Abstract
The aggregation of the two yeast proteins Sup35p and Ure2p is widely accepted as a model for explaining the prion propagation of the phenotypes [PSI+] and [URE3], respectively. Here, we demonstrate that the propagation of [URE3] cannot simply be the consequence of generating large aggregates of Ure2p, because such aggregation can be found in some conditions that are not related to the prion state of Ure2p. A comparison of [PSI+] and [URE3] aggregation demonstrates differences between these two prion mechanisms. Our findings lead us to propose a new unifying model for yeast prion propagation.
INTRODUCTION
The yeast proteins Sup35p and Ure2p, which have been proposed as being responsible for the phenotypes [PSI+] and [URE3] (Wickner, 1994), are highly tractable models for understanding the mechanism underlying prion propagation. It has been clearly demonstrated by a number of groups that the [PSI+] phenotype is associated with the aggregation and consequent non-functionality of Sup35p. The salient observations are as follows.
(i) Upon transition to its prion form, Sup35p is significantly depleted from a 100 000 g supernatant after centrifugation of a yeast extract. However, Sup35p is largely soluble under the same experimental conditions when the extract is made from a [psi–] strain (Patino et al., 1996; Paushkin et al., 1996, 1997). The prion state, as defined by the [PSI+] phenotype, thus correlates with a dramatic change in the solubility of the protein.
(ii) A fusion between the SUP35 and GFP genes demonstrate that a chimeric Sup35p–GFP protein aggregates in the cytoplasm of [PSI+] yeast strains but not in otherwise isogenic [psi–] strains (Patino et al., 1996). Furthermore, this aggregation is, as for the [PSI+] phenotype, dependent upon the activity of the molecular chaperone Hsp104 (Chernoff et al., 1995).
(iii) Sup35p purified following heterologous expression in Escherichia coli can self-assemble to form amyloid fibres (Glover et al., 1997; King et al., 1997).
(iv) Crude cell-free extracts made from [PSI+] yeast strains induce very efficient Sup35p polymerization in vitro (Glover et al., 1997; Paushkin et al., 1997).
(v) A mutation in the SUP35 gene, originally isolated and described as the PNM2 allele (Doel et al., 1994), inhibits [PSI+] propagation and encodes a protein that is poorly converted in vitro to the amyloid form (Kochneva-Pervukhova et al., 1998).
The molecular behaviour of the [URE3] determining protein Ure2p is less clear. In vitro, purified recombinant Ure2p protein forms a dimer (Perrett et al., 1999), while Thual et al. (1999) showed that the Ure2p protein can also be found as monomeric, dimeric and tetrameric forms. The ability of Ure2p to self-associate was confirmed by two-hybrid studies (Fernandez-Bellot et al., 1998). Interestingly, purified Ure2p behaves in a similar manner to Sup35p in spontaneously forming amyloid fibrils in vitro (Thual et al., 1999). The prion domain of Ure2p is sufficient to promote such polymerization in trans (Taylor et al., 1999), but also in cis when linked to the glutathione S-transferase moiety (Schlumpberger et al., 2000). Ure2p appears to be slightly more resistant to proteinase K in a [URE3] strain (Masison and Wickner, 1995). In vivo, studies with URE2–GFP fusions demonstrate a large, visible aggregation of Ure2p in a [URE3] but not a [ure3°] derivative of the strain YHE64 (Edskes et al., 1999). These data are consistent with a prion mechanism in which the [URE3] determinant would be a highly aggregated form of Ure2p.
In this communication, we present new data on the nature of the [URE3] determinant that lead us to propose an alternative model in which the prion form of Ure2p is found in a soluble state rather than in a highly aggregated state.
RESULTS
Ure2p does not form large aggregates in [URE3] strains
We constructed a yeast strain, EG12, which is suitable for genetic analysis because the URE2–GFP fusion is inserted at the URE2 locus. After plating on a selective medium containing ureidosuccinate (USA), one Usa+ clone was analysed in depth. The Usa+ phenotype could be reversed in the presence of millimolar concentration of guanidine hydrochloride, as expected for a yeast prion determinant. Furthermore, the Usa+ phenotype of the isolated spores was transmitted in a dominant way by mating. Following meiosis, this diploid gave rise to tetrads that showed the characteristic non-Mendelian segregation of the [URE3] phenotype (Aigle and Lacroute, 1975). Thus, the Usa+ phenotype of the EG12 strain showed the expected genetic behaviour for the [URE3] determinant.
