Abstract
Background.
Debates on the allocation of medical resources during the COVID-19 pandemic revealed the need for a better understanding of immunologic risk. Studies highlighted variable clinical outcomes of SARS-CoV-2 infections in individuals with defects in both adaptive and innate immunity, suggesting additional contributions from other factors. Notably, none of these studies controlled for variables linked with social determinants of health.
Objective.
To determine the contributions of determinants of health to risk of hospitalization for SARS-CoV-2 infection among individuals with inborn errors of immunodeficiencies.
Methods.
This is a retrospective, single-center cohort study of 166 individuals with inborn errors of immunity, aged two months through 69 years, who developed SARS-CoV-2 infections from March 1, 2020 through March 31, 2022. Risks of hospitalization was assessed using a multivariable logistic regression analysis.
Results.
The risk of SARS-CoV-2-related hospitalization was associated with underrepresented racial and ethnic populations (odds ratio [OR] 4.50; confidence interval [CI], 1.57 – 13.4), a diagnosis of any genetically-defined immunodeficiency (OR 3.32; CI, 1.24 – 9.43), obesity (OR 4.24; CI, 1.38 – 13.3), and neurologic disease (OR 4.47; CI, 1.44 – 14.3. COVID-19 vaccination was associated with reduced hospitalization risk (OR 0.52; CI, 0.31 – 0.81). Defects in T cell and innate immune function, immune-mediated organ dysfunction, and social vulnerability were not associated with increased risk of hospitalization after controlling for covariates.
Conclusions.
The associations between race, ethnicity, and obesity with increased risk of hospitalization for SARS-CoV-2 infection indicate the importance of variables linked with social determinants of health as immunologic risk factors for individuals with inborn errors of immunity.
Keywords: SARS-CoV-2, COVID-19, health disparities, primary immunodeficiency, inborn errors of immunity
Introduction
The COVID-19 pandemic prompted dilemmas and debates regarding the allocation of medical resources.1–3 These debates highlighted a critical gap in medicine that remains today: the paucity of established measures of immunologic risk. This contrasts with the extensive framework of risk assessment that is the bedrock of preventative medicine. The United States Department of Health and Human Services Healthy People 2030 screening recommendations include assessments for cardiovascular disease, osteoporosis, cancer, sexually transmitted diseases, mental health, diabetes, child development, pregnancy, and sensory or communication disorders.4 While these recommendations were developed pre-pandemic, COVID-19 has heightened the importance of screening for immunologic risk which is notably absent.
Current knowledge of immunologic risk is largely derived from clinical outcomes of primary or secondary immunodeficiencies. Primary immunodeficiencies, commonly referred to as inborn errors of immunity (IEIs), consist of 485 disorders caused by defects in genes important for host immunity.5 In the U.S., IEIs are estimated to affect 1 in 2000 individuals in 20206. There was a 96.3% increase in patients with IEIs from 2013 to 2021, as diagnoses of IEIs have been facilitated by genetic testing for monogenic causes of IEIs7. Studies have identified cellular and molecular mechanisms of susceptibility to SARS-CoV-2 among the four main types of inborn errors of immunity: (1) combined immunodeficiencies with diminished T and B cell function, (2) humoral immunodeficiencies impairing B cell function, (3) defective immunity affecting innate immune cells or signaling, and (4) primary immune regulatory disorders with autoimmunity as the dominant feature (Figure 1A).8,9 In the U.S., national guidelines for prioritizing anti-SARS-CoV-2 therapies highlighted defective T cell function as a high risk condition due to the established role of T cells in orchestrating serologic immunity and eradicating virus-infected cells.10 Surprisingly, patients with severe combined immunodeficiency exhibited a spectrum of outcomes with SARS-CoV-2 infections, ranging from mild to fatal. In addition to highlighting the important of innate immunity, multiple studies revealed conflicting outcomes of SARS-CoV-2 infections in individuals with other types of inborn errors of immunity,8,11–24 suggesting additional risk factors beyond mechanistically-defined immune defects. As these studies were predominantly comprised of adults and few included multivariate analyses, the relative contributions of age, comorbidities, and immunologic defects were not delineated. None of these studies controlled for race, Hispanic ethnicity, or social vulnerability. Thus, the relative contributions of molecular risk factors compared to socioeconomic disparities to SARS-CoV-2 infection outcomes remain unknown.
Figure 1. Summary of study design.
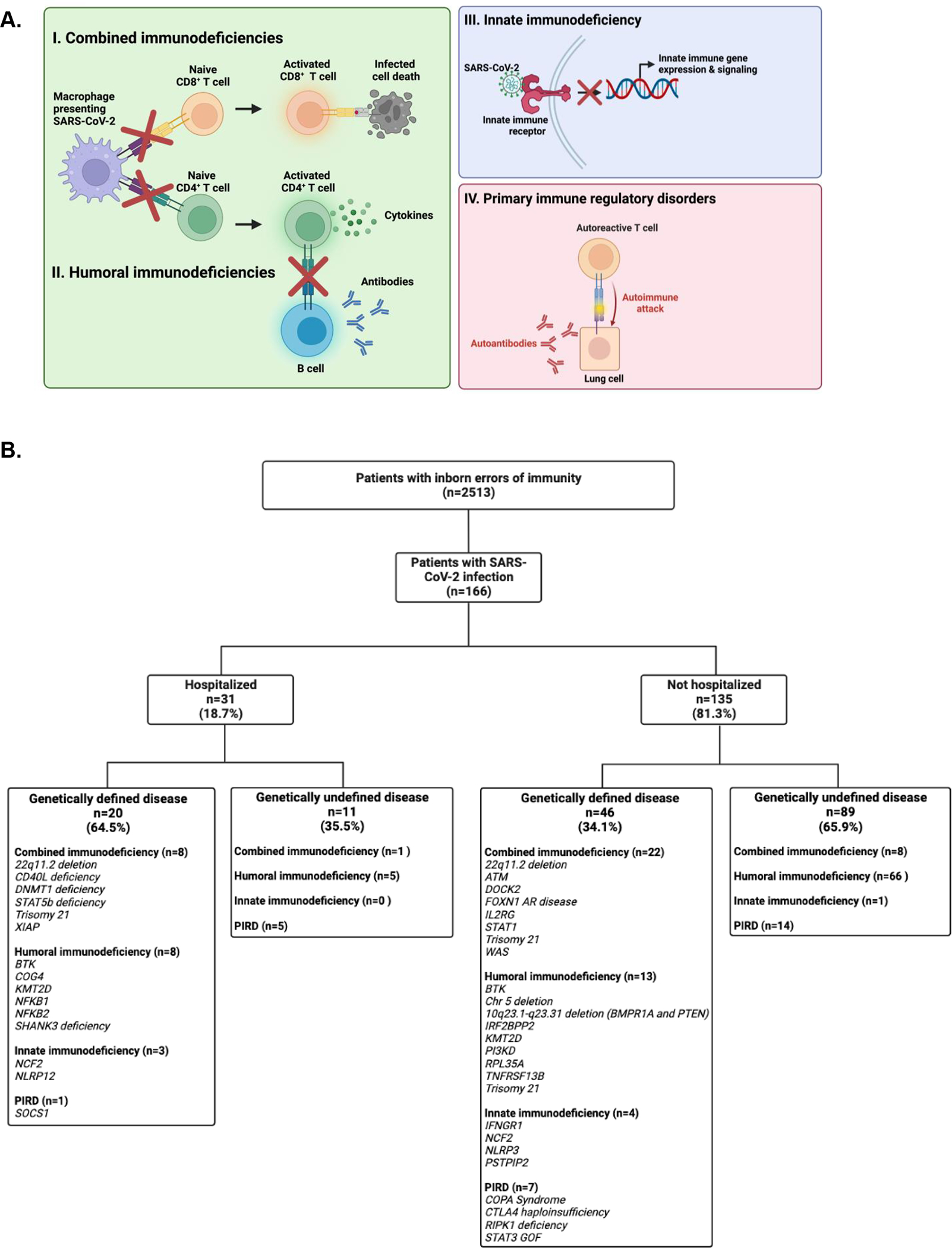
(A) Affected pathways implicated in the four main categories of inborn errors of immunity. (B) Flow diagram of study participants.
