Abstract
The p300/CBP family of transcriptional coactivators possesses multiple functional domains, including a histone acetyltransferase (HAT) and several activation domains. A number of models have been proposed to account for their roles in transcriptional activation, including interactions with basal transcription machinery and chromatin remodeling. However, individual contributions of these domains to transcriptional activation and their significance in living organisms remain unclear. We addressed the importance of the HAT activity of CBP-1, the worm ortholog of p300/CBP, in Caenorhabditis elegans with three different and complementary approaches. These include allele-specific RNA-mediated interference (RNAi), genetic rescue and the use of a specific chemical inhibitor of the p300/CBP HAT. Our findings demonstrate that HAT activity is of primary importance for CBP-1 to regulate transcription and to promote differentiation during C. elegans embryogenesis.
INTRODUCTION
Histone hyperacetylation is in general correlated with transcriptional activation (Brownell and Allis, 1996). Recently, a number of transcriptional coactivators have been identified and have been shown to possess histone acetyltransferase (HAT) activity, including the related p300 and CBP proteins (p300/CBP). p300/CBP have intrinsic as well as associated HAT activity (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Yang et al., 1996). In addition, they contain several transcriptional activation domains, as well as domains for interaction with DNA-binding transcription factors (reviewed in Goodman and Smolik, 2000). Thus, p300/CBP possess multiple functional domains, each appearing to contribute to transcriptional activation. However, the relationship between these domains and their individual contribution to transcriptional activation in vivo remain unclear.
Several models have been proposed for p300/CBP-mediated transcriptional activation. They include a direct interaction with basal transcription factors TFIIB and TBP, and an indirect interaction with the RNA polymerase II holoenzyme complex mediated by RNA helicase A (reviewed in Goodman and Smolik, 2000). In addition, analyses of p300/CBP HAT mutants in cell culture and in in vitro transcription systems provided evidence correlating HAT activity with the transcriptional activity of p300/CBP (Martinez-Balbas et al., 1998; Kraus et al., 1999). In the case of the IFN-β promoter, virus-induced histone hyperacetylation has been correlated with the ability of the activator to recruit p300 (Parekh and Maniatis, 1999). More recently, activator-dependent transcription from chromatin templates in vitro has been shown to involve histone acetylation by p300, linking p300-mediated transcriptional activation with direct histone acetylation (Kundu et al., 2000). In addition, the chemical inhibitor Lys-CoA has been shown to selectively inhibit p300 HAT activity as well as p300 HAT-dependent transcriptional activation in vitro (Lau et al., 2000). However, none of these studies have addressed the importance of p300/CBP HAT activity in a developmental context and in a living organism.
Using RNA-mediated interference (RNAi; Fire et al., 1998), we showed previously that the only Caenorhabditis elegans p300/CBP ortholog, CBP-1, is essential for differentiation. The hallmark phenotypes of cbp-1(RNAi) embryos include a complete lack of differentiation of all but neuronal cells (Shi and Mello, 1998). In addition, they contain an excess number of cells compared with the wild-type embryos at the same developmental stage, suggesting a suppressive role for CBP-1 in cell proliferation. These findings thus identify a unique set of phenotypes associated with the loss of CBP-1 during development, and suggest a fundamental role for CBP-1 in differentiation. Here we used genetic and chemical approaches to provide multiple lines of evidence that demonstrate the importance of CBP-1 HAT activity for its overall ability to regulate transcription and to promote differentiation in C. elegans. We isolated a C. elegans mutant that encodes a truncated CBP-1 with an internal deletion of the HAT domain (CBP-1Δ), and showed that CBP-1Δ can no longer promote differentiation and appears to have lost all of its biological activity. We isolated a second, biochemically null cbp-1 mutant and showed that the wild-type, but a not a mutant cbp-1 lacking the HAT activity, can rescue its embryonic lethal phenotype. To complement these genetic studies, we showed that a p300/CBP HAT-specific chemical inhibitor could result in inviable embryos with phenotypes and transcriptional profiles indistinguishable from those of the cbp-1(RNAi) embryos. Taken together, these findings demonstrate a pivotal role of HAT activity for CBP-1 to regulate transcription and differentiation in vivo.
