Abstract
One-carbon metabolism plays a crucial role in tumorigenesis as it supplies the one-carbon units necessary for nucleotide synthesis, epigenetic regulation, and redox metabolism, ensuring the rapid proliferation of cancer cells. However, their roles in prostate cancer progression remain poorly understood. In this study, we investigated the association between genetic variants in the one-carbon metabolism pathway and clinical outcomes in patients receiving androgen deprivation therapy for prostate cancer. The associations of 130 single-nucleotide polymorphisms located within 14 genes involved in the one-carbon metabolism pathway with cancer-specific survival (CSS), overall survival, and progression-free survival were assessed using Cox regression in 630 patients with prostate cancer. Subsequently, functional studies were performed using prostate cancer cell lines. After adjusting for covariates and multiple testing, MTHFD1L rs2073190 was found to be significantly associated with CSS (P = 0.000184). Further pooled analysis of multiple datasets demonstrated that MTHFD1L was upregulated in prostate cancer and increased MTHFD1L expression was positively correlated with tumor aggressiveness and poor patient prognosis. Functionally, MTHFD1L knockdown suppressed prostate cancer cell proliferation and colony formation. RNA sequencing and pathway analysis revealed that differentially expressed genes were predominantly enriched in the cell cycle pathway. In conclusion, genetic variants in MTHFD1L of one-carbon metabolism may serve as promising predictors, and our findings offer valuable insights into the underlying genetic mechanisms of prostate cancer progression.
Keywords: One-carbon metabolism, MTHFD1L, prostate cancer, survival, RNA sequencing, cell cycle
Introduction
Prostate cancer is an important health concern and the leading cause of cancer-related deaths among men. In 2020, the number of newly diagnosed cases of prostate cancer surpassed 1.4 million, resulting in approximately 375,304 deaths associated with the disease [1]. Advances in our understanding of prostate cancer and the role of androgens have led to the development of androgen deprivation therapy (ADT) [2]. ADT is the primary treatment of choice for locally advanced and metastatic prostate cancer. Despite the benefits of ADT, the development of resistance to ADT over time, known as castration-resistant prostate cancer, remains a major challenge. Under these conditions, cancer cells adapt and continue to grow despite low androgen levels [3]. Clinical predictors play crucial roles in determining the effectiveness of ADT. Various clinical factors, such as prostate-specific antigen (PSA) levels, Gleason score, tumor stage, and patient age, have been studied as potential predictors of ADT response [4]. To further improve treatment efficacy, it is necessary to incorporate genetic profiling to personalize ADT strategies for individual patients.
Cancer cells undergo metabolic reprogramming to sustain uncontrolled cell proliferation by providing energy and building blocks necessary for cellular maintenance and biosynthesis [5]. One-carbon metabolism is a well-known altered metabolic pathway in cancer. This intricate network of biochemical reactions plays a critical role during one-carbon units transfer in the form of methyl, methenyl, methylene, and formyl groups, which are vital for various cellular processes, such as molecular biosynthesis (e.g., proteins, nucleotides, phospholipids, polyamines, and creatine), regulation of nucleotide pools for genomic maintenance, epigenetic control of gene expression through DNA, RNA, and histone methylation, and redox defense mechanisms [6]. Several enzymes involved in one-carbon metabolism have been implicated in the development of cancer. Notably, methylenetetrahydrofolate dehydrogenase, cyclohydrolase, and formyltetrahydrofolate synthetase 1 (MTHFD1), a multifunctional enzyme responsible for the conversion of different forms of one-carbon-substituted tetrahydrofolate, has been associated with tumor aggressiveness and reduced patient survival in various cancers, including colorectal cancer, non-small cell lung cancer, and hepatocellular carcinoma [7-9]. Furthermore, genetic variants within one-carbon metabolism genes can functionally impact the pathway and disrupt the metabolic balance. For instance, the single-nucleotide polymorphism (SNP) G1958A in MTHFD1, which encodes the MTHFD1 R653Q variant, enhances E3 ligase binding and decreases protein stability through ubiquitination-mediated protein degradation. Expression of the MTHFD1 G1958A mutant suppressed tumorigenesis in a murine model of liver cancer [10].
However, our current understanding regarding the effect of genetic variants in the one-carbon metabolism pathway on the efficacy of ADT is limited. Hence, in this study, we investigated the relationship between 130 SNPs in 14 one-carbon metabolism pathway genes and cancer-specific survival (CSS), overall survival (OS), and progression-free survival (PFS) in a cohort of 630 patients with advanced prostate cancer who received ADT. Additionally, we assessed the biological functions of the target genes to gain insights into the potential underlying biological mechanisms that influence the survival outcomes of patients with prostate cancer.