We hypothesized that general risk factors and variables associated with health disparities such as race and ethnicity correlate with increased immunologic risk, even in a population with mechanistically defined immunodeficiencies. We performed a two-year retrospective cohort study of 166 patients with inborn errors of immunity, aged two months through 69 years of age. Of published studies to date, this is the largest and longest study of outcomes from SARS-CoV-2 infections in patients with inborn errors of immunity.
Methods
Study design and setting.
This is a retrospective, single-center cohort study at Boston Children’s Hospital. As one of the nation’s largest clinical immunology programs, the Division of Immunology provides longitudinal outpatient care for patients with inborn errors of immunity ranging from the neonatal period through 75 years of age and inpatient care for individuals until the age of 35 years. Hospitalization for those older than 35 years is coordinated at adult hospitals. Patients in our cohort had a median follow-up of 9.1 years in our immunology clinic. This study was approved by the Boston Children’s Hospital Institutional Review Board.
Study objectives, variables, and outcome.
We aimed to determine factors associated with severe SARS-CoV-2 infections in patients with inborn errors of immunity, with hospitalizations as the primary outcome. Data were abstracted from the Boston Children’s Hospital electronic medical record by clinical immunologists using a standardized data collection instrument. To reduce bias inherent in a retrospective study, variables were defined prior to study initiation: age, race, ethnicity, social vulnerability index (SVI), past medical history, and history of vaccinations and interventions for SARS-CoV-2 infections. Race and ethnicity were classified based on patient self-report in the electronic medical record. Patient zip codes were matched to census tracts for determination of SVI.25 For zip codes contained within multiple census tracts, a weighted average SVI was calculated using the American Community Survey 2014–2018 population estimates.26 Defective T cell function was defined as 30% or less of normal PHA proliferation and/or history of opportunistic infections indicative of impaired T cell function.27 We identified patients with end organ dysfunction prior to SARS-CoV-2 infection, as defined by: (1) histological evidence of immune cell infiltration in organs, or (2) laboratory or otherwise measurable evidence of impaired organ function attributed by the patient’s primary immunologist to recurrent infections, autoimmunity, or chronic inflammation. Obesity was defined based on weight categories per Centers for Disease Control and Prevention guidelines28. In patients under 20 years of age, body mass index (BMI) at the 95th percentile or above of the same age and sex was used. In patients over 20 years of age, BMI of 30 or greater was used. Patients were classified as having neurologic disease if they had active manifestations of either acute or chronic neurologic disease at the time of SARS-CoV-2 infection. This does not include neurologic complications as a result of SARS-CoV-2 infection.
Patients.
Patients eligible for this study had (1) a diagnosis of an inborn error of immunity and (2) positive nucleic acid amplification test or antigen test for SARS-CoV-2 infection between March 15, 2020 through March 31, 2022 (Figure 1B). Specifically, patients with inborn errors of immunity were first identified by an ICD-10 code (Figure E2) used for a clinical visit between January 1, 2015 and January 1, 2020, which was verified through a medical record review by this study’s clinical immunology team. Patients with positive testing for SARS-CoV-2 were identified through review of: (1) SARS-CoV-2 testing results available at Boston Children’s Hospital, (2) referrals for SARS-CoV-2 interventions, (3) and medical records. Patients were not eligible if they had a secondary or acquired immunodeficiency.
Figure E2. Severity of SARS-CoV-2 infection in patients with genetically defined inborn errors of immunity.
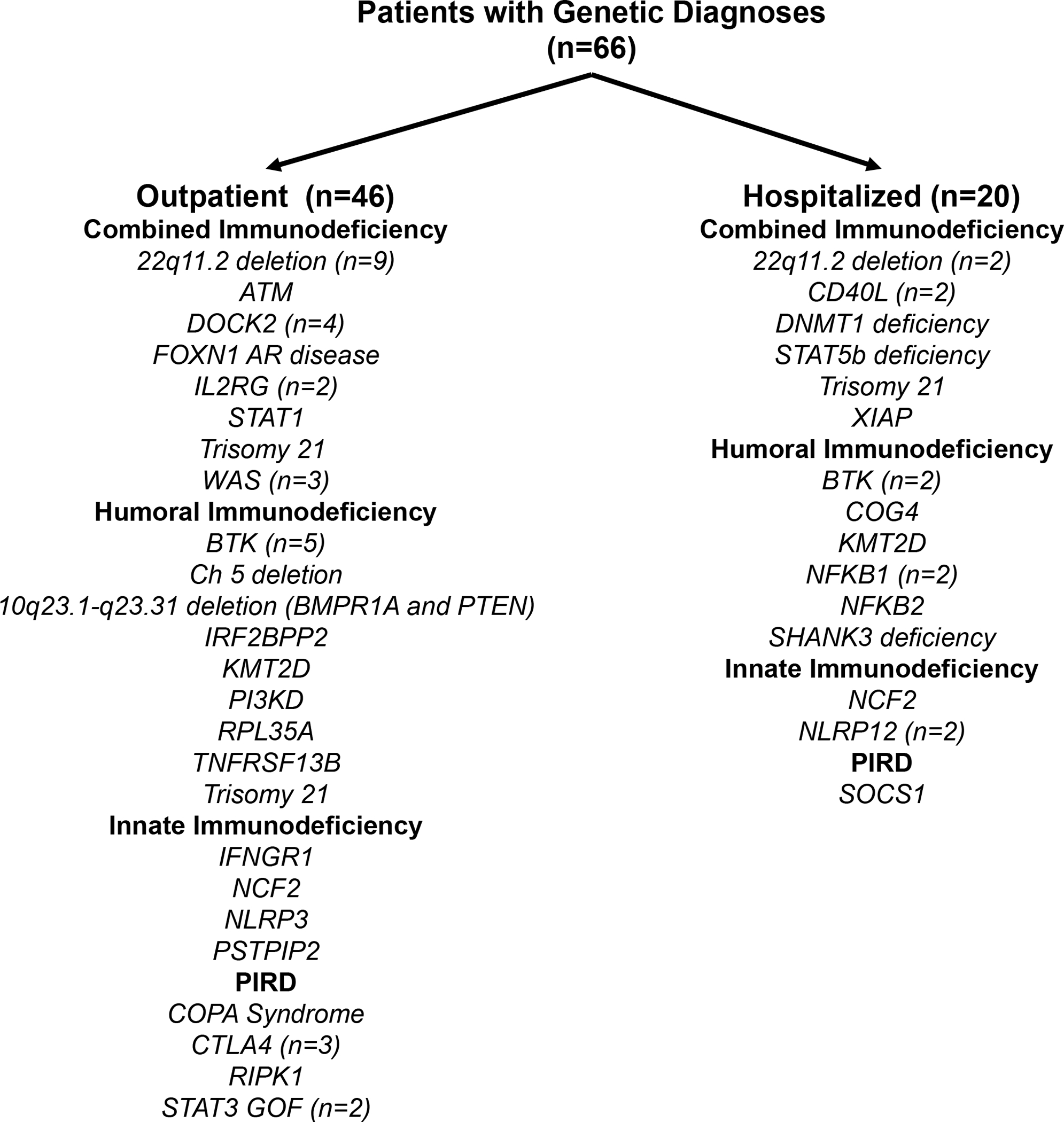
Monogenic inborn errors of immunity separated by severity of disease and categorized by phenotypic classification.
Statistical analyses.