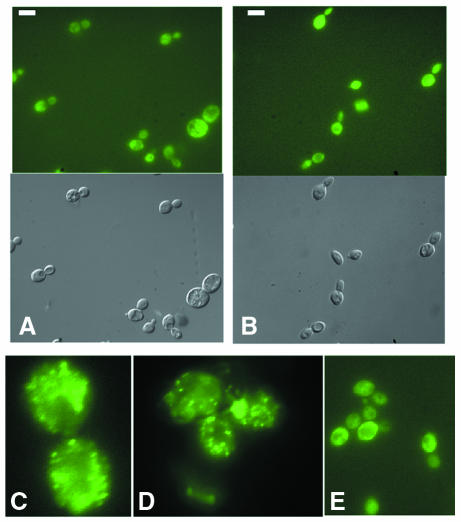
Surprisingly, Ure2p–GFP in the EG12 [URE3] cells was not present in an aggregated form that presented as fluorescent foci but was more evenly distributed in the cytoplasm, as is the case for EG12 [ure3°] (Figure 1). This result was also confirmed when a bona fide [URE3] determinant was introduced by mating EG12 [ure3°] with the strain CC34 that bears the original [URE3] determinant (data not shown). When the Ure2–GFP fusion, cloned in a single-copy plasmid, was expressed in the YHE64 [URE3] strain (Edskes et al., 1999), the specific aggregation pattern previously observed with another plasmid was found (Figure 1). However, in the EG12 [URE3] strain, we still observed an even fluorescence with no evidence of foci (Figure 1). The Ure2p–GFP construct used in this study was capable, therefore, of producing detectable clumps, as are previously described Ure2p–GFP constructs.
Fig. 1. In vivo localization of Ure2p–GFP in [URE3] and wild-type [ure3°] cells. (A) The strain shown (EG12) contains the allele URE2–GFP integrated at the URE2 locus. (B) The [ure3°] strain was generated from the EG12 [URE3] strain by growth in medium containing 5 mM guanidine hydrochloride. Bar, 10 µm. (C and D) YHE64 [URE3] transformed by pH327 (C) or pYe1l URE2–GFP (D). (E) EG12 [URE3] transformed by pYe1l URE2–GFP.
Ure2p can aggregate in wild-type cells
In vitro, Ure2p can polymerize in a concentration-dependent manner (Thual et al., 1999). Therefore, we explored the relationship between the level of expression and the aggregation, in vivo, of the Ure2p protein using the Ure2p–GFP fusion.
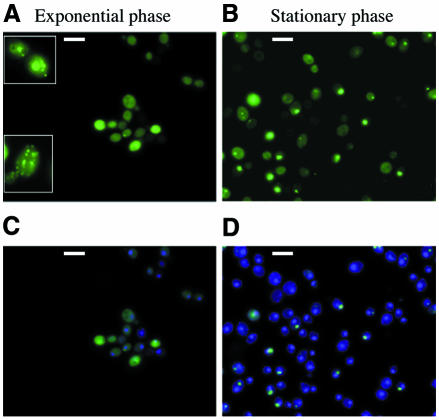
Several clones obtained with the multi-copy plasmid pYeHFc 2L V10 URE2–GFP, and verified as having a wild-type Usa– phenotype with respect to the [URE3] determinant, were analysed. Surprisingly, the majority of the cells viewed had large Ure2p–GFP aggregates in their cytoplasm (Figure 2A). In some cases, two distinct types of aggregate appeared to coexist in the cytoplasm (Figure 2A, insert).
Fig. 2. Cellular localization of Ure2p–GFP following high-level expression of the URE2–GFP allele. The EG12 strain was transformed with pYeFc 2L V10 URE2–GFP, a multi-copy plasmid bearing the URE2–GFP fusion. The phenotype of the transformed strain was verified as a wild-type (not [URE3]). The cells were observed either during exponential growth (A; insert: some cells present small or big aggregates) or in stationary phase (B). (C and D) Hoechst staining of the same cells. Bar, 10 µm.
