Abstract
In eukaryotic cells, the inheritance of both the DNA sequence and its organization into chromatin is critical to maintain genome stability. This maintenance is challenged by DNA damage. To fully understand how the cell can tolerate genotoxic stress, it is necessary to integrate knowledge of the nature of DNA damage, its detection and its repair within the chromatin environment of a eukaryotic nucleus. The multiplicity of the DNA damage and repair processes, as well as the complex nature of chromatin, have made this issue difficult to tackle. Recent progress in each of these areas enables us to address, both at a molecular and a cellular level, the importance of inter-relationships between them. In this review we revisit the ‘access, repair, restore’ model, which was proposed to explain how the conserved process of nucleotide excision repair operates within chromatin. Recent studies have identified factors potentially involved in this process and permit refinement of the basic model. Drawing on this model, the chromatin alterations likely to be required during other processes of DNA damage repair, particularly double-strand break repair, are discussed and recently identified candidates that might perform such alterations are highlighted.
The ‘access, repair, restore’ model for nucleotide excision repair
Nucleotide excision repair (NER) is a highly conserved pathway that removes bulky lesions, including UV damage such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts, from DNA. The first evidence that chromatin rearrangements occur during NER came from the in vivo observation that the nuclease accessibility of DNA is modulated during UV-induced DNA synthesis in mammalian cells (Smerdon and Lieberman, 1978). This led to the ‘access, repair, restore’ (ARR) model, which postulates how NER might function in the complex chromatin environment of a nucleus (Smerdon, 1991). The nucleosomal unit, two turns of DNA wrapped around a histone octamer, is the first level of chromatin compaction and restricts the access of trans-acting factors involved in various aspects of DNA metabolism (Wolffe, 1997). In the ARR model, initial repair steps include reactions that permit access of the repair machinery to DNA damage and later steps restore the canonical nucleosomal organization of the repaired DNA (Figure 1A; for comprehensive reviews see Moggs and Almouzni, 1999; Thoma, 1999). Here we focus on recent studies, mostly in vitro, which advance this basic model and extend it to other repair systems.
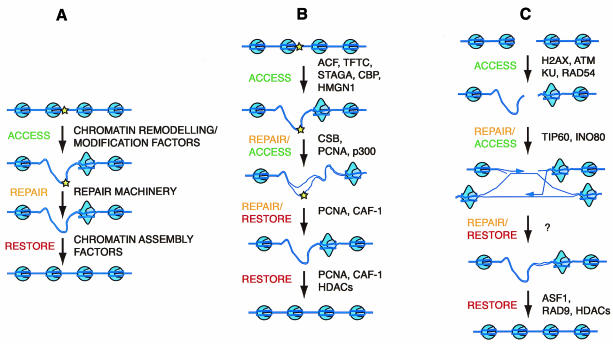
Fig. 1. (A) The original three-step ARR model for NER to explain how repair can be achieved at the nucleosomal level. To overcome inhibition by nucleosome structure (depicted in blue), the first stage in the repair process is removal or remodelling of nucleosomes to permit access to the DNA lesion (the star), here depicted for a few nucleosomes. After repair of the DNA, the original chromatin structure must be restored to ensure that epigenetic information is maintained. (B and C) An expansion of the original model to highlight factors that may be required for alterations in chromatin structure during the different enzymatic stages that comprise NER (B) and HR (C). Although only one additional stage is depicted in each case, this model is continuously evolving, and as our understanding of the interplay between chromatin dynamics and repair increases it will be adapted to include a greater degree of complexity.