Since hospitalization for acute SARS-CoV-2 infections are relatively rare until the sixth decade of life,29 we utilized two different machine learning methods to reduce the bias inherent in small sample sizes: Firth type logistic regression with intercept correction (FLIC) and Firth’s logistic regression with added covariate (FLAC)30. These methods overcome limitations of standard regression analysis for smaller datasets and also use complementary methods to correct for the inflated odds ratio sometimes generated by Firth’s logistic regression analysis30. Multicollinearity testing using variance inflation factors was used to confirm that there was no interdependence among the independent variable included in the regression analyses (Table E1).31–33 There was no missing data in this data set. Statistical analyses were performed with R Statistical Software (version 4.1.3) using the following packages: mctest33, logistf30, and rcompanion34.
Table E1.
Overall Multicollinearity Diagnostics. Multicollinearity was measured among the independent variables in the logistic regression analysis (defective T cell function, end organ dysfunction, age, obesity, vaccines, SVI, Omicron, underrepresented race and ethnicity, history of neurologic disease, history of B cell depleting factors, treatment with SARS-CoV-2 monoclonal antibody, genetic testing). None of the tests used identified significant multicollinearity and thus, all of these variables were included in the logistic regression analysis.
Role of the funding source.
The funding source had no role in the design, execution, analysis, or reporting of this study.
Results
Patient Characteristics
Among 2513 patients with a confirmed diagnosis of an inborn error of immunity, 166 had at least one SARS-CoV-2 infection during the study period (Figure 1B). Thirty-one patients were hospitalized (Table 1), most commonly for respiratory instability (Table 2). Twelve patients required critical care and one died. The median age of inpatients was 16.3 years (range 0.5 – 51.7 years), compared with 14.9 years for outpatients (range 0.2 – 68.8 years). The percentage of hospitalized patients (18.7%, with a median age of 16.3 years) is higher than that of the general pediatric and adolescent population, which has been estimated to be less than 5%.35,36 Hospitalized patients were more likely to be male, of Black, Asian/Pacific Islander, American Indian, or Alaskan Native racial backgrounds, of Hispanic ethnicity, and living in an area with a higher social vulnerability index (Table 1). Of the 36 individuals who were of Black, Asian/Pacific Islander, American Indian, or Alaskan Native racial backgrounds or Hispanic ethnicity, 42.4% were hospitalized. Sixty-four patients (38%) had COVID-19 in the period after December 31, 2021, when Omicron was reported to account for >90% of infections in Massachusetts state sequencing data37.
Table 1.
Patient characteristics.
| Hospitalized n = 31 | Outpatient n = 135 | |
|---|---|---|
| Median age, years (range) | 16.3 (0.5 – 51.7) | 14.9 (0.2 – 68.8) |
| Age subgroups, n (%) | ||
| 0.2 – 5 years | 6 (19.4) | 18 (13.3) |
| 5 – 12 years | 8 (25.8) | 36 (26.7) |
| 12.1 – 18 years | 6 (19.4) | 37 (27.4) |
| 18.1 – 29 years | 8 (25.8) | 29 (21.5) |
| 29.1 – 69 years | 3 (9.6) | 15 (11.1) |
| Biologic sex, n (%) | ||
| Male | 18 (58.1) | 73 (54.1) |
| Female | 13 (41.9) | 62 (45.9) |
| Self-reported race, n (%) | ||
| Asian/Pacific Islander | 2 (6.5) | 2 (1.5) |
| Black | 3 (9.7) | 6 (4.4) |
| American Indian or Alaska Native | 1 (3.2) | 0 |
| White | 17 (54.8) | 116 (86.0) |
| Othera | 8 (25.8) | 11 (8.1) |
| Self-reported ethnicity, n (%) | ||
| Hispanic | 8 (25.8) | 8 (5.9) |
| Non-Hispanic | 23 (74.2) | 127 (94.1) |
| Median Social vulnerability index (range) | 0.41 (0.10 – 0.88) | 0.28 (0.018 – 0.97) |
| Subtypes of inborn errors of immunity, n (%) | ||
| Humoral immune defects | 12 (37.5) | 83 (61.5) |
| Combined immune deficiency | 11 (35.5) | 30 (22.2) |
| Innate immune defect | 3 (9.7) | 4 (3.0) |
| Primary immune dysregulatory disorder | 5 (16.1) | 18 (13.3) |
| Clinical features and comorbidities, n (%) | ||
| Genetically-defined inborn error of immunity | 20 (64.5) | 46 (34.1) |
| End-organ dysfunction | 14 (45.2) | 46 (34.1) |
| Rituximab treatment at time of infection | 5 (16.1) | 6 (4.4) |
| Obesity | 10 (32.3) | 22 (16.3) |
| Neurologic disease | 8 (25.8) | 16 (11.9) |
| Chronic renal disease | 4 (12.9) | 3 (2.2) |
| Cardiovascular disease | 4 (12.9) | 13 (9.6) |
| History of organ or stem cell transplantation | 2 (6.5) | 7 (5.2) |
| Malignancy | 1 (3.2) | 2 (1.5) |
| Chronic liver disease | 1 (3.2) | 6 (4.4) |
| Trisomy 21 | 1 (3.2) | 3 (2.2) |
| COVID-19-related therapeutics n (%) | ||
| Total number of COVID-19 vaccine dosesb | ||
| 0 | 24 (77.4) | 77 (57.0) |
| 1 | 3 (9.7) | 1 (0.7) |
| 2 | 2 (6.5) | 31 (23.0) |
| 3 | 2 (6.5) | 26 (19.3) |
| Monoclonal antibodies against SARS-CoV-2c | 4 (12.9) | 31 (23.0) |
For self-reported race, “other” denotes those patients who selected “other” as their racial background.
All covid-19 vaccines were given prior to April 2022, which excluded the bivalent vaccine formulations.
Monoclonal antibodies included sotrovimab, bamlanivimab, casirivimab, imdevimab, and tixagevimab/cilgavimab.
Table 2.
Characterizations of hospitalized patients. Details of each clinical diagnusis are provided in the methods secThe numbers of patients for each category are listed, with percentages in parentheses. The one patient who died had X-linked agammaglobulinemia and died of acute respiratory distress syndrome from COVID-19. We have defined the response for hospitalization as follows: respiratory insufficiency: increased work of breathing, hypoxia, or respiratory failure; hemodynamic instability: unstable or low blood pressure requiring intervention; worsening immune dysregulation: evidence of active disease-specific organ dysfunction or cytopenias; fever and neutropenia: body temperature >38°C and absolute neutrophil count <500 cells/microliter; severe gastrointestinal symptoms: vomiting and/or diarrhea causing electrolyte derangements and/or hypotension requiring intervention. Clinical monitoring due to clinician concern was dependent on physician concern related to the patient’s baseline health status, diagnosis, and perceived risk in the setting of acute SARS CoV-2.
