated CDM–36 μM FeSO4; ◊, Pho-20 in Chelex-treated CDM; ⧫, Pho-20 in Chelex-treated CDM–36 μM FeSO4. Each point represents the mean culture density obtained from three independent cultures. The error bars represent 1 standard deviation of the mean.
Turkey serum as a source of Fe.
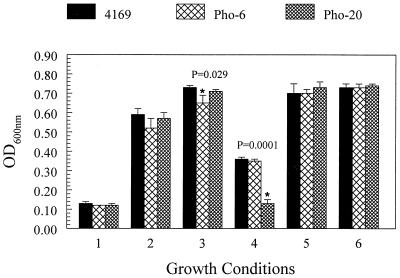
Although the previous experiments demonstrated that B. avium was capable of using free Fe in the form of FeSO4 as a nutrient for growth, B. avium encounters very little free Fe during an infection. It was important to establish whether B. avium was capable of using organic Fe as a nutrient source. Purified hemoglobin, transferrin, and other organic Fe molecules from turkey are currently unavailable from commercial sources and therefore, could not be used in growth experiments. Since it is known that oviparous birds, in general, have elevated levels of transferrin and other serum proteins (19, 52, 54), free Fe was substituted in the medium with turkey serum as a possible source of Fe. To determine the potential of B. avium to use turkey serum as a source of Fe for growth, 4169 was cultured in Chelex-treated CDM, Chelex-treated CDM–36 μM FeSO4, and in Chelex-treated CDM that had been supplemented to 1.67% with heat-inactivated turkey serum. The OD600 at stationary phase was used in this series of experiments to monitor the growth of the cultures in the different culture media. As observed in previous experiments, 4169 grew less well in Chelex-treated CDM alone than in Chelex-treated CDM that had been supplemented with 36 μM FeSO4 (Fig. 5). Growth of 4169 in serum-supplemented CDM was 460% higher than growth of 4169 in unsupplemented Chelex-treated CDM and 22% higher than growth in Chelex-treated CDM supplemented with 36 μM FeSO4. To remove free Fe that may have been present in the serum, EDDA was added to the CDM-serum medium. Although the addition of EDDA to Chelex-treated CDM–serum reduced growth of 4169 to a level below that observed in Chelex-treated CDM–serum, growth of 4169 in Chelex-treated CDM–serum–EDDA was significantly higher than growth in unsupplemented Chelex-treated CDM (P = 0.0001). To establish that the decrease in growth was due to a decrease in free Fe availability and not to undefined inhibitory factors, Fe was added back to the Chelex-treated CDM–serum–EDDA medium in the form of 50 and 86 μM FeSO4. Growth of 4169 in Chelex-treated CDM–serum–EDDA–50 μM Fe or in CDM–serum–EDDA–86 μM Fe was not significantly different from growth of the bacterium in CDM-serum (P = 0.443 and P = 0.906, respectively).
FIG. 5.
Turkey serum as a source of Fe for 4169 and the transposon mutants Pho-6 and Pho-20. Growth conditions: 1, Chelex-treated CDM; 2, Chelex-treated CDM–36 μM FeSO4; 3, Chelex-treated CDM–1.7% turkey serum; 4, Chelex-treated CDM–1.7% turkey serum–50 μM EDDA; 5, Chelex-treated CDM–1.7% turkey serum–50 μM EDDA–50 μM FeSO4; 6, Chelex-treated CDM–1.7% turkey serum–50 μM EDDA–86 μM FeSO4. Each bar represents the mean stationary-phase culture density obtained from three independent cultures. The error bars represent 1 standard deviation of the mean. An asterisk at the top of a bar indicates that the mean OD600 of the mutant strain is significantly different from the mean OD600 of 4169 for the particular growth condition (P ≤ 0.05). P values are noted for each asterisk.