Pioneering biochemical studies, using crude cell extracts to perform NER, showed that repair synthesis is strongly reduced on UV-damaged DNA pre-assembled into nucleosomes (Wang et al., 1991) or UV-irradiated simian virus 40 (SV40) minichromosomes (Sugasawa et al., 1993), compared with naked DNA. A detailed study of repair at specific sites in a mononucleosome further demonstrated that CPD removal is inhibited at most nucleosomal positions and is not influenced by rotational setting or by the presence of histone tails (Liu and Smerdon, 2000). Combined analysis of genetic defects in the human UV-sensitivity disorder xeroderma pigmentosum (XP) and biochemical assays have led to characterization of the NER pathway in impressive detail. This process involves sequential damage recognition, open complex formation, oligonucleotide excision and DNA synthesis, and the complete set of polypeptides required for NER in vitro has now been identified (de Laat et al., 1999). Such a purified NER system has been used recently to investigate damage repair within nucleosomal contexts. The efficiency of damage excision carried out by purified NER factors is lower on both UV-irradiated SV40 minichromosomes (after long incubation times) and reconstituted mononucleosomes than on naked DNA (Araki et al., 2000; Hara et al., 2000). The use of a dinucleosome as substrate further demonstrated that excision of 6-4 photoproducts by purified factors is strongly inhibited by this chromatin structure even when the lesion is located within the linker DNA (Ura et al., 2001). Proteins that are required before excision include XPA, replication protein A (RPA) and the XPC–hHR23B (homologous to yeast RAD23) complex (Volker et al., 2001). Consistent with inhibition occurring at this stage, the affinities of both XPA and XPC for DNA are decreased 5-fold on nucleosomal compared with naked DNA (Hara et al., 2000).
If, then, an access mechanism is required before NER can take place, what processes are involved? In the same way that some transcription factor complexes gain access to their binding sites, repair in vivo may involve recruitment of chromatin remodelling machines or histone modifying enzymes (Meijer and Smerdon, 1999), and also architectural factors. Indeed, sequence analyses, in vitro studies and yeast genetics have begun to identify factors that connect chromatin dynamics with repair (see Figure 2). ATP-dependent chromatin remodelling factors contain an SNF2 family ATPase subunit, which also contains a helicase motif (Vignali et al., 2000). One such remodelling factor that can perform an access role during NER (at least in vitro) is the ATP-utilizing chromatin assembly and remodelling factor (ACF), which contains the SNF2 family member imitation switch (ISWI) and Acf1. This complex can catalyse nucleosome movement, and addition of recombinant ACF to a purified in vitro NER system largely relieves the inhibition of repair by dinucleosomal structures (Ura et al., 2001). Interestingly, another SNF2 family member, yeast Rad16, is also required for NER, although its biochemical function is currently unclear (Eisen et al., 1995). A further connection between remodelling and NER is highlighted by studies of transcription coupled repair (TCR). TCR is a sub-pathway of NER targeted to the transcribed strands of active genes, defects in which lead to the human UV-sensitivity disorder Cockayne syndrome (van Gool et al., 1997). During TCR some access requirements may be bypassed due to the chromatin being remodelled for transcription (Kingston and Narlikar, 1999). However, studies of the CSB (Cockayne syndrome B) protein and its yeast homologue Rad26 suggest that further remodelling may be involved during TCR; both these proteins are in the SNF2 family (Eisen et al., 1995) and recombinant CSB was recently shown to alter the digestion profile of nucleosomal arrays in vitro, suggesting that this protein does indeed have remodelling activity (Citterio et al., 2000). It is possible that CSB remodels chromatin or other DNA–protein contacts in the area of the damage, and further enhances the recruitment of the downstream NER factors (Tijsterman and Brouwer, 1999).
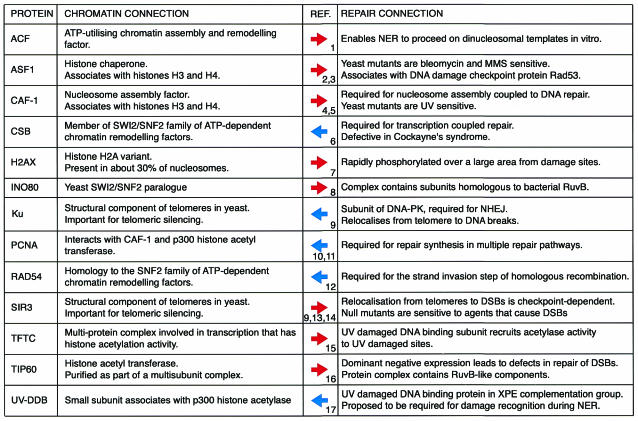
Fig. 2. Proteins that connect repair and chromatin. This table lists, in alphabetical order, factors that link repair and chromatin dynamics. Each protein’s roles in repair and other chromatin processes are highlighted. The central arrow represents the direction in which the connection was discovered: left to right for known chromatin proteins for which a DNA repair role was subsequently discovered, and right to left in the alternative case. The central numbers refer to the references in which this connection was made, and correspond to: 1, Ura et al., 2001; 2, Tyler et al., 1999; 3, Emili et al., 2001; 4, Gaillard et al., 1996; 5, Kaufman et al., 1997; 6, Citterio et al., 2000; 7, Rogakou et al., 1998; 8, Shen et al., 2000; 9, Martin et al., 1999; 10, Moggs et al., 2000; 11, Hasan et al., 2001; 12, Eisen et al., 1995; 13, McAinsh et al., 1999; 14, Mills et al., 1999; 15, Brand et al., 2001; 16, Ikura et al., 2000; 17, Datta et al., 2001.