| Medical service n=18 | Intensive care n=13 | |
|---|---|---|
| Age, n (%) | ||
| 0.2 – 5 years | 5 (27.7) | 1 (7.7) |
| 5 – 12 years | 3 (16.7) | 5 (38.4) |
| 12.1 – 18 years | 3 (16.7) | 3 (23.1) |
| 18.1 – 29 years | 5 (27.7) | 3 (23.1) |
| 29.1 – 69 years | 2 (11.1) | 1 (7.7) |
| Biologic sex, n (%) | ||
| Male | 12 (66.6) | 6 (46.1) |
| Female | 6 (33.3) | 7 (53.9) |
| Self-reported race, n (%) | ||
| Asian/Pacific Islander | 0 (0) | 2 (15.3) |
| Black | 3 (16.7) | 0 (0) |
| American Indian ur Alaska Native | 0 (0) | 1 (7.7) |
| White | 11 (61.1) | 6 (46.3) |
| Othera | 4 (22.2) | 4 (30.7) |
| Self-reported ethnicity, n (%) | ||
| Hispanic | 5 (27.8) | 3 (23.1) |
| Nun-Hispanic | 13 (72.2) | 10 (76.9) |
| Median Social vulnerability index (range) | 0.41 (0.1 – 0.88) | 0.41 (0.1 – 0.79) |
| Reasons for hospitalization, n (%) | ||
| Respiratory instability | 6 (33.3) | 7 (53.8) |
| Hemudynamic instability | 1 (5.6) | 3 (23.1) |
| Wursening immune dysregulatiun | 3 (16.7) | 1 (7.7) |
| Fever and neutropenia | 2 (11.1) | 0 (0) |
| Severe gastrointestinal symptoms | 4 (22.2) | 0 (0) |
| Multisystem Inflammatory Syndrume in Children | 0 (0) | 2 (15.4) |
| Clinical munituring due tu clinician cuncern | 2 (11.1) | 0 (0) |
Humoral immunodeficiencies were the most common type of inborn error of immunity, followed by combined immunodeficiencies (Table 1), which reflects the relative prevalence of these disorders in the U.S7. Among inpatients, 64.5% had a genetically defined immune disorder attributable to a genetic variant confirmed by clinical-grade DNA sequencing, compared to 34.1% of outpatients. Genetic sequencing was completed more frequently in patients with a combined immunodeficiency or innate defects than those with humoral defects or PIRD, higher social vulnerability index (SVI), and a history of end organ dysfunction prior to SARS-CoV-2 infection (Table E2 and Table E3). Organ dysfunction caused by recurrent infections and/or autoimmunity is a complication of inborn errors of immunity that may be treated with B cell-depleting therapies.38 Patients requiring hospitalization for SARS-CoV-2 infections were more likely to have organ dysfunction and B cell-depleting therapies within one year prior to infection, compared to outpatients. Among comorbidities known to be associated with severe COVID-19,39 obesity, and neurologic disease were the most common and occurred more frequently in inpatients than outpatients (Table 1).
Table E2.
Proportions of sequenced patients within each type of IEI. To compare the proportions of sequenced individuals among the four types of IEIs, pairwise permutation t tests with corrections for multiple testing was performed.31,32,33
| Number of patients sequenced (%) | |
|---|---|
| Innate n=7 | 7 (100)▫▫ |
| Combined n=41 | 34 (82.9)▫▫ |
| PIRD n=23 | 11 (47.8)▫ |
| Humoral n=95 | 23 (24.2) |
The number of sequenced individuals with combined immunodeficiency exceeded those with PIRD (adjusted p = 6.870e–03) and humoral deficiencies (adjusted p = 1.335e–09).
The number of sequenced individuals with innate defects was comparable to those with combined immunodeficiency (adjusted p = 0.242). The number of sequenced individuals with innate individuals exceeded those with PIRD (adjusted p = 2.292e–02) and humoral defects (adjusted p = 7.128e–05).
The number of sequenced individuals with PIRD exceeded those with humoral deficiencies (adjusted p = 3.055e–02).
Table E3.
Factors associated with genetic sequencing. We used two machine learning methods to identify factors associated with any genetic sequencing test (targeted gened or panel sequencing, whole exome sequencing, and chromosomal microarray).
| Firth’s logistic regression with added covariate (FLAC) | OR | lower 0.95 | upper 0.95 | p | |
|---|---|---|---|---|---|
| Age of COVID | 0.979 | 0.951 | 1.005 | 1.10×10−1 | |
| SVI | 9.953 | 1.340 | 86.513 | 2.42×10−2 * | |
| Underrepresented race and/or ethnicity | 0.926 | 0.354 | 2.437 | 8.74×10−1 | |
| End organ dysfunction | 4.538 | 2.122 | 10.223 | 6.62×10−5 *** | |
| Combined or innate deficiency | 8.838 | 3.220 | 29.556 | 5.73×10−6 *** | |
| Firth type logistic regression with intercept correction (FLIC) | OR | lower 0.95 | upper 0.95 | p | |
| Age of COVID | 0.978 | 0.950 | 1.00 | 1.08×10−1 | |
| SVI | 9.87 | 1.30 | 89.02 | 2.62×10−2 * | |
| Underrepresented race and/or ethnicity | 0.913 | 0.346 | 2.43 | 8.54×10−1 | |
| End organ dysfunction | 4.52 | 2.11 | 10.24 | 7.34×10−5 *** | |
| Combined or innate deficiency | 8.74 | 3.16 | 29.82 | 7.36×10−6 *** |
Use of SARS-CoV-2-directed interventions
FDA emergency authorization for COVID-19 vaccines was granted in December 2020, but distribution was hampered by limitations in supply, logistics, and public trust.40 The majority of inpatients (74.2%) and outpatients (57.0%) were infected with SARS-CoV-2 before COVID-19 vaccination. Additionally, 35 patients received post-exposure treatment with monoclonal antibodies SARS-CoV-2 as they were asymptomatic but tested positive, in accordance with FDA recommendations,four of whom required subsequent hospitalization due to worsening disease.
Risk factors associated with hospitalization among patients with inborn errors of immunity
Among demographic factors, Black, Asian/Pacific Islander, American Indian, or Alaskan Native racial background and/or Hispanic ethnicity was significantly associated with an increased risk of hospitalization (OR 4.50; CI, 1.58 – 13.4), while social vulnerability index was not. Among the comorbidities associated with severe COVID-19 in the general population, obesity (OR 4.21; CI, 1.38 – 13.3) and a history of neurologic disease (OR 4.47; CI, 1.44 – 14.0), were associated with a significantly increased risk of hospitalization. The median age for the hospitalized and outpatient cohorts were 16.3 years and 14.9 years, respectively. Our regression analysis identified a marginally increased risk of hospitalization (OR 1.04; CI, 1.01 – 1.08) with older age. Notably, the risk of hospitalization was increased in patients with any genetically defined immunodeficiency (OR 3.32; CI, 1.23 – 9.43). This was unlikely to be due to differences in the proportions of patients sequenced among IEI subtypes as defects affecting either T cell or innate immunity, the two IEIs that were most frequently sequenced, were not associated with an increased risk of hospitalization. There was also no increased risk of hospitalization with a history of immune-associated organ dysfunction, B cell-depleting agents within one year of infection, or infection after December 31, 2021, at which point the Omicron variant accounted for over 90% of SARS-CoV-2 infections in Massachusetts37. B cell-depleting agents were associated with increased mortality from COVID-19 in the general population, but the effect of B cell depletion on SARS-CoV-2 infection outcomes in patients with IEI was unknown41. In this cohort, a history of treatment with B cell-depleting therapies within one year of infection did not meet the threshold for significance (OR 4.97, CI 0.95 – 27.0, p = 0.057), indicating a need for larger studies of infectious risks associated with B cell depletion in patients with IEIs. With respect to SARS-CoV-2 interventions, an increased number of COVID-19 vaccination doses was associated with reduced hospitalization risk (OR 0.52; CI, 0.31 – 0.81), while treatment with SARS-CoV-2 monoclonal antibody was not statistically significant. A second logistic regression model identified the same risk factors with similar adjusted ORs (Figure E1).
Figure E1. Secondary analysis of adjusted risk of SARS-CoV-2-related hospitalization in individuals with inborn errors of immunity.