RESULTS AND DISCUSSION
Allele-specific RNAi suggests potential importance of the CBP-1 HAT domain
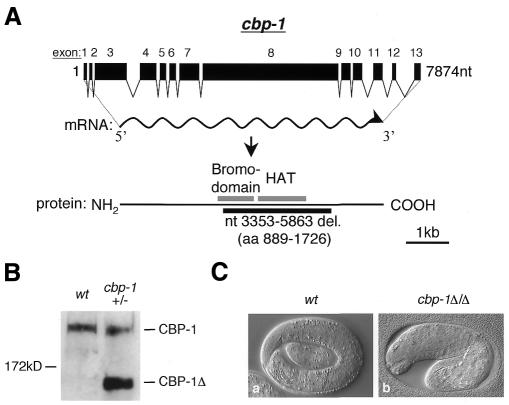
To explore the role of CBP-1 HAT activity in vivo, we used a PCR-based screening approach (Jansen et al., 1997; Barstead, 1999) and isolated a cbp-1 mutant with an internal, in-frame deletion of a region inclusive of the HAT and bromo domains [Figure 1A, CBP-1Δ amino acids (aa) 889–1726] (Bannister and Kouzarides, 1996; Ogryzko et al., 1996). The truncated protein still contains both the N- and C-terminal activation domains and is predicted to be 132 kDa in size. Western blotting and immunostaining with CBP-1 polyclonal antibodies raised against the N- and C-terminal regions of CBP-1 (see Methods) showed stable expression of CBP-1Δ in cbp-1 heterozygous mutants (Figures 1B, 2B and data not shown). The fact that these antibodies recognized CBP-1Δ suggests that the overall CBP-1 structure is not grossly altered by the deletion. The cbp-1 homozygous mutants (bm1) are arrested at an embryonic stage similar to what is called the ‘two-fold’ stage (Riddle et al., 1997), characterized by the completion of differentiation and the beginning of body morphogenesis (Figure 1C, panel b). Based on the recent analysis of a truncated mouse CBP (aa 1–1084), which acted dominant negatively (Oike et al., 1999), CBP-1Δ is predicated to function as a dominant negative as well. However, the cbp-1 heterozygotes are by and large wild type, and the reason for this is unclear. Taken together, these findings suggest that while maternally provided CBP-1 is sufficient to support embryonic differentiation, zygotic supply of CBP-1 is essential for continued embryonic development. In addition, they also suggest that CBP-1Δ cannot support embryonic development beyond the ‘two-fold’ stage as does the wild-type CBP-1. However, it is unclear whether maternally provided CBP-1Δ can promote differentiation during early stages of embryogenesis.
Fig. 1. A cbp-1 deletion mutant displays an embryonic lethal phenotype. (A) Isolation of a cbp-1 deletion mutant. An internal in-frame deletion mutant of cbp-1 was isolated from a DEB-mutagenized C. elegans library by PCR. The cbp-1 gene and the predicted 227 kDa protein are shown schematically. The HAT and bromo domains and the deleted region (aa 889–1726) in the mutant are indicated. (B) The truncated CBP-1 is stably expressed in cbp-1 heterozygous mutant animals. The wild-type and the truncated CBP-1 proteins (CBP-1Δ) are labeled on the right. The genotypes of the animals analyzed are indicated at the top. (C) Animals homozygous for cbp-1Δ die at the 1.5- to 2-fold stage of C. elegans embryonic development. The Nomarski images of a wild-type (a) and homozygous mutant animal (b) are shown. The wt embryo is at the pretzel stage while the cbp-1 mutant embryo died at the ∼1.5- to 2-fold stage (b).