The covalent modification of histones may also contribute to the mechanism whereby damaged sites can be accessed. Significantly, in vitro, histone H3 acetyl-transferase activity can be directed to sites of UV damage via the TBP-free TAFII complex (TFTC), which contains both SAP130, a subunit that specifically binds damaged DNA, and the histone acetyl-transferase hGCN5 (Brand et al., 2001). Furthermore, CREB binding protein (CBP) and p300 histone acetyl-transferases can interact with the small subunit of the XP-E damage-specific DNA binding protein (DDB), and the human STAGA complex (SPT3–TAFII31–GCN5L acetylase) interacts with the large subunit of DDB (Datta et al., 2001; Martinez et al., 2001). Given these findings, it is tempting to speculate that physical interactions between histone acetyl-transferase subunits and DDBs represent a mechanism by which the cell can achieve localized modification at damage sites, perhaps leading to more efficient recruitment of NER factors.
Intriguingly, proliferating cell nuclear antigen (PCNA) has also been shown to interact with the p300 histone acetyl-transferase in vitro and to co-localize with p300 in vivo (Hasan et al., 2001). PCNA is required for the repair synthesis step of NER, which is further downstream in the pathway than damage recognition. Hence, as depicted in Figure 1B, there are at least two distinct stages of the NER process at which chromatin modifying factors could be recruited. It will certainly be interesting to investigate whether other repair proteins associate with chromatin modifying factors and whether specific chromatin alterations are required in a progressive, and perhaps hierarchical, manner during NER.
In addition to nucleosomal organization, the impact of the folding of arrays into higher-order structures and the necessity of modulating such structures during repair must also be considered. High mobility group proteins with nucleosomal binding domains (HMGN proteins; the group previously called HMG-14/-17) are thought to participate in regulating such higher-order folding (Bustin, 1999). Interestingly, cells derived from HMGN1 knockout mice have recently been shown to be defective in UV damage repair (M. Bustin, personal communication).
After DNA repair has occurred, the original chromatin structure must be restored to ensure that there is no loss of epigenetic information. One factor potentially involved in this latter stage of NER is the three-subunit complex chromatin assembly factor 1 (CAF-1), which has an important role in replication-dependent chromatin assembly (Ridgway and Almouzni, 2000; Verreault, 2000). Studies in vitro have shown that this complex is required for the assembly of nucleosomes around a repair site in a PCNA-dependent manner (Gaillard et al., 1996; Moggs et al., 2000). Furthermore, CAF-1 and PCNA are recruited to chromatin in UV-damaged cells (Martini et al., 1998). The fact that Saccharomyces cerevisiae cells lacking CAF-1 function (cac mutants) are sensitive to UV light is consistent with this complex being important during NER (Kaufman et al., 1997; Game and Kaufman, 1999). CAF-1 is associated specifically with acetylated forms of histones H3 and H4 (Verreault et al., 1996), which are rapidly deacetylated by unknown factors once in the newly assembled chromatin (Mello and Almouzni, 2001). Interestingly, the p48 subunit of CAF-1 associates with a variety of histone deacetylase activities (Ridgway and Almouzni, 2000), which could be important during the final restoration stage. The work highlighted here suggests many molecules that can now be incorporated into an enriched version of the ARR model for NER (Figure 1B). A future challenge is to demonstrate specific roles for these factors during NER in vivo.