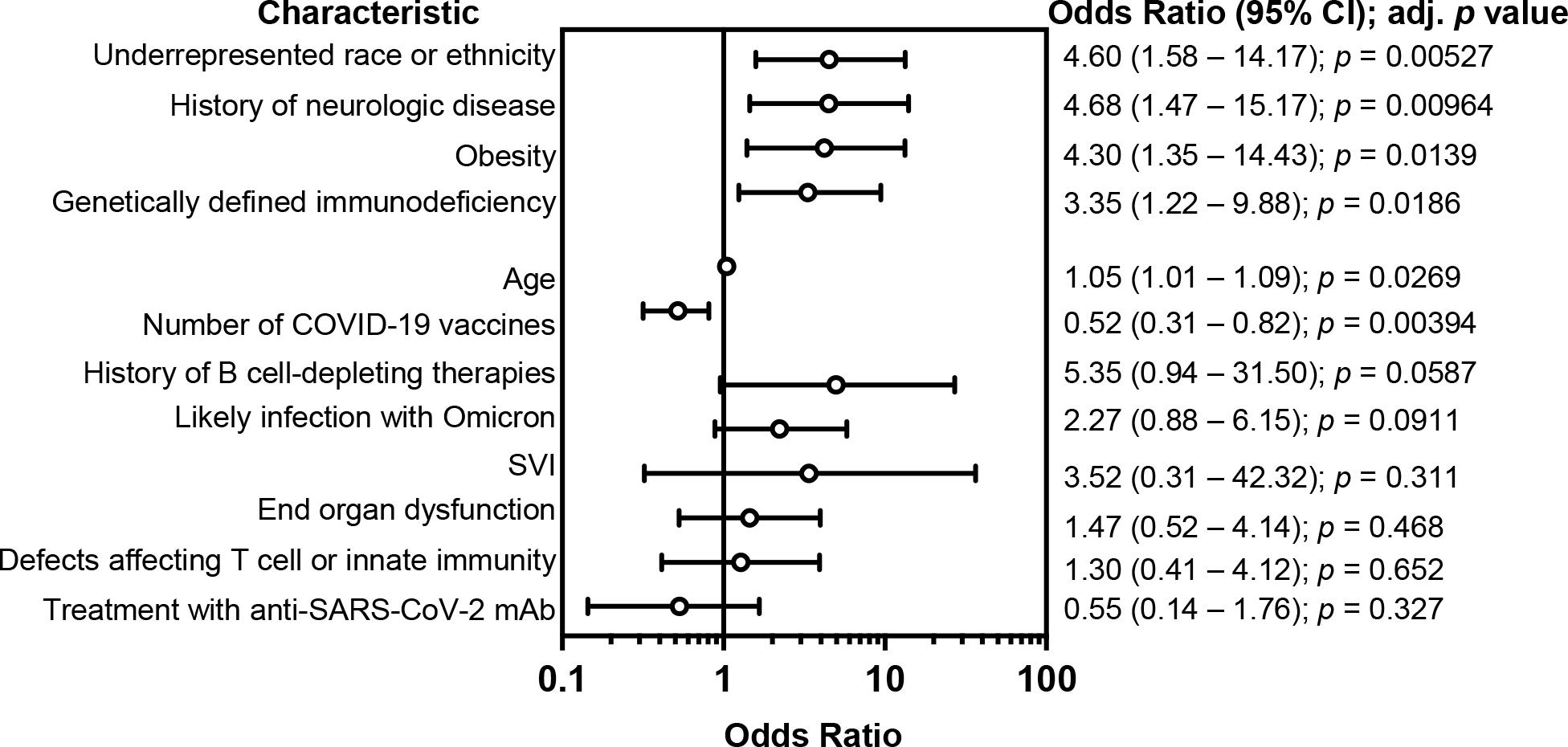
Odds ratios and 95% confidence intervals (CI) derived from a standard logistic regression analysis model, after adjusting for all covariates listed. Likely infection with Omicron refers to individuals who tested positive after December 31, 2021, when the Omicron variant accounted for at least 90% of SARS-CoV-2 infections in Massachusetts based on wastewater sequencing reports37.
Discussion
Although inborn errors of immunity are rare diseases, they serve as real-world disease models that enable delineation of specific pathways contributing to immunologic risk. While rare disease research focuses on disease-specific principles toward a goal of personalized medicine, common risk factors for disease can be overlooked. This study complements work done by others, 8,11–23 as the first published study to examine the effects of race, ethnicity, and social vulnerability index among individuals with IEIs.
We found that obesity was associated with increased odds of hospitalization, concordant with SARS-CoV-2 outcomes in the general pediatric and adult populations.42,43 Guidelines for clinical management of inborn errors of immunity highlight low body weight as a predictor of immunologic compromise.23,27 However, the prevalence of obesity in pediatric and adult patients with inborn errors of immunity is now comparable to that of the general population.44 During the H1N1 influenza pandemic, obesity was also associated with increased morbidity and mortality in the general population.45 Adipose tissue can serve as a reservoir for viral replication, the priming of inflammatory myeloid and lymphoid cells, and a source of inflammatory cytokines and adipokines.46 Future studies are needed to determine how obesity influences clinical outcomes other than SARS-CoV-2 infections in patients with inborn errors of immunity.
Here, we show that underrepresented racial and ethnic groups, but not SVI, are associated with increased odds of hospitalization. The lack of association between SVI and SARS-CoV-2-related hospitalizations in our study may arise from the availability of medical coverage for children and adolescents living in Massachusetts, which is the highest in the U.S.: only 1.2% of children in Massachusetts are uninsured, compared to a national average of 5.2%.47 Approximately one third of children in Massachusetts receive healthcare through the state’s Medicaid program providing school-based medical care.47 Additionally, this study was conducted at a tertiary medical center providing immunology subspecialty care. However, the persistent disparities in healthcare are underscored by the 5.2-fold odds of hospitalization associated with Black, Black, Asian/Pacific Islander, American Indian, or Alaskan Native race and/or Hispanic ethnicity in this study. This is concordant with studies in the general pediatric population showing increased hospitalizations and deaths from SARS-CoV-2 infections among non-Hispanic Black or Hispanic children compared to White children.48,49
Race and ethnicity have been linked with disadvantages in SDOH for individuals with specific conditions, including cardiovascular disease50 and diabetes.51 Studies analyzing data have shown an association between mortality from COVID-19 and adverse SDOH for Black individuals in U.S counties with greater levels of adverse SDOH.52 In contrast, counties with adverse SDOH less than the median of all counties studied had no association between mortality rates from COVID-19 and the percentage of Black residents. Specific to the pediatric population, there are significant racial and ethnic differences across all categories of SDOH, including economic stability, education access and quality, health care access and quality, neighborhood and built environment, and social and community context.52 Within our cohort, race and ethnicity were associated with hospitalizations independently of SVI, indicating the need for additional studies to delineate how underlying inequities and structural racism affect other outcomes in patients with inborn errors of immunity.
Our findings highlight the role for genetic sequencing in assessing immunologic risk among individuals with diverse types of IEIs. In our study, patients with a genetically defined immunodeficiency had 3.3-fold greater odds of hospitalization. Our regression analysis identified no increased risk of hospitalization for patients with defects affecting T cell and innate immunity, despite the higher rates of genetic sequencing in these patients. Thus, the association between genetic risk and hospitalization was not due to over- or under-representation of any specific IEI in the sequenced group of patients. Additionally, the reasons for hospitalization indicate that our findings were not attributable solely to clinical monitoring of patients with genetic diagnoses (Table 2). In light of these findings, we posit that individuals with genetically confirmed immunodeficiencies have more severe immunologic disease and thus, a higher risk of severe SARS-CoV-2 infections. This hypothesis is supported by published studies showing that the likelihood of a genetic diagnosis is higher in patients with severe clinical presentations.23 We included organ dysfunction in the regression analyses as quantitative measure of disease severity independent of IEI subtype.38 Although organ dysfunction was present in 45.2% of inpatients and 34.1% of outpatients, it was not associated with increased hospitalization risk in our cohort after controlling for multiple covariates in two different machine learning models (Fig. 2 and Fig. E1). We acknowledge that organ dysfunction does not fully encapsulate disease severity. First, our study participants are predominantly children, adolescent, and young adults and the correlation between organ dysfunction and disease severity may be more robust with age. Secondly, disease severity encompasses factors that are not possible to accurately measure in a retrospective analysis, such as changes in quality of life, or days of school and/or work missed. Thus, increasing access to genetic sequencing may enable better identification of children, adolescents, and young adults with increased immunologic risk. Further studies are needed to identify the biologic correlates of severe disease identified by genetic diagnoses. Identification of genetic disorders will be made possible only through equitable access to genetic testing, a goal that remains elusive in the U.S. due to gaps in clinician knowledge, limited and inconsistent insurance coverage of genetic tests, and the scarcity of community-based or patient-centered educational resources in genomic medicine.53
Figure 2. Adjusted risk of SARS-CoV-2-related hospitalization in individuals with inborn errors of immunity.