The degree of aggregation of the Ure2p–GFP was enhanced in stationary phase cells, where the majority of cells contain visible aggregates (Figure 2B); in the exponential-phase cells, only ∼10% of the cells contained aggregates (Figure 2A). In stationary phase cells, no aggregates could be observed in the non-transformed [URE3] EG12 strain (data not shown).
The Ure2p prion-forming domain enhances the aggregation
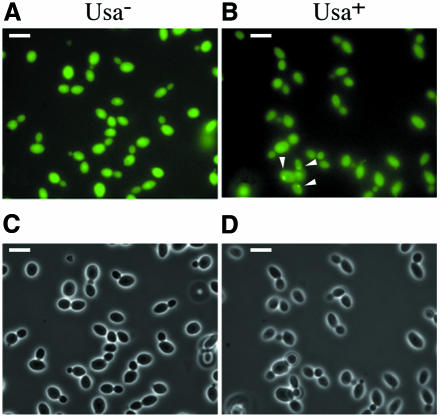
When overexpressed, the N-terminal domain of Ure2p strongly increases the rate of [URE3] formation (Masison and Wickner, 1995). When overexpressed in the EG12 strain, the N-terminal domain of Ure2p increased the rate of [URE3] appearance 1000-fold. The frequency of Usa+ clones found under these conditions was ∼10–3. However, the transformed cells do not show any differences in the pattern of GFP fluorescence (Figure 3A). Isolated [URE3] clones were examined for evidence of protein aggregation. Interestingly, the typical pattern of GFP aggregation was only occasionally observed in these cells. However, the majority of cells that formed a given clonal population did not contain any aggregates (Figure 3B). These results indicate that, in [URE3] colonies, Ure2p is more prone to aggregate but remains evenly distributed in more than 99% of the cells. This finding further demonstrates that the aggregation of Ure2p observed with the GFP assay cannot be responsible for the [URE3] phenotype per se.
Fig. 3. Trans effect of the prion-forming domain on Ure2p–GFP aggregation. The strain EG12 was transformed by a plasmid that overexpresses the prion-forming domain of Ure2p. (A) The transformed cells. (B) Clones of the EG12 strain selected for the Usa+ phenotype in which the characteristic aggregation pattern (indicated by arrows) is found in less than 1% of the cells. (C and D) Nomarski of the same cells. Bar, 10 µm.
The presence of the prion isoform does not affect the Ure2p solubility
We next examined the level of Ure2p aggregation in total cell-free lysates using a differential centrifugation assay. As a control, we also tested the solubility of Sup35p, the prion determinant of the [PSI+] phenotype.
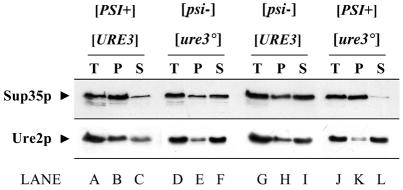
To allow for direct comparability between the aggregation behaviour of the two prion proteins, four isogenic strains were constructed that differed only by the presence or absence of the [PSI+] and/or [URE3] determinants. As previously described (Patino et al., 1996), Sup35p was largely found in the 100 000 g supernatant when the crude extract was prepared from a [psi–] strain (Figure 4, lane F). The presence or the absence of [URE3] in these strains did not influence the distribution of Sup35p in the different fractions (Figure 4, cf. lanes F and I).
Fig. 4. Physical state of Ure2p and Sup35p analysed by differential centrifugation. Total crude extract of the four isogenic strains CW7a ([PSI+] [URE3]), CW10a ([psi–] [ure3°]), CW8a ([psi–] [URE3]) and CW9a ([PSI+] [ure3°]) were fractionated by centrifugation at 100 000 g. An identical volume of total extract (T), precipitated (P) and soluble (S) fractions was loaded onto a 10% SDS–PAGE. After transfer onto nitrocellulose membranes, the proteins were detected using affinity-purified Sup35p (upper panel) and Ure2p (lower panel) antibodies.