To determine if the transposon insertions into the Fe-regulated genes affected the ability of Pho-6 and Pho-20 to use turkey serum as a source of Fe, the growth of the mutants was compared to the growth of 4169 in serum-supplemented media. 4169 and the mutants were cultured in Chelex-treated CDM, in Chelex-treated CDM–FeSO4, and in Chelex-treated CDM that had been supplemented with 1.67% heat-inactivated turkey serum. The results from these experiments showed that 4169, Pho-6, and Pho-20 grew equally well in Chelex-treated CDM and in Chelex-treated CDM–FeSO4 (Fig. 5). 4169 and Pho-20 grew equally well in Chelex-treated CDM–serum, while Pho-6 had slightly diminished growth in that medium (P = 0.029). A dramatic decrease in growth was seen when Pho-20 was cultured in CDM-serum-EDDA (P = 0.0001). To confirm that the decrease in growth of Pho-20 was due to the removal of free Fe by EDDA supplementation, FeSO4 was added back to the CDM-serum-EDDA medium. Additions of free Fe to the medium restored the growth of Pho-20 to a level that was indistinguishable from that of 4169 in the same medium (P = 0.389).
DISCUSSION
Studies have established that bacterial pathogens require an external source of Fe to successfully colonize and multiply within an infected animal (53). Although abundant amounts of Fe are found in animals, the element is sequestered in forms that are difficult for bacteria to acquire. In serum, Fe is complexed with transferrin and hemopexin. On mucosal surfaces, Fe is bound to lactoferrin. Free Fe is very toxic (51) and is usually sequestered in the cytoplasm in heme-containing molecules and bound to enzymes as cofactors. Because of the highly efficient nature of these Fe-scavenging molecules, the concentration of free Fe in an animal is kept below the minimal concentration needed for most bacterial pathogens to flourish (53). In response to the selective pressure, bacterial pathogens obtained or evolved molecular mechanisms for acquiring the Fe from the host’s Fe-binding systems. In general, those molecular mechanisms are expressed only when the bacterium encounters an Fe-deficient environment. When B. avium was cultured under conditions in vitro that mimicked the Fe-deficient conditions of an infected host, the bacterium responded by expressing five OMPs having apparent molecular masses of 95, 92, 91.5, 84, and 51 kDa. OMPs of similar sizes were not observed in cells grown in Fe-rich medium. The current data do not establish that all five FeRPs have roles in Fe uptake. It is equally feasible that one or more of the Fe-responsive proteins are involved in other aspects of virulence. For example, Fe limitation is used as an environmental cue by Corynebacterium diphtheriae. Diphtheria toxin is expressed only when the bacterium is confronted with Fe limitation, a condition commonly encountered when the bacterium colonizes the host’s respiratory mucosa (42, 49). While diphtheria toxin is essential for virulence, it has no direct role in Fe acquisition.
Genes in B. avium that encoded Fe-regulated, extracytoplasmic proteins were targeted for mutagenesis by use of the transposon TnphoA, a derivative of Tn5 that was used successfully in B. avium to isolate hemagglutination-negative mutants (32). A protocol for detecting mutants with TnphoA insertions using the chromogenic indicator BCIP has been described previously (34, 50), and mutants of B. avium that responded to Fe limitation by expressing a strong alkaline phosphatase activity were obtained with this standard BCIP assay. To obtain mutants with meager responses would have required prolonged incubation in the chromogenic indicator, and preliminary experiments showed that B. avium did not remain viable for more than 1 h when the standard assay was employed. To expand the usefulness of the standard BCIP assay, the conditions were changed to optimize long-term survival of the cells. When the concentration of the Tris buffer in the standard assay was decreased from 1.0 M to 0.15 M, viable cells could be obtained 24 h after incubation in the BCIP reagent. With this modified technique, a panel of 15 mutants that had transposon insertions in Fe-responsive genes was identified. Of the panel, we chose two mutants with the strongest BCIP activities to investigate. Quantitative assays with NPP established that Pho-6 and Pho-20 responded to Fe stress by inducing expression of alkaline phosphatase activity from PhoA gene fusions. Compared to 4169, the transposon insertions did not affect the rate of growth of Pho-6 or Pho-20 in Chelex-treated CDM or in Chelex-treated CDM–FeSO4. It was clear from these results that the transposon mutations did not abolish the ability of the Pho-6 or Pho-20 to acquire free Fe from the medium.