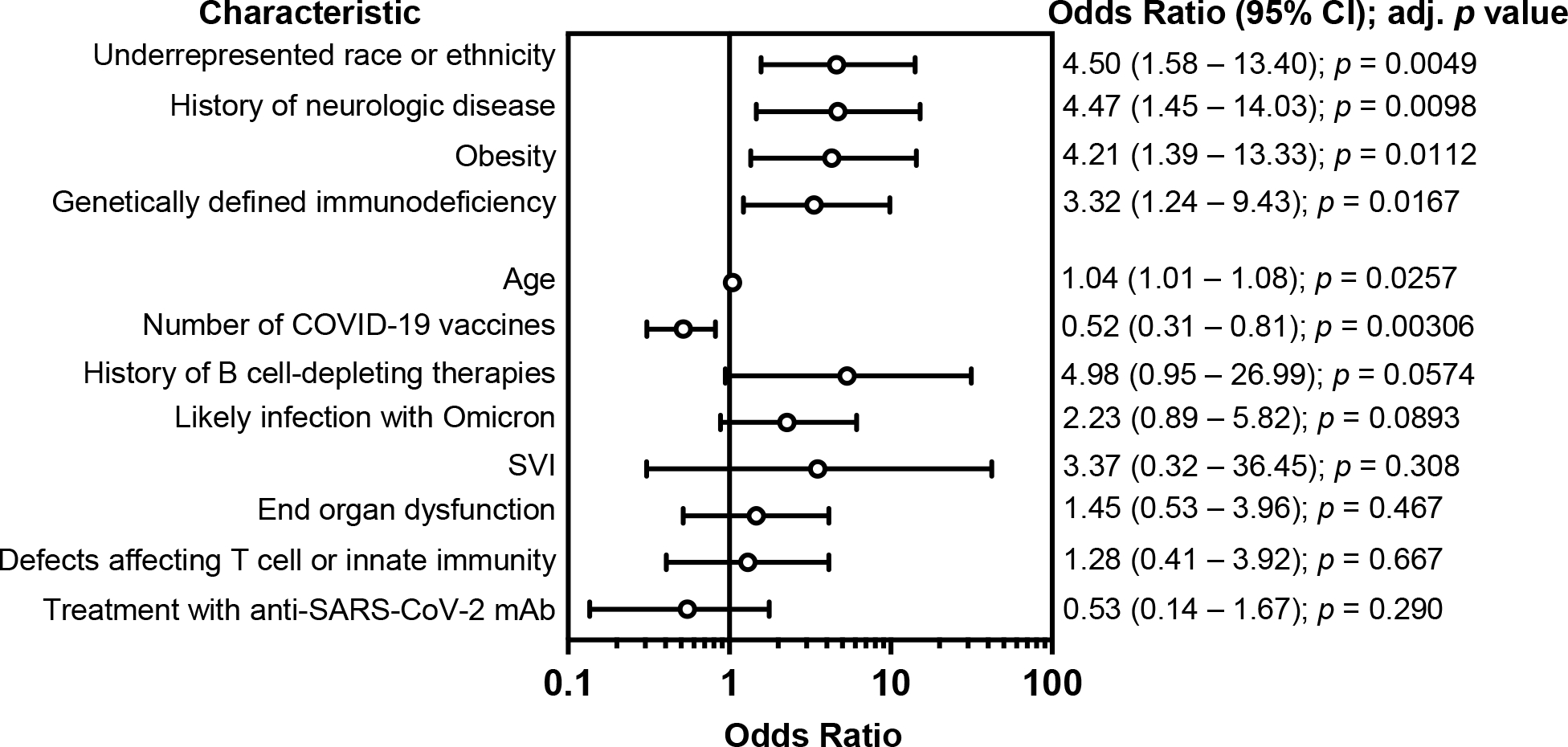
Odds ratios and 95% confidence intervals (CI) for each respective determinant of health, as calculated by logistic regression with covariate analysis. Likely infection with Omicron refers to individuals who tested positive after December 31, 2021, when the Omicron variant accounted for at least 90% of SARS-CoV-2 infections in Massachusetts based on wastewater sequencing reports.37
Despite the declining costs of genetic sequencing and the emergence of precision therapies for genetically defined immune disorders54,55, genetic tests are not routinely covered by most insurance companies.56 Genetic diagnoses in our cohort were made feasible by an institutional program that enables clinical whole exome sequencing at no cost to patients, which enabled access for patients with high SVI (Table E3). Even with this support for sequencing, the process includes several barriers. Thus, providers are possibly prioritizing those with potentially more severe disease: end organ dysfunction, combined immunodeficiencies, and innate disorders. Although a higher proportion of patients with defective T cell or innate immunity required hospitalization compared to those with humoral or primary immune dysregulatory disorders, we did not find a significant association between hospitalization and defective T cell or innate immunity after controlling for covariates. This may be due, at least in part, to national newborn screening programs for severe combined immunodeficiency that enable early diagnosis and treatment.57 Thus, most children with the most severe combined immunodeficiencies will have undergone hematopoietic stem cell transplantation before entry into daycare and school settings with potentially high levels of viral transmission. Additionally, the unprecedented speed in vaccine development during the COVID-19 pandemic reduced morbidity and mortality anticipated among individuals with immunodeficiencies. We found that increasing numbers of COVID-19 vaccinations were associated with a significantly reduced risk of hospitalization, further providing evidence that the school-aged and older individuals in this cohort had partial retention of immune function. This is concordant with the largest study of mRNA vaccine efficacy in individuals with congenital immunodeficiencies, which showed that the majority of patients with combined immunodeficiencies mounted an intact vaccine-specific T cell response.58 Although impaired T cell function is a commonly used measure of susceptibility to viral infections, our study highlights the limitations of applying this principle without acknowledgement of other determinants for health. Genetic diagnoses, obesity, comorbid conditions, and race modulate the immunologic risk often attributed solely to the mechanisms underlying congenital immunodeficiencies.
By using two different regression models, we adjusted for variations in SARS-CoV-2 directed interventions as some patients received vaccines and monoclonal antibodies while others did not. These differences were largely due to the availability of these agents during time of infection and governmental recommendations regarding eligibility. Similar models have been used to account for confounding factors in COVID-19 studies.59,60 Furthermore, the single center design of our study minimized variability in clinical practice. Patients with primary immunodeficiencies and SARS-CoV-2 infection were managed using institution-approved guidelines and algorithms to standardize modalities of care.
Our study has limitations. This is a single center, retrospective study and inborn errors of immunity are rare diseases. It is infeasible to definitively determine the total number of patients with SARS-CoV-2 infections because SARS-CoV-2 test availability was limited at the start of the pandemic, patients may not report all positive home antigen tests to healthcare providers, and asymptomatic SARS-CoV-2 infections are underreported. Additionally, there are limitations of using end organ dysfunction and defective T cell function as measures of disease severity. While these are quantifiable measures of disease, disease severity includes other features that are not possible to measure in a retrospective study, such as quality of life. Our study lays the groundwork for future prospective studies of immunologic risk that can include more comprehensive measures of disease severity. Given the limitations inherent in studies of rare diseases with small sample sizes, our study sought to determine the contributions of common risk factors that are often overlooked in rare diseases due to the “N of 1” approach typical of personalized medicine.