Materials and methods
Evaluation of patient response
The present study included 630 patients diagnosed with prostate cancer who underwent ADT at three medical centers in Taiwan: the National Taiwan University Hospital, Kaohsiung Medical University Hospital, and Kaohsiung Veterans General Hospital. Detailed information regarding this study can be found in previous publications [11,12]. Approval for the study was granted by the Institutional Review Board of Kaohsiung Medical University Hospital (KMU-HIRB-2013132) in accordance with the Good Clinical Practice guidelines. Written informed consent was obtained from all the participants. Clinically relevant data pertaining to patients’ clinicopathological characteristics were extracted from their medical records. CSS was defined as the duration from the initiation of ADT to the last follow-up or death specifically caused by cancer. OS was defined as the duration from the initiation of ADT to death from any disease. PFS was defined as the duration from the initiation of ADT to either disease progression or death from cancer-related causes. Over a median follow-up duration of 165.8 months, 518 (82.6%) patients experienced disease progression, 414 (65.7%) died, and 314 (49.8%) died of cancer [13]. PSA level, clinical stage, Gleason score at diagnosis, PSA nadir, and time to PSA nadir were all found to be significant predictors of CSS, OS, and PFS during ADT. However, age at diagnosis was only associated with PFS and OS.
Selection and genotyping of SNPs
Haplotype-tagged SNPs spanning 14 major genes associated with the one-carbon metabolism pathway were selected. These genes included betaine-homocysteine S-methyltransferase (BHMT), choline dehydrogenase (CHDH), dihydrofolate reductase (DHFR), methylenetetrahydrofolate dehydrogenase, cyclohydrolase, formyltetrahydrofolate synthetases (MTHFD1, MTHFD1L, and MTHFD2), methylenetetrahydrofolate reductase (MTHFR), 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR), 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR), phosphatidylethanolamine N-methyltransferase (PEMT), replication factor C subunit 1 (RFC1), serine hydroxymethyltransferases (SHMT1 and SHMT2), and thymidylate synthetase (TYMS). The selection process employed the Haploview 4.2 tagger algorithm, utilizing data from the 1000 Genomes Project for Han Chinese individuals in Beijing, China and Southern Han Chinese [14]. Genomic DNA was extracted from peripheral lymphocytes and genotyping was performed using the Affymetrix Axiom Genotyping Array system (Thermo Fisher Scientific, Waltham, MA, USA) at the National Centre for Genome Medicine in Taiwan, following previously described procedures [15]. SNPs that did not meet the following criteria were excluded from further analyses: genotyping call rates less than 0.9, minor allele frequency less than 0.05, and deviation from the Hardy-Weinberg equilibrium less than 0.001. Consequently, 130 SNPs were retained for subsequent analysis.
Bioinformatic analysis
The functional relevance of MTHFD1L rs2073190 was assessed using the HaploReg [16]. To investigate the relationship between MTHFD1L expression and various factors, such as tissue type, Gleason score, stage, and survival outcomes in prostate cancer, all publicly available prostate gene expression datasets from Gene Expression database of Normal and Tumor tissues 2 [17], Oncopression [18], Prostate Cancer Transcriptome Atlas [19], and The Cancer Genome Atlas Prostate Adenocarcinoma (TCGA PRAD) were used.
Western blot analysis
PC-3 and C4-2 cell lines were treated with specific MTHFD1L short interfering RNAs (siRNAs; Sigma-Aldrich, St. Louis, MO, USA) using the T-Pro NTR II transfection reagent (T-Pro Biotechnology, JT97-N002M, New Taipei City, Taiwan) for three days. Following treatment, cell lysates were collected and boiled in 6× Laemmli sample buffer at 95°C for 10 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Merck Millipore, Darmstadt, Germany). After blocking with 5% milk in Tris-buffered saline with Tween 20 (TBST) buffer, the membranes were incubated with primary antibodies against MTHFD1L (1:1000, 14999S, Cell Signaling Technology, Danvers, Massachusetts, USA) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:10000, Genetex, Inc., Irvine, CA, USA) overnight at 4°C. Subsequently, the membranes were washed thrice with TBST and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody at room temperature for 1 h. The membranes were then incubated with HRP substrate (WesternBright ECL HRP Substrate, Advansta, San Jose, CA, USA), and the resulting signal was detected using an AmershamTM Imager 600 (GE Healthcare, Chicago, IL, USA).
RNA extraction and RNA sequencing
Total RNA was extracted from control and siMTHFD1L B-treated PC-3 and C4-2 cells using the Qiagen RNeasy Mini Kit (Qiagen #74104, Hilden, Germany), following the manufacturer’s guidelines. The quality and quantity of purified RNA were assessed using a Bioanalyzer 2100 with an RNA 6000 LabChip kit (Agilent Technology, Santa Clara, CA, USA). All RNA sample preparations were performed in accordance with the official Illumina protocol. The SureSelect XT HS2 mRNA Library Preparation Kit (Agilent Technology, Santa Clara, CA, USA) was used for library construction, and size selection was achieved using AMPure XP beads (Beckman Coulter, Brea, CA, USA). Subsequent sequencing was conducted using Illumina sequencing-by-synthesis technology (Illumina, San Diego, CA, USA). Sequencing data (FASTQ reads) were generated by Welgene Biotech’s pipeline using Illumina’s base-calling program bcl2fastq v2.20. The sequencing depth exceeded 30 million reads for each sample, and alignment to GRCh37 was performed with default parameters using TopHat 2.0.13. The assembly was conducted using Cufflink 2.2.1, with Ensembl v75 annotations. Transcript abundance was quantified in fragments per kilobase of exons per million mapped fragments. The sequencing results were available in the Gene Expression Omnibus database (accession number GSE249098).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Complementary DNA was synthesized using the Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The qRT-PCR was conducted using a StepOnePlus™ Real-Time PCR System with SYBR™ Green PCR Master Mix (Applied Biosystems, Beverly, MA, USA). Data analysis was performed following the comparative 2-ΔΔCt method and normalized to GAPDH expression.