Comparison of the OMP profile of Pho-6 to the OMP profile of 4169 demonstrated that the transposon insertion in Pho-6 did not alter expression of the five FeRPs. While it is possible that the mutated gene in Pho-6 encodes an OMP other than one of the five FeRPs that was not detected in the silver-stained SDS-PAGE gels, it is equally feasible that the mutant gene in Pho-6 encodes an Fe-regulated periplasmic protein, a peripheral cytoplasmic membrane protein, or a membrane-spanning integral cytoplasmic membrane protein. Fe-regulated proteins of other bacterial species have been found in those cellular compartments (8, 14, 40). Cell fractionation studies with anti-PhoA antibodies were not successful in determining the cellular location of the hybrid protein in Pho-6. No immunoreactive protein was detected in the immunoblots (data not shown). These data suggested that the PhoA hybrid protein in Pho-6 was either unstable or was expressed in amounts below the level of detection afforded by the anti-PhoA immunoblots. Current experiments are focused on using alkaline phosphatase activity as a means of identifying the cellular location of the hybrid protein in fractionated Pho-6 cells.
Lactoferrin-binding proteins and transferrin-binding proteins have been purified from the outer membranes of B. pertussis and B. bronchiseptica (35). From those data, it was reasonable to assume that B. avium can express proteins with similar binding activities that would bind one or more types of organic Fe molecules. While lactoferrin would likely be available to the bacterium during early stages of infection, the bacterium would likely be exposed to other organic Fe sources during later stages (3). Inflammatory responses at the site would likely stimulate passive release of hemoglobin and transferrin from the serum into the respiratory mucosa. Breakdown of tracheal tissues and cells during acute stages of infection presumably would release intracellular ferricomplexes to the mucosal surface. Experiments investigating the growth of Pho-20 and 4169 in turkey serum suggested that the 95- and 84-kDa FeRPs may be directly involved in acquiring Fe from organic Fe-binding molecules such as transferrin, lactoferrin, and hemoglobin. Additional evidence to support that hypothesis was that the two FeRPs are within the range of molecular masses observed for bacterial receptors for organic Fe-binding molecules, including the transferrin-binding protein Tbp1 (30), the lactoferrin-binding protein (48), and the hemoglobin-binding protein of N. meningitidis, as well as the hemoglobin-binding protein of H. ducreyi (15). It will be interesting to determine if the 95- and the 84-kDa FeRPs have binding affinities for purified turkey transferrin, lactoferrin, or hemoglobin. The three organic Fe-binding molecules are not currently available from commercial sources for binding experiments. We are purifying transferrin from turkey serum to test whether B. avium OMPs bind the molecule (52).
It is not clear why at least one new OMP derived from the phoA gene fusion was not found in the outer membrane preparations of Pho-20. One explanation is that the hybrid protein was rapidly degraded. An alternative possibility was that the transposon inserted within a gene encoding a minor FeRP that was not detectable by silver staining. Consistent with either model is that immunoblotting with a mouse anti-PhoA antibody did not detect a hybrid protein in Pho-20. A third possibility is that the transposon inserted into a multicistronic Fe-regulated operon. In this model, the insertion occurred within a gene encoding a periplasmic or cytoplasmic membrane protein that was located upstream of the genes encoding the 84- and 95-kDa FeRPs. Cloning and sequencing of the mutant gene in Pho-20 will be necessary to determine which of the three models is correct.
The responsiveness of the Fe-regulated genes of B. avium to Fe starvation was consistent with a model of genetic repression that has been described in a variety of species. Fur is an Fe-responsive repressor that when complexed with Fe binds to an operator sequence located near the promoter of Fur-regulated genes (13, 24). Binding of FurFe to the operator inhibits expression of the Fur-regulated gene by interfering with transcription. In the absence of Fe, Fur undergoes an allosteric change that greatly decreases the repressor’s affinity for the operator sequence. Recently, the fur gene of B. pertussis was identified by complementing deregulated siderophore mutants of B. bronchiseptica (4, 6). We have strong evidence that B. avium encodes sequences homologous to the fur gene of B. pertussis and that a Fur-like repressor controls expression of a number of Fe-responsive genes (unpublished data). Repressor titration studies are being conducted in the laboratory to determine whether the genes interrupted by the transposon insertions in Pho-6 and Pho-20 are regulated by the fur gene of B. avium.