The morbidity and mortality caused by the COVID-19 pandemic transformed immunologic risk from an abstract concept into a real-world axis that influences daily decisions about school, work, and travel. The findings of this study indicate the importance of considering factors beyond molecular mechanisms as measures of immunologic risk and suggest that this risk can be modified by initiatives that address long-standing challenges with obesity and healthcare disparities.
Highlights:
What is already known about this topic? Outcomes of SARS-CoV-2 infections in individuals with inborn errors of immunity (IEI) are highly variable.
What does this article add to our knowledge? Prior studies of patients with IEI have not controlled for race or social vulnerability. We found that hospitalizations for SARS-CoV-2 were associated with race, ethnicity, obesity, and neurologic disease in individuals with IEIs. Specific types of immunodeficiency, organ dysfunction, and social vulnerability were not associated with increased risk of hospitalization.
How does this study impact current management guidelines? Current guidelines for the management of IEIs focus on risk conferred by genetic and cellular mechanisms. This study highlights the importance of considering variables linked with social determinants of health and common comorbidities as immunologic risk factors.
Acknowledgements
This study was supported by the National Institutes of Health (T32AI007512 to H.C.O., A.A.N., B.L., D.H.; K23AI143962 to L.M.B., R01DK130465 and R01AI139633-04S1 to J.C.), the Wallace Family Fund and Perkins Fund (to J.C.), and the Department of Pediatrics at King Abdulaziz University, Jeddah, KSA (to S.B.H.).
Abbreviations:
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- COVID-19
Coronavirus disease 2019
- IEI
inborn errors of immunity
- OR
odds ratio
- CI
confidence interval
Footnotes
Disclosures: No authors have disclosures relevant to the content of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alan A. Nguyen, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Saddiq B. Habiballah, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Brenna LaBere, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Megan Day-Lewis, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Megan Elkins, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Amer Al-Musa, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Anne Chu, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Jennifer Jones, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Ari J. Fried, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Douglas McDonald, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
David P. Hoytema van Konijnenburg, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Shira Rockowitz, Research Computing, Information Technology, Boston Children’s Hospital, Boston, MA; The Manton Center for Orphan Disease Research, Boston Children’s Hospital, Boston, Massachusetts.
Piotr Sliz, Research Computing, Information Technology, Boston Children’s Hospital, Boston, MA; The Manton Center for Orphan Disease Research, Boston Children’s Hospital, Boston, Massachusetts; Division of Molecular Medicine, Boston Children’s Hospital, Boston, Massachusetts.
Hans C. Oettgen, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Lynda C. Schneider, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Andrew MacGinnitie, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Lisa M. Bartnikas, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Craig D. Platt, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
Toshiro K. Ohsumi, Be Biopharma, Boston, Massachusetts.
Janet Chou, Division of Immunology, Boston Children’s Hospital and Harvard Medical School, Boston, Massachusetts.
References
- 1.Emanuel EJ, Persad G, Upshur R, Thome B, Parker M, Glickman A, et al. Fair Allocation of Scarce Medical Resources in the Time of Covid-19. N Engl J Med. 2020;382(21):2049–55. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon ME, Truog RD, Ubel PA. Allocating Resources Across the Life Span During COVID-19—Integrating Neonates and Children Into Crisis Standards of Care Protocols. JAMA Pediatrics. 2021;175(4):347–8. [DOI] [PubMed] [Google Scholar]
- 3.Bledsoe TA, Jokela JA, Deep NN, Snyder Sulmasy L. Universal Do-Not-Resuscitate Orders, Social Worth, and Life-Years: Opposing Discriminatory Approaches to the Allocation of Resources During the COVID-19 Pandemic and Other Health System Catastrophes. Ann Intern Med. 2020;M20–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Healthy People 2030 | health.gov.
- 5.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42(7):1473–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prince B, Lecerf K, Mustillo P, Scherzer R. Incidence of Primary Immunodeficiency Disorders at a Tertiary Care Immunology Clinic. Journal of Allergy and Clinical Immunology. 2020;145(2, Supplement):AB31. [Google Scholar]
- 7.Quinn J, Modell V, Orange JS, Modell F. Growth in diagnosis and treatment of primary immunodeficiency within the global Jeffrey Modell Centers Network. Allergy Asthma Clin Immunol. 2022;18(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucciol G, Tangye SG, Meyts I. Coronavirus disease 2019 in patients with inborn errors of immunity: lessons learned. Curr Opin Pediatr. 2021;33(6):648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Bastard P, COVID Human Genetic Effort, Cobat A, Casanova J-L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603(7902):587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prioritization of Therapeutics. COVID-19 Treatment Guidelines. [Google Scholar]
- 11.Meyts I, Bucciol G, Quinti I, Neven B, Fischer A, Seoane E, et al. Coronavirus disease 2019 in patients with inborn errors of immunity: An international study. J Allergy Clin Immunol. 2021;147(2):520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karakoc Aydiner E, Bilgic Eltan S, Babayeva R, Aydiner O, Kepenekli E, Kolukisa B, et al. Adverse COVID-19 outcomes in immune deficiencies: Inequality exists between subclasses. Allergy. 2022;77(1):282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castano-Jaramillo LM, Yamazaki-Nakashimada MA, O’Farrill-Romanillos PM, Muzquiz Zermeño D, Scheffler Mendoza SC, Venegas Montoya E, et al. COVID-19 in the Context of Inborn Errors of Immunity: a Case Series of 31 Patients from Mexico. J Clin Immunol. 2021;41(7):1463–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esenboga S, Ocak M, Akarsu A, Bildik HN, Cagdas D, Iskit AT, et al. COVID-19 in Patients with Primary Immunodeficiency. J Clin Immunol. 2021;41(7):1515–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goudouris ES, Pinto-Mariz F, Mendonça LO, Aranda CS, Guimarães RR, Kokron C, et al. Outcome of SARS-CoV-2 Infection in 121 Patients with Inborn Errors of Immunity: A Cross-Sectional Study. J Clin Immunol. 2021;41(7):1479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho H, Mathew S, Peluso MJ, Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy Clin Immunol Pract. 2021;9(1):490–493.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcus N, Frizinsky S, Hagin D, Ovadia A, Hanna S, Farkash M, et al. Minor Clinical Impact of COVID-19 Pandemic on Patients With Primary Immunodeficiency in Israel. Front Immunol. 2020;11:614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milito C, Lougaris V, Giardino G, Punziano A, Vultaggio A, Carrabba M, et al. Clinical outcome, incidence, and SARS-CoV-2 infection-fatality rates in Italian patients with inborn errors of immunity. J Allergy Clin Immunol Pract. 2021;9(7):2904–2906.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milota T, Sobotkova M, Smetanova J, Bloomfield M, Vydlakova J, Chovancova Z, et al. Risk Factors for Severe COVID-19 and Hospital Admission in Patients With Inborn Errors of Immunity - Results From a Multicenter Nationwide Study. Front Immunol. 2022;13:835770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moazzen N, Ahanchian H, Aelami MH, Asiyon H, Astaneh M, Naeimi AM, et al. COVID-19 in children with inborn errors of immunity: clinical scenarios. Am J Clin Exp Immunol. 2021;10(3):77–85. [PMC free article] [PubMed] [Google Scholar]
- 21.Shields AM, Burns SO, Savic S, Richter AG, UK PIN COVID-19 Consortium. COVID-19 in patients with primary and secondary immunodeficiency: The United Kingdom experience. J Allergy Clin Immunol. 2021;147(3):870–875.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Impact of SARS-CoV-2 Infection in Patients with Inborn Errors of Immunity: the Experience of the Italian Primary Immunodeficiencies Network (IPINet) - [DOI] [PMC free article] [PubMed]
- 23.Abolhassani H, Delavari S, Landegren N, Shokri S, Bastard P, Du L, et al. Genetic and immunologic evaluation of children with inborn errors of immunity and severe or critical COVID-19. J Allergy Clin Immunol. 2022;150(5):1059–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cousins K, DeFelice N, Jeong S, Feng J, Lee ASE, Rotella K, et al. SARS-COV-2 infections in inborn errors of immunity: A single center study. Frontiers in Immunology. 2022;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC/ATSDR Social Vulnerability Index (SVI). 2022.