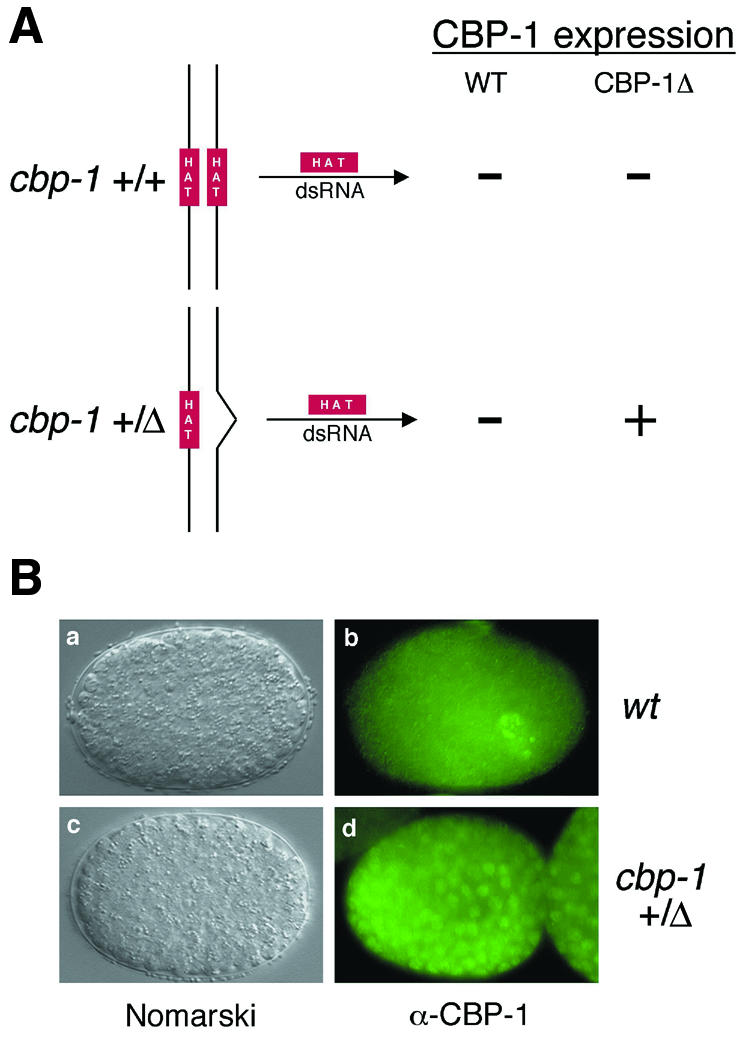
Fig. 2. Allele-specific RNAi. (A) Schematic diagram of the allele-specific RNAi approach. HAT dsRNA is predicted to interfere only with the expression from the wt but not the mutant cbp-1 allele. (B) CBP-1Δ lacking the HAT domain is incapable of promoting differentiation. dsRNA corresponding to the HAT domain deleted in the cbp-1Δ was injected into wt (cbp-1 +/+) and heterozygous mutant animals (cbp-1 +/Δ). Nomarski analysis shows identical RNAi phenotypes (a and c). Wild-type CBP-1 expression was abrogated in wt (b) and cbp-1 heterozygous (d) animals by HAT dsRNA, while the expression of CBP-1Δ is unaffected (compare d with b).
We used an allele-specific RNAi approach to determine whether CBP-1Δ possesses any transcriptional or biological activity. Double-stranded (ds) RNA corresponding to the HAT region that is absent from the mutant cbp-1 allele was used for RNAi. As shown in Figure 2A, the ds ‘HAT RNA’ is predicted to inhibit expression only from the wild-type but not the mutant cbp-1 allele in the heterozygous mothers. As shown in Figure 2B, the ds HAT RNA inhibited wild-type CBP-1 expression in wild-type C. elegans embryos (panel b), resulting in embryos that displayed phenotypes characteristic of cbp-1(RNAi) embryos (panel a; Shi and Mello, 1998). The affected embryos derived from the cbp-1 heterozygous mothers continued to express the HAT-less CBP-1Δ protein, as shown by immunostaining using CBP-1 polyclonal antibodies (Figure 2B, panel d), but nevertheless displayed phenotypes indistinguishable from those of the cbp-1(RNAi) embryos (Shi and Mello, 1998) (Figure 2B, compare panel c with a). These include an excess number of cells as well as a complete lack of differentiation of all non-neuronal lineage analyzed by Nomarski microscopy (Figure 2B, panels a and c; data not shown). Another hallmark of the cbp-1(RNAi) embryos is the presence of an excess number of cells that display features of neuronal differentiation (Shi and Mello, 1998). Using immunostaining, we found that about twice the normal complement of cells in the above embryos expressed the neuron-specific protein MEC-7 (Hamelin et al., 1992), as described previously for the cbp-1(RNAi) embryos (Shi and Mello, 1998; data not shown). Taken together, these findings suggest that CBP-1Δ is functionally null and that the HAT region may be essential for CBP-1 function.