Different repair processes and associated chromatin rearrangements
The ARR model has proven useful for analysing chromatin rearrangements associated with the complex NER pathway. Other repair pathways are likely to have distinct chromatin modulation requirements due to mechanistic differences at the DNA level. The effectiveness of simpler repair processes may not be altered by the presence of nucleosomes; for example, DNA ligase I can reseal a single-strand break embedded in a nucleosomal substrate in vitro (Chafin et al., 2000). In this case the nucleosomal organization may even act favourably by keeping the ends to be rejoined in close proximity. On the other hand, the light-dependent reversion of UV damage by photolyases is inhibited by the presence of nucleosomes both in vivo and in vitro (Suter et al., 1997; Schieferstein and Thoma, 1998). It is not yet clear what chromatin rearrangements may occur during post-replicative repair to correct mismatch errors that arise during DNA synthesis. However, epistasis analysis in yeast has shown that Rad6 and Rad5 are required for this repair system; Rad6 can ubiquitylate histones and is also involved in telomeric silencing (Jentsch et al., 1987; Huang et al., 1997), and Rad5 is in the SNF2 family (Eisen et al., 1995), suggesting that the ARR model may also be valid for this pathway.
DNA double-strand breaks (DSBs) can be created by a wide variety of endogenous and exogenous agents including ionizing radiation (IR). The repair of this highly genotoxic lesion has been studied in considerable detail and can be undertaken by two major pathways: homologous recombination (HR) and non-homologous end joining (NHEJ) (van Gent et al., 2001). Both processes are likely to require chromatin alterations, the former for strand invasion and branch migration, and the latter for end processing. As in the case of NER, recent progress has identified chromatin remodelling, modifying and restoration factors that could be involved during DSB repair (Figures 1C and 2). One of the earliest detectable events in cells treated with IR is the rapid and extensive phosphorylation of the histone variant H2AX (Rogakou et al., 1998, 1999). This phosphorylation precedes the formation of IR-dependent foci containing repair proteins such as Rad51 or Rad50 (Paull et al., 2000), and the exact function of these foci remains to be elucidated. H2AX phosphorylation also occurs during other cellular processes that involve DSB formation, such as meiosis and V(D)J recombination, suggesting that it may be a general signal for the existence of a break (Modesti and Kanaar, 2001). In yeast, a similar phosphorylation site in histone H2A is essential for normal repair of DSBs by NHEJ (Downs et al., 2000). Members of the phosphatidylinositol-3-OH kinase related kinase (PIKK) family of checkpoint signalling proteins have been implicated in this response in yeast and human cells. This modification could alter chromatin structure to permit the access or action of repair factors, or it may function as a signal amplification event, perhaps helping to recruit not only repair or downstream signalling factors, but also additional chromatin modulating activities.
Given that histone acetylation is used to modulate chromatin structure during transcription and NER, it is not surprising to find that it may also be utilized during DSB repair. Recently, the histone acetyl-transferase TIP60 (TBP interacting protein) was purified as a component of a large multisubunit complex, which, in addition to acetyl-transferase activity, had DNA helicase activity. Two subunits of this complex had similarity to RuvB, the bacterial Holliday junction helicase, and these were found to possess ATPase activity, which may be important for helicase function (Ikura et al., 2000). Thus, acetyl-transferase activity is tightly associated with proteins that may perform the branch migration step of HR. Importantly, HeLa cells expressing a dominant-negative form of TIP60 lacking acetyl-transferase activity could not efficiently repair DSBs. Furthermore, these cells were defective in triggering apoptosis, suggesting that there are connections between chromatin alterations and downstream signalling events.
Not only covalent modifications, but also ATP-dependent chromatin remodelling have been implicated in DSB repair. A search for S. cerevisiae homologues of Drosophila ISWI identified Ino80, which was purified as part of a complex with chromatin remodelling, ATPase and helicase activities (Shen et al., 2000). Like the TIP60 acetyl-transferase, this Ino80 remodelling complex contained proteins homologous to RuvB, and cells lacking Ino80 were sensitive to DNA-damaging agents. Taken together, these results clearly suggest that chromatin alterations may be an intrinsic component of the branch migration step of HR (Figure 1C). It seems possible that the TIP60 and Ino80 protein complexes achieve chromatin alterations by utilizing some aspect of their helicase activity; the large topological changes that helicases confer on DNA molecules may provide a mechanism by which DNA–protein contacts can be altered. Indeed, in vitro, the spontaneous migration of an artificial Holliday junction is inhibited by positioning a nucleosome on the substrate DNA and this inhibition can be relieved by the addition of a bacterial RuvAB complex (Grigoriev and Hsieh, 1998). Other steps of HR may also be targets of chromatin remodelling activity; Rad54, which is required at the strand pairing stage, is in the SNF2 family (Eisen et al., 1995), although no associated remodelling activity has been identified to date.