In the strain lacking both the [URE3] and [PSI+] determinants, Ure2p was found in the supernatant fraction, and this partition was not changed by the presence of [URE3] (Figure 4, compare lanes E and F with H and I). Thus, the [URE3] determinant does not seem to be the consequence of the aggregation of soluble Ure2p into a ‘pelletable’ 100 000 g fraction, i.e. a high-molecular-weight aggregate. Interestingly, the presence of [PSI+] in a [URE3] strain leads to a partial aggregation of Ure2p (Figure 4, compare lanes B and C with H and I).
DISCUSSION
Previous cellular studies using Ure2p–GFP fusions have suggested that the [URE3] determinant is a highly aggregated form of Ure2p. Using a fusion of full-length Ure2p with GFP, we selected a bona fide [URE3] mutant of the EG12 strain, which fulfils all the genetic criteria for the prion determinant. However, when analysed by fluorescence microscopy, this [URE3] mutant did not exhibit any large aggregates. This was not due to the inability of our GFP construct to monitor the presence of Ure2p–GFP aggregates per se, since these were found in YHE64 [URE3] strain transformed with this construct (Edskes et al., 1999). This lack of visible aggregation was also observed in a recent study that focused on an unusual allele of URE2 that induces [URE3] at high frequency (Fernandez-Bellot et al., 2000).
Aggregation was observed in the EG12 strain when the Ure2p–GFP protein was overexpressed; but, when individual colonies were tested for the [URE3] phenotype, no correlation between the aggregation of Ure2p–GFP and the presence of the [URE3] prion phenotype could be established. Furthermore, expression of the Ure2p prion-forming domain greatly increased the frequency of appearance of foci. However, when individual colonies were tested for the presence of [URE3] and the aggregation, it appeared that the majority of Usa+ cells were devoid of aggregated Ure2p–GFP.
Although Sup35p is subject to a dramatic change in its solubility in [PSI+] strains, Ure2p showed roughly the same degree of solubility whether or not [URE3] was present in a strain devoid of [PSI+]. This finding does not rule out the possibility that Ure2p adopts subtle changes in its state of association in [URE3] cells, but our data demonstrate that [URE3] may not be the consequence of a polymerization mechanism that leads to the formation of large, insoluble forms of the protein that act to seed prion propagation.
Speculation
Based on our findings, we propose a new scheme for the replication of the [URE3] determinant (Figure 5). This model is compatible with the nucleated conformational conversion model proposed by Serio et al. (2000) and is based upon the assumption that both the macro-aggregation of Ure2p and [URE3] propagation are, in some way, related but do not necessarily correspond to the same state of Ure2p. The conditions under which Ure2p aggregation may be observed are also favourable for the formation of [URE3], as is the case when overexpressing Ure2p or its N-terminal prion-forming domain. We propose that Ure2p exists in equilibrium with a Ure2p* isoform. This Ure2p* isoform would self-aggregate more easily than Ure2p itself, giving rise to either amorphous aggregates or to more organized structures such as the amyloid fibres that have been observed in vitro (Taylor et al., 1999; Thual et al., 1999). In our model, the Ure2p* species could also give rise to another Ure2p species (Ure2p[URE3]) or, alternatively, it could be Ure2p[URE3] itself. This isoform, which does not sediment at 100 000 g centrifugation, would correspond to the form of the prion protein that has the characteristic self-propagating properties and could comprise a smaller non-pelletable aggregate. When Ure2p is overexpressed, the overall concentration of Ure2p increases, leading to a higher level of proteins taking up the Ure2p* state. As a consequence, the protein would either aggregate or remain infectious and essentially soluble. Once this state is reached, the cells become [URE3]. It is important to note that, in this case, the mechanism of [URE3] formation is not due to the formation of a large structure that can be observed with the GFP-based assay. However, these structures may be observed more often than in the wild-type cells, since they would be in equilibrium with Ure2p*.