A wild-type, but not a mutant cbp-1 gene carrying a HAT point mutation, rescues cbp-1 embryonic phenotypes
A recent study shows that although the bromo domain of p300, the mammalian homolog of CBP-1, is not sufficient to mediate chromatin binding, it nevertheless plays an important role in the efficiency with which p300 is able to access chromatin (Manning et al., 2001). Since the deletion in CBP-1Δ includes both the HAT and the bromo domain, the allele-specific RNAi experiment described above cannot rule out the possibility that the observed defects are in part due to a compromised ability of CBP-1Δ to bind chromatin. To investigate further the requirement of CBP-1 HAT activity in development, we isolated a second cbp-1 deletion mutant (YS4), which carries a deletion spanning part of its promoter and the first four exons [nucleotides (nt) –805 to +1665]. Immunostaining showed that CBP-1 proteins are absent in the homozygous animals, suggesting that this deletion mutation abolished cbp-1 expression (data not shown). The homozygous animals are arrested at similar stages of embryonic development to the animals (YS2) carrying the internal deletion of cbp-1 (bm1) described above. We used heterozygous YS4 animals for genetic rescue with either a wild-type or a mutant cbp-1 gene encoding a HAT-defective CBP-1 protein (see below).
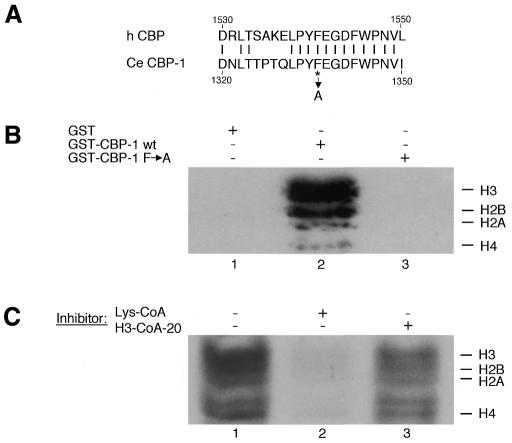
The putative HAT region of CBP-1 is highly homologous to that of mammalian p300 and CBP (Shi and Mello, 1998). Amino acids that have been shown to be critical for the p300 and CBP HAT activity (Bannister and Kouzarides, 1996; Kraus et al., 1999) are all conserved in CBP-1. Although the activity was weaker than that of human p300 assayed under the same conditions, GST–CBP-1 HAT (aa 803–1620) displayed significant HAT activity and acetylated all four histones in vitro (Figure 3B, lane 2). As a control, we found GST alone had no HAT activity (Figure 3B, lane 1). Previously, a point mutation of F to A at position 1541 of human CBP was shown to abrogate its HAT activity (Martinez-Balbas et al., 1998). We generated the same mutation in CBP-1 (Figure 3A) and found that it also abolished CBP-1 HAT activity (Figure 3B, lane 3). Furthermore, this HAT-defective mutant failed to rescue in all 16 transgenic lines assayed while the wild-type cbp-1 gene reproducibly rescued the zygotic lethality of cbp-1 (YS4) homozygotes (see Methods). Immunostaining with CBP-1 antibodies indicated that the mutant CBP-1 protein was expressed in the non-rescued lines (data not shown). Taken together, these findings strongly suggest that the HAT activity is critical for the biological functions of CBP-1 during C. elegans development.