Evidence from yeast suggests that higher-order structural changes may also occur during DSB repair. In S. cerevisiae Ku, a silent information regulator (SIR) and a myosin-like protein (Mlp2) are required for the maintenance of normal telomeric chromatin structures. Lack of any of these proteins results in sensitivity to agents that cause DSBs (Martin et al., 1999; Galy et al., 2000). Intriguingly, Ku and SIR relocate from telomeres to DSBs, and this relocalization is dependent on the checkpoint protein and PIKK family member Mec1 (Martin et al., 1999; McAinsh et al., 1999; Mills et al., 1999). However, the kinetics of this relocalization makes it unlikely that SIR proteins participate in the initial repair response. It may be that such proteins act to alter higher-order structures at positions where repair is not initially successful, perhaps as a signal or to prevent unwanted DNA transactions.
As for NER, the final stage of the ARR model for DSB repair involves restoration of the original chromatin structure. Genetic studies in yeast suggest that the chromatin assembly factor Asf1 (anti-silencing function) may be involved in this process. Asf1 is a histone chaperone that can synergize with CAF-1 for chromatin assembly during DNA replication or NER (Tyler et al., 1999; J. Mello, H. Sillje, D. Roche and E. Nigg, personal communication). However, Asf1 may also function independently of CAF-1 because yeast strains lacking this protein are sensitive to DSB inducing agents such as bleomycin and methyl methane sulfonate. Although the exact connection between Asf1 and the DSB repair process has not been identified, it is possible that Asf1 may operate during the chromatin restoration stage after repair processing is complete. In a further development, the yeast Asf1 protein has been shown to associate with a DNA damage-dependent checkpoint protein, Rad53 (Emili et al., 2001; Hu et al., 2001). However, interaction between human Asf1 and the human Rad53 homologue Chk2 has yet to be identified. Interestingly though, other human checkpoint proteins, ATM (mutated in ataxia telangiectasia, a human radiation sensitivity and cancer predisposition syndrome), ATR (ATM-related) and Rad9 have been shown to interact with deacetylases, for example HDAC1, and deacetylation activity is likely to be necessary for the restoration event (Kim et al., 1999; Schmidt and Schreiber, 1999; Cai et al., 2000). These checkpoint proteins act at an early stage of the cellular decision to delay cell cycle progression or trigger apoptosis in response to DNA damage and are present in radiation-induced nuclear foci (Burtelow et al., 2000; Tibbetts et al., 2000; Andegeko et al., 2001). The biological significance of these interactions between chromatin regulators and checkpoint proteins in the cellular DNA damage response remains to be identified.
Conclusions/perspectives
In summary, the fields of repair and chromatin dynamics have begun to come together to enhance our comprehension of repair in a nuclear environment. With the identification of proteins that link repair and chromatin dynamics, as highlighted in Figure 2, the original linear ARR model is constantly evolving (Figure 1). Detailed study of the biochemical properties and physical partners of these factors suggests the existence of complex networks of chromatin modifying, remodelling and assembly factors, signalling pathways and repair proteins. Building on the currently fragmented knowledge of these networks will require the integration of multiple strategies, but will lead to a better understanding of genome function and the cellular response to DNA damage.

Geneviève Almouzni & Catherine M. Green
Acknowledgments
Acknowledgements
We thank Raymond Poot and Ethel Moustacchi for critical reading of the manuscript and apologize to those whose work we have not been able to discuss due to space limitations. C.M.G. has been supported by a TMR network grant (ERBF MRX CT-98-0191) to G.A. followed by an EU Marie Curie Fellowship (HPMF-CT-2000-00976). G.A.’s laboratory is supported by la Ligue Nationale contre le Cancer (Equipe labellisée la Ligue), Euratom (FIGH-CT1999-00010) and the Commissariat à l’Energie Atomique (LRC no. 26).