Current B. avium vaccines are comprised either of an attenuated strain of B. avium (7) or of killed cells (Intervet, Inc., Millsboro, Del.). While vaccination with the attenuated strain appears to ameliorate symptoms of disease, it apparently does not protect the birds from colonization (27). Colonized, asymptomatic birds have the potential to spread the disease to unvaccinated animals. It is likely that bacteria during early stages of colonization will respond to the low-Fe microenvironment of the respiratory mucosa by expressing proteins required for Fe uptake. Analysis of the outer membranes of B. avium 4169, 838, W, 950, 955, and 75 demonstrated that each of the strains expressed five FeRPs of similar molecular masses. These data are consistent with the five FeRPs being common constituents of the B. avium outer membrane, and as such, they are potential vaccine candidates. FeRPs of other bacterial species have been shown to elicit strong immunoreactive responses. Sera from six patients infected with N. meningitidis had antibodies to the 70- and 90-kDa FeRPs that reacted with FeRPs of the same size in heterologous strains (2, 5). Monoclonal antibodies against the ferripyochelin receptor of Pseudomonas aeruginosa reacted with heterologous serotypes, and the antireceptor antibodies were protective in a mouse challenge model (46). Antibodies against iron-regulated proteins provided protective immunity in lambs against challenge by Pasteurella haemolytica A2 (22). At this point, we have yet to establish whether the FeRPs of B. avium are antigenic or whether the proteins have common surface-exposed epitopes. Future studies of B. avium will investigate the immunogenicity of the five FeRPs to determine whether FeRPs from different strains have common antigenic features that will provide protective immunity to turkeys against colonization and infection by heterologous strains.
Siderophores and siderophore biosynthesis genes have been identified in B. pertussis (23, 28, 36) and in the closely related species B. bronchiseptica (21, 36). We were unable to detect siderophores in culture supernatants of Fe-starved B. avium by using standard biochemical assays (43), and preliminary hybridization experiments indicated that genes homologous to the biosynthesis genes of B. bronchiseptica (20, 21) do not occur in B. avium (data not shown). B. avium and B. pertussis express a variety of virulence factors having similar functions and phenotypes. Both species produce a dermonecrotic toxin, a tracheal cytotoxin, and a hemagglutinin (11, 17). Both species have a tropism for the ciliated epithelium of the upper respiratory tract, and colonization of their respective hosts leads to inflammation, loss of epithelial cells, anorexia, sneezing, and rhinorrhea. Tracheal rales characterize both turkey bordetellosis and whooping cough in humans (26). Although B. pertussis and B. avium share a number of similar virulence determinants, only B. pertussis expresses a pertussis toxin and an adenylate cyclase toxin. It is interesting to note that in the absence of siderophore production and synthesis of both toxins, B. avium produces symptoms in birds that are very similar to the symptoms observed in humans infected with B. pertussis. The roles of pertussis toxin, adenylate cyclase toxin, and siderophore biosynthesis in human infection need further investigation.
B. pertussis is an obligate human pathogen, and research into pathogenesis of the bacterium is hampered by the lack of a relevant animal challenge model. Neither mice nor rats when challenged with B. pertussis develop symptoms that are identical to those observed in human infection. The use of human volunteers to investigate the pathogenesis of B. pertussis is not a viable option because of the severity of the disease. In contrast to B. pertussis, challenge studies to investigate the pathogenesis of B. avium can be done with turkeys, the intrinsic host for the bacterium. Since infection with B. avium produces symptoms in turkeys that are similar to the symptoms observed in humans with whooping cough, we propose that B. avium is an excellent model system with which to study the pathogenesis of B. pertussis. Future studies will use B. avium mutants in animal challenge experiments to directly determine the importance of the Fe-regulated genes in virulence.
ACKNOWLEDGMENT
This work was supported by funds made available to T.D.C. from the School of Medicine and Biomedical Sciences at the State University of New York at Buffalo.