Fig. 3. Analysis of the effect of Lys-CoA on CBP-1 HAT activity. (A) Partial HAT domain sequence alignment between human CBP and C. elegans CBP-1. Partial HAT sequences of human CBP (top) and C. elegans CBP-1 (bottom) are taken from Martinez-Balbas et al. (1998) and Shi and Mello (1998). The F residue mutated to A is marked by an asterisk. (B) Wild-type, but not CBP-1 with a point mutation in the HAT domain, acetylates histones. Equal amounts of purified wild-type and mutant GST–CBP-1 HAT (aa 803–1620) proteins were assayed for HAT activity using purified histones as substrates. GST–CBP-1 (lane 2), but not the CBP-1 point mutant (lane 3), acetylated all four histones. (C) Lys-CoA inhibits CBP-1 HAT activity. Lys-CoA, a CBP/p300-specific HAT inhibitor, inhibits CBP-1 HAT activity at 10 µM (lane 2). A P/CAF-specific HAT inhibitor, H3-CoA-20, does not (lane 3).
Lys-CoA results in inviable embryos with phenotypes indistinguishable from those of cbp-1 null embryos
As a complement to the genetic approaches described above, we turned to a chemical approach to analyze the requirement for CBP-1 HAT activity during early embryogenesis. It has been shown recently that Lys-CoA can selectively inhibit mammalian p300 HAT activity, while a related compound, H3-CoA-20, preferentially inhibits P/CAF HAT activity in vitro (Lau et al., 2000). We first asked whether these chemicals have similar effects on CBP-1. Consistent with the mammalian data, Lys-CoA, but not H3-CoA-20, inhibited the ability of CBP-1 HAT to acetylate histones (Figure 3C, compare lanes 2 and 3).
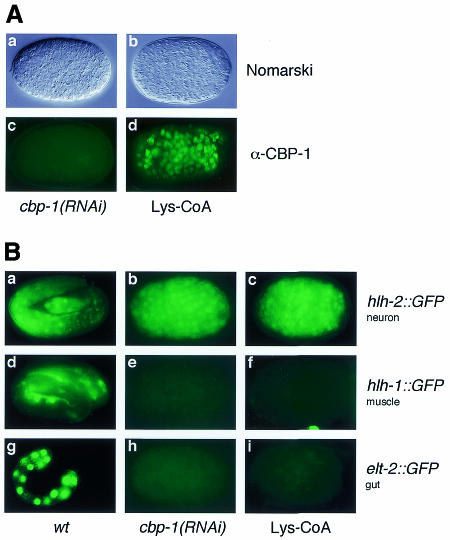
We next determined the effect of these chemicals on C. elegans embryonic development. Injection of Lys-CoA but not H3-CoA-20 into the gonad arms of hermaphrodite mothers resulted in embryonic arrest of F1 animals. Strikingly, ∼90% of the embryos laid during the first 3 h after recovery displayed phenotypes identical to those of the cbp-1(RNAi) embryos, including an excess number of cells and a complete lack of differentiation of all but neuronal cell types (Figure 4A, panel b; data not shown). This is not due to inhibition of CBP-1 expression since CBP-1 proteins were readily detectable in the embryos affected by Lys-CoA (Figure 4A, panel d). For comparison, cbp-1(RNAi) embryos are also shown (Figure 4A, panels a and c). After the first 3 h, the injected animals continued to produce dead embryos for another 6 h, but they displayed a wide range of phenotypes, including cbp-1 zygotic-like phenotypes (data not shown). This is probably due to partial inhibition of CBP-1 HAT activity in these embryos as a result of Lys-CoA breakdown and/or dilution. As a specificity control, we found that the P/CAF inhibitor H3-CoA-20 had no effect on worm development (data not shown). Interestingly, inhibition of Ce-P/CAF (Y47G6A.6) expression by RNAi also had no effect on C. elegans development (M. Victor and Y. Shi, unpublished results), consistent with the result of the chemical inhibitor experiment. Taken together, embryos affected by Lys-CoA display cbp-1 null phenotypes without affecting CBP-1 expression. The most likely explanation is that the observed null phenotype is a result of the inhibition of CBP-1 HAT activity in vivo.