References
- Andegeko Y., Moyal, L., Mitelman, L., Tsarfaty, I., Shiloh, Y. and Rotman, G. (2001) Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem., 276, 38224–38230. [DOI] [PubMed] [Google Scholar]
- Araki M. et al. (2000) Reconstitution of damage DNA excision reaction from SV40 minichromosomes with purified nucleotide excision repair proteins. Mutat. Res., 459, 147–160. [DOI] [PubMed] [Google Scholar]
- Brand M., Moggs, J.G., Oulad-Abdelghani, M., Lejeune, F., Dilworth, F.J., Stevenin, J., Almouzni, G. and Tora, L. (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J., 20, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtelow M.A., Kaufmann, S.H. and Karnitz, L.M. (2000) Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J. Biol. Chem., 275, 26343–26348. [DOI] [PubMed] [Google Scholar]
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R.L., Yan-Neale, Y., Cueto, M.A., Xu, H. and Cohen, D. (2000) HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem., 275, 27909–27916. [DOI] [PubMed] [Google Scholar]
- Chafin D.R., Vitolo, J.M., Henricksen, L.A., Bambara, R.A. and Hayes, J.J. (2000) Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J., 19, 5492–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E., Van Den Boom, V., Schnitzler, G., Kanaar, R., Bonte, E., Kingston, R.E., Hoeijmakers, J.H. and Vermeulen, W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol., 20, 7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Bagchi, S., Nag, A., Shiyanov, P., Adami, G.R., Yoon, T. and Raychaudhuri, P. (2001) The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res., 486, 89–97. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers, N.G. and Hoeijmakers, J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Downs J.A., Lowndes, N.F. and Jackson, S.P. (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature, 408, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Sweder, K.S. and Hanawalt, P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A., Schieltz, D.M., Yates, J.R.,III and Hartwell, L.H. (2001) Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell, 7, 13–20. [DOI] [PubMed] [Google Scholar]
- Gaillard P.H., Martini, E.M., Kaufman, P.D., Stillman, B., Moustacchi, E. and Almouzni, G. (1996) Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell, 86, 887–896. [DOI] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin, J.C., Scherthan, H., Doye, V., Rascalou, N. and Nehrbass, U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Game J.C. and Kaufman, P.D. (1999) Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics, 151, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev M. and Hsieh, P. (1998) Migration of a Holliday junction through a nucleosome directed by the E. coli RuvAB motor protein. Mol. Cell, 2, 373–381. [DOI] [PubMed] [Google Scholar]
- Hara R., Mo, J. and Sancar, A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Hassa, P.O., Imhof, R. and Hottiger, M.O. (2001) Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature, 410, 387–391. [DOI] [PubMed] [Google Scholar]
- Hu F., Alcasabas, A.A. and Elledge, S.J. (2001) Asf1 links Rad53 to control of chromatin assembly. Genes Dev., 15, 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Kahana, A., Gottschling, D.E., Prakash, L. and Liebman, S.W. (1997) The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6693–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T., Ogryzko, V.V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. and Nakatani, Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath, J.P. and Varshavsky, A. (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature, 329, 131–134. [DOI] [PubMed] [Google Scholar]
- Kaufman P.D., Kobayashi, R. and Stillman, B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- Kim G.D., Choi, Y.H., Dimtchev, A., Jeong, S.J., Dritschilo, A. and Jung, M. (1999) Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J. Biol. Chem., 274, 31127–31130. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar, G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Liu X. and Smerdon, M.J. (2000) Nucleotide excision repair of the 5S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem., 275, 23729–23735. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche, T., Suka, N., Grunstein, M. and Gasser, S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- Martinez E., Palhan, V.B., Tjernberg, A., Lymar, E.S., Gamper, A.M., Kundu, T.K., Chait, B.T. and Roeder, R.G. (2001) Human staga complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol., 21, 6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Roche, D.M., Marheineke, K., Verreault, A. and Almouzni, G. (1998) Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol., 143, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A.D., Scott-Drew, S., Murray, J.A. and Jackson, S.P. (1999) DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol., 9, 963–966. [DOI] [PubMed] [Google Scholar]