REFERENCES
- 1.Ala’Aldeen D A. Transferrin receptors of Neisseria meningitidis: promising candidates for a broadly cross-protective vaccine. J Med Microbiol. 1996;44:237–243. doi: 10.1099/00222615-44-4-237. [DOI] [PubMed] [Google Scholar]
- 2.Ala’Aldeen D A, Wall R A, Borriello S P. Immunogenicity and cross-reactivity of the 70-kDa iron-regulated protein of Neisseria meningitidis in man and animals. J Med Microbiol. 1990;32:275–281. doi: 10.1099/00222615-32-4-275. [DOI] [PubMed] [Google Scholar]
- 3.Arp L H, Cheville N F. Tracheal lesions in young turkeys infected with Bordetella avium. Am J Vet Res. 1984;45:2196–2200. [PubMed] [Google Scholar]
- 4.Beall B W, Sanden G N. Cloning and initial characterization of the Bordetella pertussis fur gene. Curr Microbiol. 1995;30:223–226. doi: 10.1007/BF00293637. [DOI] [PubMed] [Google Scholar]
- 5.Black J R, Dyer D W, Thompson M K, Sparling P F. Human immune response to iron-repressible outer membrane proteins of Neisseria meningitidis. Infect Immun. 1986;54:710–713. doi: 10.1128/iai.54.3.710-713.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke D S, Jensen M M. Immunization against turkey coryza by colonization with mutants of Alcaligenes faecalis. Avian Dis. 1980;24:726–733. [PubMed] [Google Scholar]
- 8.Chin N, Frey J, Change C F, Chang Y F. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1996;143:1–6. doi: 10.1111/j.1574-6968.1996.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 9.Connell T D, Black W J, Kawula T H, Barritt D S, Dempsey J A, Kverneland K, Jr, Stephenson A, Schepart B S, Murphy G L, Cannon J G. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol Microbiol. 1988;2:227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 10.Connell T D, Martone A J, Holmes R K. A new mobilizable cosmid vector for use in Vibrio cholerae and other Gram− bacteria. Gene. 1995;153:85–87. doi: 10.1016/0378-1119(94)00804-2. [DOI] [PubMed] [Google Scholar]
- 11.Cookson B T, Goldman W E. Tracheal cytotoxin: a conserved virulence determinant of all Bordetella species. J Cell Biochem. 1987;11B:124. [Google Scholar]
- 12.Cornelissen C N, Sparling P F. Binding and surface exposure characteristics of the gonococcal transferrin receptor are dependent on both transferrin-binding proteins. J Bacteriol. 1996;178:1437–1444. doi: 10.1128/jb.178.5.1437-1444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai P J, Angerer A, Genco C A. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J Bacteriol. 1996;178:5020–5023. doi: 10.1128/jb.178.16.5020-5023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Dhaenens L, Szczebara F, Husson M O. Identification, characterization, and immunogenicity of the lactoferrin-binding protein from Helicobacter pylori. Infect Immun. 1997;65:514–518. doi: 10.1128/iai.65.2.514-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkins C. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect Immun. 1995;63:1241–1245. doi: 10.1128/iai.63.4.1241-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagerland J A, Myers R K, Arp L H. Uptake of ferritin and Bordetella avium in bronchus-associated lymphoid tissue of turkeys. Vet Immunol Immunopathol. 1994;40:367–377. doi: 10.1016/0165-2427(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 17.Gentry-Weeks C R, Cookson B T, Goldman W E, Rimler R B, Porter S B, Curtiss R., III Dermonecrotic toxin and tracheal cytotoxin, putative virulence factors of Bordetella avium. Infect Immun. 1988;56:1698–1707. doi: 10.1128/iai.56.7.1698-1707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry-Weeks C R, Hultsch A L, Kelly S M, Keith J M, Curtiss R., III Cloning and sequencing of a gene encoding a 21-kilodalton outer membrane protein from Bordetella avium and expression of the gene in Salmonella typhimurium. J Bacteriol. 1992;174:7729–7742. doi: 10.1128/jb.174.23.7729-7742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George R, Barber D L, Schneider W J. Characterization of the chicken oocyte receptor for low and very low density lipoproteins. J Biol Chem. 1987;262:16838–16847. [PubMed] [Google Scholar]
- 20.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 21.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Analysis of the alcABC operon encoding alcaligin biosynthesis enzymes in Bordetella bronchiseptica. Gene. 1997;194:19–24. doi: 10.1016/s0378-1119(97)00094-2. [DOI] [PubMed] [Google Scholar]