Fig. 4. Phenotype and promoter activity analyses of Lys-CoA-affected embryos. (A) Lys-CoA injection results in a cbp-1(RNAi)-like phenotype. Injection of wt animals with either cbp-1 dsRNA or Lys-CoA results in identical phenotypes by Nomarski microscopy (compare a and b). In contrast, unlike the cbp-1(RNAi) embryo (c), the Lys-CoA-affected embryo continued to express CBP-1 (d). (B) Lys-CoA de-represses a neuronal promoter but inhibits gut- and muscle-specific promoters. The expression patterns of hlh-2::GFP, hlh-1::GFP and elt-2::GFP in wild-type animals are shown in the left panels (a, d and g). After interference with CBP-1 expression using RNAi (the middle panels) or inhibition of CBP-1 HAT activity by Lys-CoA (the right panels), the same surplus number of neuronal cells (hlh-2::GFP positive) was observed in either cbp-1(RNAi) or Lys-CoA-treated embryos (b and c). In contrast, the activities of the muscle- and endoderm-specific promoters, hlh-1::GFP and elt-2::GFP, are completely abrogated in cbp-1(RNAi) and Lys-CoA-treated embryos (e, f and h, i).
Regulation of CBP-1 target promoters by Lys-CoA
We next investigated the effect of Lys-CoA on transcription by analyzing three cell type-specific promoters linked to a green fluorescent protein (GFP) reporter. RNAi inhibition of CBP-1 induces ectopic expression of HLH-2::GFP in non-neuronal cells (Figure 4B, compare panels a and b; Shi and Mello, 1998), a promoter whose activity is restricted to neurons late in C. elegans embryogenesis (Krause et al., 1997). In contrast, inhibition of CBP-1 by RNAi eliminated the expression of HLH-1::GFP and ELT2::GFP (Figure 4B, compare panels e and h with d and g), promoters that are active in muscle and endodermal cells, respectively (Krause et al., 1990; Fukushige et al., 1998). Significantly, all these three promoters were affected by Lys-CoA identically. As shown in Figure 4B, the expression of HLH1::GFP and ELT2::GFP is completely abolished in Lys-CoA-affected embryos displaying cbp-1(RNAi)-like phenotypes (panels f and i). In addition, Lys-CoA induced ectopic expression of HLH2::GFP, a phenomenon characteristic of cbp-1(RNAi) embryos (Figure 4B, panel c). Taking the phenotypic and promoter analyses together, these findings demonstrate that inhibition of CBP-1 HAT activity is equivalent to the removal of CBP-1 from the embryos, suggesting that HAT activity of CBP-1 is essential for transcription and all biological functions of CBP-1 during C. elegans embryogenesis. The findings highlight the primary importance of the HAT activity for CBP-1 to function as a transcriptional co-factor that promotes differentiation in vivo. In its absence, CBP-1 is non-functional despite the presence of all other functional domains. However, our results do not exclude important roles that other domains may play in CBP-1-dependent transcription after the HAT activity of CBP-1 is engaged.
In summary, we have provided multiple lines of evidence demonstrating that CBP-1 HAT activity is crucial for CBP-1 function in the living organism C. elegans. We have shown by allele-specific RNAi that CBP-1 lacking the HAT domain is functionally null. We demonstrated that a HAT-defective CBP-1 mutant failed to rescue the zygotic phenotypes of a cbp-1 null mutant. Finally, we provided evidence that Lys-CoA, which selectively inhibits CBP-1 HAT activity, also incapacitates the ability of CBP-1 to regulate transcription and to promote differentiation. Thus, a combined genetic and chemical biology approach has provided evidence that HAT is indispensable for every aspect of CBP-1 activity in C. elegans development. At the molecular level, it is possible that the HAT activity has to be in place first before other biochemical activities of p300/CBP can engage in aspects of transcriptional activation. This is likely to be true for the p300/CBP family proteins in other organisms as well, i.e. the HAT activity provides a fundamental activity that is essential for their transcription and biological functions.