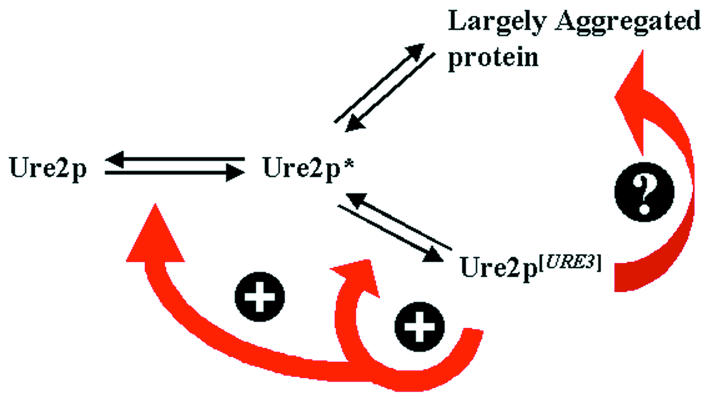
Fig. 5. Model for [URE3] generation. The cellular, functional Ure2p exists in equilibrium with an altered intermediate Ure2p*. This latter species would give rise either to a new stable non-functional Ure2p[URE3] prion protein or, alternatively, to an aggregated form of Ure2p. These two states distinguish themselves by their biophysical characteristics, since Ure2p[URE3] is found in the soluble fraction after centrifugation. However, the soluble Ure2p[URE3] cannot be functional, since the phenotype corresponds to the inactivation of Ure2p. The ‘dead-end product’ observed using Ure2p–GFP would be formed by the macro-aggregation of this species and only be observed in some yeast strains. The detectable macro-aggregates would be the consequence, rather than the cause, of the prion phenotype.
The balance between the soluble and the aggregated forms of the Ure2p protein in a given strain is probably influenced by the genetic background of that strain. A pathway could exist (see Figure 5) that would give rise to large Ure2p aggregates with previously formed Ure2p[URE3]. Such a ‘dead-end product‘ would be formed less efficiently in the genetic background used in our study (see also Fernandez-Bellot et al., 2000) compared to the strains described by others (Edskes et al., 1999).
METHODS
Strains. The following strains of Saccharomyces cerevisiae were used in this study.
CC30: MATα, trp1-1, ade2-1, leu2-3,112, his3-11,15, ΔURA2::HIS3
CC34a: MATα, trp1-1, ade2-1, leu2-3,112, his3-11,15, ΔURA2::HIS3, [URE3]
CC34α: MATα, trp1-1, ade2-1, leu2-3,112, his3-11,15, ΔURA2::HIS3, [URE3]
EG12 MATα; Δtrp1, ade2-1, leu2-3, 112, his3-11,15 Δura2::HIS3, URE2::GFP-TRP1
BMA64–1b MATa; Δtrp1, ade2-1, leu2-3, 112, his3-11,15
W9: MATa, SUQ5, ade2-1; his5-2; lys1-1; ΔURA2 [PSI+]
CW7a MATa, SUQ5, ade2-1, trp1-1, leu2-3,112, his5-2; lys1-1; Δ URA2::HIS3 [URE3] [PSI+]
CW8a MATa, SUQ5, ade2-1, trp1-1, leu2-3,112, his5-2; lys1-1; Δ URA2::HIS3 [URE3]
CW9a MATa, SUQ5, ade2-1, trp1-1, leu2-3,112, his5-2; lys1-1; Δ URA2::HIS3 [PSI+]
CW10a MATa, SUQ5, ade2-1, trp1-1, leu2-3,112, his5-2; lys1-1; Δ URA2::HIS3
Construction of the EG12 strain. A DNA fragment containing the GFP open reading frame and the S. cerevisiae TRP1 gene was amplified by polymerase chain reaction (PCR) with primers containing homologous parts of the sequence of the URE2 gene: primer 68 (5′aaccttcttttcctccttcttctttctttcttgtttttaaagcagcctCTTAAATAAATACTACTC) and primer 117 (5′GAAGACCCGCGGTCATCAAGGCATTGCGTGGTGAATACCCATACGACgtccc), using the pJM plasmid as the backbone vector (J.M. Rouillard, unpublished data). A MATa transformant was selected and crossed with the CC30 (MATα) strain. After sporulation, two spores of different mating type were selected among the spores carrying the TRP1 gene, giving the EG12 strain.