- 26.Bureau UC. American Community Survey 2014–2018 5-Year Estimates Now Available. Census.gov.
- 27.Bonilla FA, Khan DA, Ballas ZK, Chinen J, Frank MM, Hsu JT, et al. Practice parameter for the diagnosis and management of primary immunodeficiency. J Allergy Clin Immunol. 2015;136(5):1186–1205.e1–78. [DOI] [PubMed] [Google Scholar]
- 28.BMI for Children and Teens. Centers for Disease Control and Prevention. 2023. [Google Scholar]
- 29.Cases, Data, and Surveillance. Centers for Disease Control and Prevention. 2020. [Google Scholar]
- 30.Puhr R, Heinze G, Nold M, Lusa L, Geroldinger A. Firth’s logistic regression with rare events: accurate effect estimates and predictions? Stat Med. 2017;36(14):2302–17. [DOI] [PubMed] [Google Scholar]
- 31.Vatcheva KP, Lee M, McCormick JB, Rahbar MH. Multicollinearity in Regression Analyses Conducted in Epidemiologic Studies. Epidemiology (Sunnyvale). 2016;6(2):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenland S, Daniel R, Pearce N. Outcome modelling strategies in epidemiology: traditional methods and basic alternatives. Int J Epidemiol. 2016;45(2):565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imdadullah M, Aslam M, Altaf S. The R Journal: mctest: An R Package for Detection of Collinearity among Regressors. The R Journal. 2016;8(2):495–505. [Google Scholar]
- 34.Mangiafico S rcompanion: Functions to Support Extension Education Program Evaluation. 2023.
- 35.Molteni E, Sudre CH, Canas LS, Bhopal SS, Hughes RC, Antonelli M, et al. Illness duration and symptom profile in symptomatic UK school-aged children tested for SARS-CoV-2. The Lancet Child & Adolescent Health. 2021;5(10):708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forrest CB, Burrows EK, Mejias A, Razzaghi H, Christakis D, Jhaveri R, et al. Severity of Acute COVID-19 in Children <18 Years Old March 2020 to December 2021. Pediatrics. 2022;149(4):e2021055765. [DOI] [PubMed] [Google Scholar]
- 37.Broad COVID-19 Testing Dashboard.
- 38.Delmonte OM, Castagnoli R, Calzoni E, Notarangelo LD. Inborn Errors of Immunity With Immune Dysregulation: From Bench to Bedside. Front Pediatr. 2019;7:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.People with Certain Medical Conditions. Centers for Disease Control and Prevention. 2023. [Google Scholar]
- 40.Jean-Jacques M, Bauchner H. Vaccine Distribution—Equity Left Behind? JAMA. 2021;325(9):829–30. [DOI] [PubMed] [Google Scholar]
- 41.Jones JM, Faruqi AJ, Sullivan JK, Calabrese C, Calabrese LH. COVID-19 Outcomes in Patients Undergoing B Cell Depletion Therapy and Those with Humoral Immunodeficiency States: A Scoping Review. Pathog Immun. 2021;6(1):76–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584(7821):430–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldstein LR, Tenforde MW, Friedman KG, Newhams M, Rose EB, Dapul H, et al. Characteristics and Outcomes of US Children and Adolescents With Multisystem Inflammatory Syndrome in Children (MIS-C) Compared With Severe Acute COVID-19. JAMA. 2021;325(11):1074–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruffner MA, Sullivan KE. Complications associated with underweight primary immunodeficiency patients: prevalence and associations within the USIDNET Registry. J Clin Immunol. 2018;38(3):283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Kerkhove MD, Vandemaele KAH, Shinde V, Jaramillo-Gutierrez G, Koukounari A, Donnelly CA, et al. Risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med. 2011;8(7):e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanneganti T-D, Dixit VD. Immunological complications of obesity. Nat Immunol. 2012;13(8):707–12. [DOI] [PubMed] [Google Scholar]
- 47.The Number of Uninsured Children is on the Rise. Center For Children and Families. 2019. [Google Scholar]
- 48.Moreira A, Chorath K, Rajasekaran K, Burmeister F, Ahmed M, Moreira A. Demographic predictors of hospitalization and mortality in US children with COVID-19. Eur J Pediatr. 2021;180(5):1659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antoon JW, Grijalva CG, Thurm C, Richardson T, Spaulding AB, Teufel RJ, et al. Factors Associated With COVID-19 Disease Severity in US Children and Adolescents. J Hosp Med. 2021;16(10):603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker RJ, Strom Williams J, Egede LE. Influence of Race, Ethnicity and Social Determinants of Health on Diabetes Outcomes. Am J Med Sci. 2016;351(4):366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalsania AK, Fastiggi MJ, Kahlam A, Shah R, Patel K, Shiau S, et al. The Relationship Between Social Determinants of Health and Racial Disparities in COVID-19 Mortality. J Racial Ethn Health Disparities. 2022;9(1):288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monroe P, Campbell JA, Harris M, Egede LE. Racial/ethnic differences in social determinants of health and health outcomes among adolescents and youth ages 10–24 years old: a scoping review. BMC Public Health. 2023;23(1):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Academies of Sciences E, Division H and M, Policy B on HS, Health R on G and P. Introduction and Overview. In: Understanding Disparities in Access to Genomic Medicine: Proceedings of a Workshop. National Academies Press (US); 2018. [PubMed] [Google Scholar]
- 54.Arlabosse T, Booth C, Candotti F. Gene Therapy for Inborn Errors of Immunity. J Allergy Clin Immunol Pract. 2023;11(6):1592–601. [DOI] [PubMed] [Google Scholar]
- 55.Ballow M, Leiding JW. Precision Medicine in the Treatment of Primary Immune Deficiency Patients With Disorders of Immune Dysregulation. Clin Rev Allergy Immunol. 2022;63(1):1–8. [DOI] [PubMed] [Google Scholar]
- 56.Reuter CM, Kohler JN, Bonner D, Zastrow D, Fernandez L, Dries A, et al. Yield of whole exome sequencing in undiagnosed patients facing insurance coverage barriers to genetic testing. J Genet Couns. 2019;28(6):1107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lankester AC, Albert MH, Booth C, Gennery AR, Güngör T, Hönig M, et al. EBMT/ESID inborn errors working party guidelines for hematopoietic stem cell transplantation for inborn errors of immunity. Bone Marrow Transplant. 2021;56(9):2052–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Leeuwen LPM, GeurtsvanKessel CH, Ellerbroek PM, de Bree GJ, Potjewijd J, Rutgers A, et al. Immunogenicity of the mRNA-1273 COVID-19 vaccine in adult patients with inborn errors of immunity. J Allergy Clin Immunol. 2022;149(6):1949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adir Y, Humbert M, Saliba W. COVID-19 risk and outcomes in adult asthmatic patients treated with biologics or systemic corticosteroids: Nationwide real-world evidence. J Allergy Clin Immunol. 2021;148(2):361–367.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan W-J, Liang W-H, Shi Y, Gan L-X, Wang H-B, He J-X, et al. Chronic Respiratory Diseases and the Outcomes of COVID-19: A Nationwide Retrospective Cohort Study of 39,420 Cases. J Allergy Clin Immunol Pract. 2021;9(7):2645–2655.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]