- 22.Gilmour N J, Donachie W, Sutherland A D, Jones G E, Quirie M. Vaccine containing iron-regulated proteins of Pasteurella haemolytica A2 enhances protection against experimental pasteurellosis in lambs. Vaccine. 1991;9:137–140. doi: 10.1016/0264-410x(91)90271-7. [DOI] [PubMed] [Google Scholar]
- 23.Gorringe A R, Woods G, Robinson A. Growth and siderophore production by Bordetella pertussis under iron-restricted conditions. FEMS Microbiol Lett. 1990;54:101–105. doi: 10.1016/0378-1097(90)90265-r. [DOI] [PubMed] [Google Scholar]
- 24.Hantke K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli. Mol Gen Genet. 1984;197:288–294. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 25.Hellwig D H, Arp L H. Identification of Bordetella avium antigens recognized after experimental inoculation in turkeys. Am J Vet Res. 1990;51:1188–1191. [PubMed] [Google Scholar]
- 26.Hinz K-H, Glunder G, Luders H. Acute respiratory disease in turkey poults caused by Bordetella bronchiseptica-like bacteria. Vet Rec. 1978;103:262–263. doi: 10.1136/vr.103.12.262. [DOI] [PubMed] [Google Scholar]
- 27.Jackwood M W, Saif Y M. Efficacy of a commercial turkey coryza vaccine (Art-Vax,TM) in turkey poults. Avian Dis. 1985;29:1130–1139. [PubMed] [Google Scholar]
- 28.Kang H Y, Brickman T J, Beaumont F C, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Legrain M, Mazarin V, Irwin S W, Bouchon B, Quentin-Millet M J, Jacobs E, Schryvers A B. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene. 1993;130:73–80. doi: 10.1016/0378-1119(93)90348-7. [DOI] [PubMed] [Google Scholar]
- 31.Leyh R, Griffith R W. Characterization of the outer membrane proteins of Bordetella avium. Infect Immun. 1992;60:958–964. doi: 10.1128/iai.60.3.958-964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyh R D, Griffith R W, Arp L H. Transposon mutagenesis in Bordetella avium. Am J Vet Res. 1988;49:687–692. [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 34.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menozzi F D, Gantiez C, Locht C. Identification and purification of transferrin- and lactoferrin-binding proteins of Bordetella pertussis and Bordetella bronchiseptica. Infect Immun. 1991;59:3982–3988. doi: 10.1128/iai.59.11.3982-3988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore C H, Foster L-A, Gerbig D G, Jr, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 38.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 39.Rimler R B, Simmons D G. Differentiation among bacteria isolated from turkeys with coryza (rhinotracheitis) Avian Dis. 1983;27:491–500. [PubMed] [Google Scholar]
- 40.Rohrback M R, Braun V, Köster W. Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FjuD with the integral membrane protein FhuB. J Bacteriol. 1995;177:7186–7193. doi: 10.1128/jb.177.24.7186-7193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo T A, Singh G. An extraintestinal, pathogenic isolate of Escherichia coli (O4/K54/H5) can produce a group 1 capsule which is divergently regulated from its constitutively produced group 2, K54 capsular polysaccharide. J Bacteriol. 1993;175:7617–7623. doi: 10.1128/jb.175.23.7617-7623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt M P, Holmes R K. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 44.Simmons D G, Rose L P, Fuller F J, Maurer L C, Luginbuhl G H. Turkey coryza: lack of correlation between plasmids and pathogenicity of Bordetella avium. Avian Dis. 1986;30:593–597. [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 46.Sokol P A, Woods D E. Monoclonal antibodies to Pseudomonas aeruginosa ferripyochelin-binding protein. Infect Immun. 1986;53:621–627. doi: 10.1128/iai.53.3.621-627.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stainer D W, Scholte M J. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;5:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 48.Stojiljkovic I, Hwa V, de Saint Martin L, O’Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 49.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 50.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δ fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieira A V, Schneider W J. One-step chromatographic method for the purification of avian serotransferrin. Protein Expr Purif. 1993;4:110–113. doi: 10.1006/prep.1993.1016. [DOI] [PubMed] [Google Scholar]
- 53.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White H B., III Vitamin-binding proteins in the nutrition of the avian embryo. J Exp Zool Suppl. 1987;1:53–63. [PubMed] [Google Scholar]