METHODS
Strains, culture and isolation of two cbp-1 deletion mutants. N2 wild-type worms were cultured as described previously (Brenner, 1974). lit-1(t1534) unc-32(e189)/qC1 III; him-3(e1147) IV served as a source for qC1. Transgenic animals carrying HLH-1::GFP, HLH-2::GFP and ELT-2::GFP were gifts from M. Krause and J. McGhee.
A mutant library representing 1 million genomes was generated by mutagenesis using 1,2,3,4-diepoxybutane (DEB) (Epstein and Shakes, 1995) and screened for deletions in cbp-1 by PCR as described (Jansen et al., 1997; Barstead, 1999). bm-1 (LGIII) carries an internal deletion (nt 3353–5863) while bm-2 (LGIII) carries a 2470 bp deletion spanning nt –851 to +1665 of the cbp-1 gene. Strains YS2 and YS4 refer to the two lines of animals carrying either the internal (YS2) or the N-terminal deletion (YS4) in the cbp-1 gene that were back-crossed five times and balanced by qC1. In both cases, the cbp-1 mutant allele is marked with dpy-18, a nearby recessive genetic marker. The Dpy phenotype was used as an indicator for homozygosity of the cbp-1 locus. The Dpy animals are easily identifiable and their presence indicates a successful rescue.
Antibody production, worm extract preparation and western blotting. The regions corresponding to aa 133–277 (N-terminal) and 1775–1942 (C-terminal) of CBP-1 were PCR amplified and subcloned into pGEX-T2. The resulting purified GST fusion proteins were used for the generation of polyclonal antibodies in rabbits. Specificity of the sera was verified by peptide competition and by RNAi. To prepare worm extracts, worms were resuspended in lysis buffer (10 mM sodium phosphate pH 7.4, 8 M urea, 2 mM EDTA, 1% SDS and 1% β-mercaptoethanol) and frozen in liquid nitrogen. After thawing, they were sonicated and the debris removed by centrifugation. Proteins from the crude worm extracts were separated by electrophoresis and analyzed with specific antibodies.
RNAi, chemical injection and analysis of resulting embryos. dsRNAs were synthesized using Bluescript-based DNA templates and T3 and T7 polymerases (RiboMAX kit, Promega). Lys-CoA and H3-CoA-20 were injected into the gonads of young adults, resulting in a final concentration of ∼10 µM. After injection of either the dsRNA or the chemicals, animals were allowed to recover overnight and eggs laid were analyzed for phenotype and/or the presence of GFP signal by fluorescence microscopy. Immunostaining and Nomarski analysis were carried out as described (Shi and Mello, 1998).
Generation of a point mutation in the HAT domain and in vitro HAT assay. Using the Stratagene Site-Directed Mutagenesis Kit, the conserved F residue at position 1331 in the HAT domain (Shi and Mello, 1998) was changed to alanine. To generate GST–HAT fusion proteins, a 2.4 kb XhoI fragment of cbp-1 (aa 803–1620) containing either the wild-type or the mutant HAT domain was cloned into a GST vector. GST fusion proteins were purified and used for histone acetylation assays as described (Gu and Roeder, 1997).
Genetic rescue. A 13.5 kb cbp-1 genomic fragment carrying either the wild-type or the mutant cbp-1 gene (HAT point mutation) was used for genetic rescue as described (Mello et al., 1991). This genomic fragment includes the entire cbp-1 gene plus a 4.35 kb promoter and a 1.3 kb 3′ sequence beyond the stop codon. Rescue was scored by the presence of fertile, dumpy animals (cbp-1; dpy-18 homozygous). Worms transformed with the wild-type cbp-1 gene exhibited rescue in 10 out of 20 lines assayed. In most rescued lines, animals reached adulthood but produced dead embryos that resemble the cbp-1 maternal effect lethal phenotype (Shi and Mello, 1998). Failure to rescue the maternal function of CBP-1 was expected and was most likely due to transgene silencing in the germline, which is common in C. elegans (Kelly and Fire, 1998).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Fred Winston, Keith Blackwell, Azad Bonni and Grace Gill for critical reading of the manuscript, Phil Cole, Jim McGhee, Michael Krause, Gary Ruvkun, Yuji Kohara and Alan Coulson for reagents, and Bob Barstead and Gary Moulder for their didactic role in the generation and screening of chemically mutagenized C. elegans libraries. This work was supported by a grant from the NIH (GM58012) to Y.S.