- Meijer M. and Smerdon, M.J. (1999) Accessing DNA damage in chromatin: insights from transcription. BioEssays, 21, 596–603. [DOI] [PubMed] [Google Scholar]
- Mello J.A. and Almouzni, G. (2001) The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev., 11, 136–141. [DOI] [PubMed] [Google Scholar]
- Mills K.D., Sinclair, D.A. and Guarente, L. (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- Modesti M. and Kanaar, R. (2001) DNA repair: spot(light)s on chromatin. Curr. Biol., 11, R229–R232. [DOI] [PubMed] [Google Scholar]
- Moggs J.G. and Almouzni, G. (1999) Chromatin rearrangements during nucleotide excision repair. Biochimie, 81, 45–52. [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Grandi, P., Quivy, J.P., Jonsson, Z.O., Hubscher, U., Becker, P.B. and Almouzni, G. (2000) A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol., 20, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T.T., Rogakou, E.P., Yamazaki, V., Kirchgessner, C.U., Gellert, M. and Bonner, W.M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895. [DOI] [PubMed] [Google Scholar]
- Ridgway P. and Almouzni, G. (2000) CAF-1 and the inheritance of chromatin states: at the crossroads of DNA replication and repair. J. Cell Sci., 113, 2647–2658. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch, D.R., Orr, A.H., Ivanova, V.S. and Bonner, W.M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem., 273, 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon, C., Redon, C. and Bonner, W.M. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol., 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieferstein U. and Thoma, F. (1998) Site-specific repair of cyclobutane pyrimidine dimers in a positioned nucleosome by photolyase and T4 endonuclease V in vitro. EMBO J., 17, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D.R. and Schreiber, S.L. (1999) Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry, 38, 14711–14717. [DOI] [PubMed] [Google Scholar]
- Shen X., Mizuguchi, G., Hamiche, A. and Wu, C. (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature, 406, 541–544. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. (1991) DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol., 3, 422–428. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. and Lieberman, M.W. (1978) Nucleosome rearrangement in human chromatin during UV-induced DNA-repair synthesis. Proc. Natl Acad. Sci. USA, 75, 4238–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Masutani, C. and Hanaoka, F. (1993) Cell-free repair of UV-damaged simian virus 40 chromosomes in human cell extracts. I. Development of a cell-free system detecting excision repair of UV-irradiated SV40 chromosomes. J. Biol. Chem., 268, 9098–1004. [PubMed] [Google Scholar]
- Suter B., Livingstone-Zatchej, M. and Thoma, F. (1997) Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J., 16, 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F. (1999) Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J., 18, 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts R.S., Cortez, D., Brumbaugh, K.M., Scully, R., Livingston, D., Elledge, S.J. and Abraham, R.T. (2000) Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev., 14, 2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M. and Brouwer, J. (1999) Rad26, the yeast homolog of the Cockayne syndrome B gene product, counteracts inhibition of DNA repair due to RNA polymerase II transcription. J. Biol. Chem., 274, 1199–1202. [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Adams, C.R., Chen, S.R., Kobayashi, R., Kamakaka, R.T. and Kadonaga, J.T. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature, 402, 555–560. [DOI] [PubMed] [Google Scholar]
- Ura K., Araki, M., Saeki, H., Masutani, C., Ito, T., Iwai, S., Mizukoshi, T., Kaneda, Y. and Hanaoka, F. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J., 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., Hoeijmakers, J.H. and Kanaar, R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- van Gool A.J., van der Horst, G.T., Citterio, E. and Hoeijmakers, J.H. (1997) Cockayne syndrome: defective repair of transcription? EMBO J., 16, 4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A. (2000) De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev., 14, 1430–1438. [PubMed] [Google Scholar]
- Verreault A., Kaufman, P.D., Kobayashi, R. and Stillman, B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Vignali M., Hassan, A.H., Neely, K.E. and Workman, J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M. et al. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Wu, X.H. and Friedberg, E.C. (1991) Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem., 266, 22472–22478. [PubMed] [Google Scholar]
- Wolffe A.P. (1997) Chromatin: Structure and Function. Academic Press, New York, NY.