Construction of isogenic strains bearing different combinations of the [URE3] and [PSI+] prion determinants. The strain CW7 carrying both the [PSI+] and [URE3] determinants was constructed by crossing the strains CC34α and W9. One spore showing both the [URE3] and [PSI+] prion phenotypes was then selected. This strain, designated CW7, was subjected to growth on 2.5 or 5 mM guanidine hydrochloride to produce three new isogenic strains that differed only by the presence of the [PSI+] or [URE3] determinants.
Construction of plasmids. The multi-copy plasmid pYeHFc 2L V10 URE2 was derived from the plasmid pYeHFc 2L URE2 originally described by Komar et al. (1998) as follows. A BamHI–BglII fragment containing the PGK1 promoter was obtained from the pYeV10 vector (Cullin and Pompon, 1988) and inserted into the unique BamHI restriction site. The prion domain (1–65) of URE2 was PCR amplified and cloned in pYeHFc 2L.
The BamHI–AatII cassette, containing a GFP fragment, obtained from pYeGC2U (J.M. Rouillard, unpublished data) was then inserted in pYeHFC 2L V10 URE2. The amino-acid sequence of the Ure2p–GFP fusion protein expressed by these vectors is given below, with the Ure2 protein sequence indicated in bold and the GFP sequence underlined.
MMNNNGNQVSNLSNALRQVNIGNRNSNTTTDQSNINFEFSTGVNNNNNNNSSSNNNNVQNNNSGRNGSQNNDNENNIKNTLEQHRQQQQAFSDMSHVEYSRITKFFQEQPLEGYTLFSHRSAPNGFKVAIVLSELGFHYNTIFLDFNLGEHRAPEFVSVNPNARVPALIDHGMDNLSIWESGAILLHLVNKYYKETGNPLLWSDDLADQSQINAWLFFQTSGHAPMIGQALHFRYFHSQKIASAVERYTDEVRRVYGVVEMALAERREALVMELDTENAAAYSAGTTPMSQSRFFDYPVWLVGDKLTIADLAFVPWNNVVDRIGINIKIEFPEVYKWTKHMMRRPAVIKALRGGGRYPYDVPDYAMSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFGYGVQCFARYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYKAA
The single-copy plasmid pYeHFc 1L URE2 was constructed by gap repair, replacing the 2µ-LEU2 DNA fragment of pYeHFc 2L V10 URE2 with an ARS CEN-LEU2 insert.
Microscopy techniques. Cells were usually cultured in 10 ml of an appropriate liquid growth medium. Three millilitres of the medium were removed and 3 µl of a solution of 4 µg/µl bis-benzimide (Hoechst, Sigma) were added. After incubation for 15 min at 30°C, cells were pelleted and washed twice in sterile water. The cell pellets were then resuspended in a 50 µl Dabco solution [218 mM diazabicyclo 2-2-2 octane (Sigma), 25% (v/v) PBS buffer, 75% (v/v) glycerol]. Cells were photographed using a DMRB microscope (Leica, Germany) with a PL APO 63 objective.
Protein extraction and fractionation. The four isogenic strains CW7a, CW8a, CW9a and CW10a, carrying various combinations of the [PSI+] and [URE3] determinants, were grown to a cell density of ∼5 × 106 cells/ml in YPD. Cells were then resuspended (to a density of 3.3 × 108 cells/ml) in 10 mM phosphate buffer pH 7.5, 250 mM NaCl, 2 mM phenylmethylsulfonyl and one tablet of protease inhibitor cocktail (Boehringer) and disrupted with glass beads at 4°C. The resulting total crude extract was fractionated by centrifugation for 15 min at 100 000 g at 4°C using a TL100 centrifuge (Beckman). The resulting supernatant was recovered, and an equal volume of buffer was added to the pellet. After incubation for 10 min at 95–100°C in 2× standard SDS–PAGE loading buffer, identical volumes (15 µl) of each fraction were loaded onto a 10% SDS–polyacrylamide gel. For western blot analysis, the proteins were transferred onto a nitrocellulose membrane and detected with affinity purified polyclonal antibodies raised against full-length, recombinant Sup35p and Ure2p, respectively.