REFERENCES
- Bannister A.J. and Kouzarides, T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Barstead R.J. (1999) Reverse genetics. In Hope, I.A. (ed.), C. elegans: A Practical Approach. Oxford University Press, New York, NY, pp. 97–118.
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E. and Allis, C.D. (1996) Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev., 6, 176–184. [DOI] [PubMed] [Google Scholar]
- Epstein H.F. and Shakes, D.C. (eds) (1995) Methods in Cell Biology. Academic Press, San Diego, CA.
- Fire A.S.X., Montgomery, M.K., Kostas, S., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference in C. elegans mediated by double stranded RNA. Nature, 39, 806–810. [DOI] [PubMed] [Google Scholar]
- Fukushige T., Hawkins, M.G. and McGhee, J.D. (1998) The TATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol., 198, 286–302. [PubMed] [Google Scholar]
- Goodman R.H. and Smolik, S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Gu W. and Roeder, R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Hamelin M., Scott, I.M., Way, J.C. and Culotti, J.G. (1992) The mec-7 β-tubulin gene of Caenorhabditis elegans is expressed primarily in the touch receptor neurons. EMBO J., 11, 2885–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Hazenclonk, E., Thijssen, K.L. and Plasterk, R.H.A. (1997) Reverse genetics by chemical mutagenesis in Caenorhabidits elegans. Nature Genet., 17, 119–121. [DOI] [PubMed] [Google Scholar]
- Kelly W.G. and Fire, A. (1998) Chromatin silencing and the maintenance of a functional germline in Caenorhabitis elegans. Development, 125, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L., Manning, E.T. and Kadonaga, J.T. (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol., 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M. et al. (1997) A C. elegans E/Daughterless bHLH protein is present during neuronal but not striated muscle development. Development, 124, 2179–2189. [DOI] [PubMed] [Google Scholar]
- Krause M.W., Fire, A., Harrision, S.W., Priess, J. and Weintraub, H. (1990) CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell, 63, 907–922. [DOI] [PubMed] [Google Scholar]
- Kundu T.K., Palhan, V.B., Wang, Z., An, W., Cole, P.A. and Roeder, R.G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell, 6, 551–561. [DOI] [PubMed] [Google Scholar]
- Lau O.D. et al. (2000) HATs off: selective synthetic inhibitors of the histone acetyltransferase p300 and PCAF. Mol. Cell, 5, 589–595. [DOI] [PubMed] [Google Scholar]
- Manning E.T., Ikehara, T., Ito, T., Kadonaga, J.T. and Kraus, W.L. (2001) p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol., 21, 3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M., Bannister, A.J., Martin, K., Haus-Seuffert, P., Meisterernst, M. and Kouzarides, T. (1998) The acetyltransferase activity of CBP stimulates transcription. EMBO J., 10, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer, J.M., Stinchcomb, D. and Ambros, V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V., Schiltz, R.L., Russanova, V., Howard, B.H. and Nakatani, Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Oike Y. et al. (1999) Truncated CBP protein leads to classical Rubinstein–Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet., 8, 387–396. [DOI] [PubMed] [Google Scholar]
- Parekh B.S. and Maniatis, T. (1999) Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Riddle D.L., Blumenthal, T., Meyer, B.J. and Priess, J.R. (eds) (1997) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Shi Y. and Mello, C. (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev., 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-J., Ogryzko, V.V., Nishikawa, J.-I., Howard, B.H. and Nakatani, Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]