Acknowledgments
ACKNOWLEDGEMENTS
Many thanks to Dr C. Mann (CEA, Saclay) for the microscopy facilities. We are grateful to R.B. Wickner and H. Edskes for generously providing strains and plasmids. The work was supported by a grant from Action Concertée Coordonnée Science du Vivant no. 10 (9510001) to C.C., an EC Contract (no. BI104-98-6045, Maintenance and transmission of yeast prions: a model system) to C.C. and M.F.T. and grants from the BBSRC and The Wellcome Trust to M.F.T.
REFERENCES
- Aigle M. and Lacroute, F. (1975) Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet., 136, 327–335. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist, S.L., Ono, B., Inge-Vechtomov, S.G. and Liebman, S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Cullin C. and Pompon, D. (1988) Synthesis of functional mouse cytochromes P-450 P1 and chimeric P-450 P3-1 in the yeast Saccharomyces cerevisiae. Gene, 65, 203–217. [DOI] [PubMed] [Google Scholar]
- Doel S.M., McCready, S.J., Nierras, C.R. and Cox, B.S. (1994) The dominant PNM2-mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics, 137, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H.K., Gray, V.T. and Wickner, R.B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA, 96, 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet, E., Baudin-Baillieu, A., Gaumer, S., Komar, A.A. and Cullin, C. (1998) Characterization of two interacting domains of Ure2p, a prion-like protein of yeast. Biochem. J., 338, 403–407. [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Bellot E., Guillemet, E. and Cullin, C. (2000) The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J., 19, 3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Kowal, A.S., Schirmer, E.C., Patino, M.M., Liu, J.J. and Lindquist, S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- King C.Y., Tittmann, P., Gross, H., Gebert, R., Aebi, M. and Wuthrich, K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA, 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Paushkin, S.V., Kushnirov, V.V., Cox, B.S., Tuite, M.F. and Ter-Avanesyan, M.D. (1998) Mechanism of inhibition of ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J., 17, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A., Guillemet, E., Reiss, C. and Cullin, C. (1998) Enhanced expression of the yeast Ure2 protein in Escherichia coli: the effect of synonymous codon substitutions at a selected place in the gene. Biol. Chem., 379, 1295–1300. [PubMed] [Google Scholar]
- Masison D. and Wickner, R.B. (1995) Prion-inducing domain of yeast ure2p and protease resistance of ure2p in prion-containing cells. Science, 270, 93–95. [DOI] [PubMed] [Google Scholar]
- Patino M.M., Liu, J.J., Glover, J.R. and Lindquist, S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov, V.V., Smirnov, V.N. and Ter-Avanesyan, M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov, V.V., Smirnov, V.N. and Ter, A.M. (1997) In vitro propagation of the prion-like state of yeast Sup35 protein. Science, 277, 381–383. [DOI] [PubMed] [Google Scholar]
- Perrett S., Freeman, S.J., Butler, P.J. and Fersht, A.R. (1999) Equilibrium folding properties of the yeast prion protein determinant Ure2. J. Mol. Biol., 290, 331–345. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M., Wille, H., Baldwin, M.A., Butler, D.A., Herskowitz, I. and Prusiner, S.B. (2000) The prion domain of yeast Ure2p induces autocatalytic formation of amyloid fibers by a recombinant fusion protein. Protein Sci., 9, 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio T.R., Cashikar, A.G., Kowal, A.S., Sawicki, G.J., Moslehi, J.J., Serpell, L., Arnsdorf, M.F. and Lindquist, S.L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science, 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Taylor K.L., Cheng, N., Williams, R.W., Steven, A.C. and Wickner, R.B. (1999) Prion domain initiation of amyloid formation in vitro from native Ure2p. Science, 283, 1339–1343. [DOI] [PubMed] [Google Scholar]
- Thual C., Komar, A.A., Bousset, L., Fernandez-Bellot, E., Cullin, C. and Melki, R. (1999) Structural characterization of Saccharomyces cerevisiae prion-like protein ure2. J. Biol. Chem., 274, 13666–13674. [DOI] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae.Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]