Cell cycle analysis
PC-3 and C4-2 cells were seeded into 10-cm dishes and treated with individual MTHFD1L siRNAs (siMTHFD1L A-D) for 3 days. The cells were then collected, adjusted to a fixed number of cells, fixed with 5 ml ice-cold 70% ethanol, and placed at -20°C overnight. For cell cycle analysis, the cells were washed with cold phosphate-buffered saline twice and stained with propidium iodide (PI) solution (containing 20 µg/ml PI, 0.1% Triton X-100, and 200 µg/ml RNase A) for 30 min at room temperature. The samples were analyzed by Attune™ NxT Acoustic Focusing Cytometer (Invitrogen, Waltham, MA, USA) and quantified using FlowJo software.
Proliferation and colony formation assay
The day prior to siRNA treatment, PC-3 and C4-2 cells were seeded in 96-well plates (approximately 1,000 cells/well). The proliferation assay was performed on the 2nd and 3rd day after siRNA treatment. Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) solution was applied to each well and incubated at 37°C for 1 h and the signal was detected by measuring absorbance at 450 nm using an Epoch Microplate Spectrophotometer (BioTek Instruments, Winooski, VT, USA). For the colony formation assay, PC-3 and C4-2 cells were grown in 12-well plates (approximately 500 cells/well) and treated with different siRNAs (siMTHFD1L A-D) for 7 days. Then, the colonies were fixed and stained with crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The total number of colonies was counted and analyzed using the ImageJ software [20].
Migration assay
Cell migration activities were evaluated using both wound-healing and transwell assays. In the transwell assay, PC-3 and C4-2 cells underwent MTHFD1L siRNA treatment for 2 days before being transferred to wells covered with a permeable membrane (pore size: 8.0 µm, Corning, NY, USA). Each well, containing 5×104 cells, was cultured in 200 µl of serum-free medium on the upper layer of inserts. Subsequently, 750 µl of Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum was added to the bottom layer of inserts. After 48 h, the migrated cells were fixed with 100% methanol and stained with a 1% crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA). Images were captured under a light microscope, and the number of migrated cells was quantified using ImageJ. For the wound-healing assay, PC-3 or C4-2 cells were treated with MTHFD1L siRNAs for 2 days and then seeded into 12-well plates. Once the cells reached approximately 90% confluence, a scratch was introduced in each well using a 200 µl pipette tip, and images were taken to document the initial statuses. Wound-healing activities were recorded after 24 h for comparison. Changes in the gap area, indicative of wound-healing activities, were quantified using ImageJ.
Statistical analysis
Statistical analyses were conducted using Statistical Product and Service Solutions version 19.0.0 (IBM, Armonk, NY, USA). A two-sided significance level of P < 0.05 was considered statistically significant. To control for the false discovery rate, multiple testing corrections were applied with a threshold of q values below 0.05 [21]. A pooled analysis was performed using Review Manager (Cochrane, London, UK), and a random-effects model was employed to account for potential heterogeneity among the studies.
Results
To assess the potential link between one-carbon metabolism and prostate cancer progression, we analyzed the correlation between 130 SNPs in 14 genes associated with the one-carbon metabolism pathway and three crucial clinical outcomes: PFS, OS, and CSS. Among these SNPs, five SNPs (rs77987602 and rs1650697 in DHFR and rs2073067, rs175866, and rs2073190 in MTHFD1L) were associated with PFS, six SNPs (rs6801605 in CHDH, rs506500 and rs558133 in BHMT, and rs2073190, rs487637, and rs61116487 in MTHFD1L) were associated with OS, and eight SNPs (rs6801605 in CHDH, rs3733781 in MTRR, rs506500, rs558133, and rs585800 in BHMT, and rs175866, rs2073190, and rs487637 in MTHFD1L) were associated with CSS (P < 0.05, Figure 1). However, after implementing multiple testing corrections (q < 0.05), only SNP rs2073190 in MTHFD1L exhibited a significant association with CSS. Specifically, each additional minor allele G of rs2073190 was associated with a 38% increased risk of cancer-specific mortality (hazard ratio [HR] = 1.38, 95% confidence interval [CI] = 1.15-1.64, P = 0.000385, q = 0.033, Table 1). Moreover, even after adjusting for clinical factors in multivariate analysis, MTHFD1L rs2073190 maintained its significance as an independent prognostic factor for CSS (P = 0.000184). Notably, individuals with the risk allele G of MTHFD1L rs2073190 exhibited poorer OS and PFS in both univariate and multivariate analyses (P ≤ 0.009, Table 1).
Figure 1.
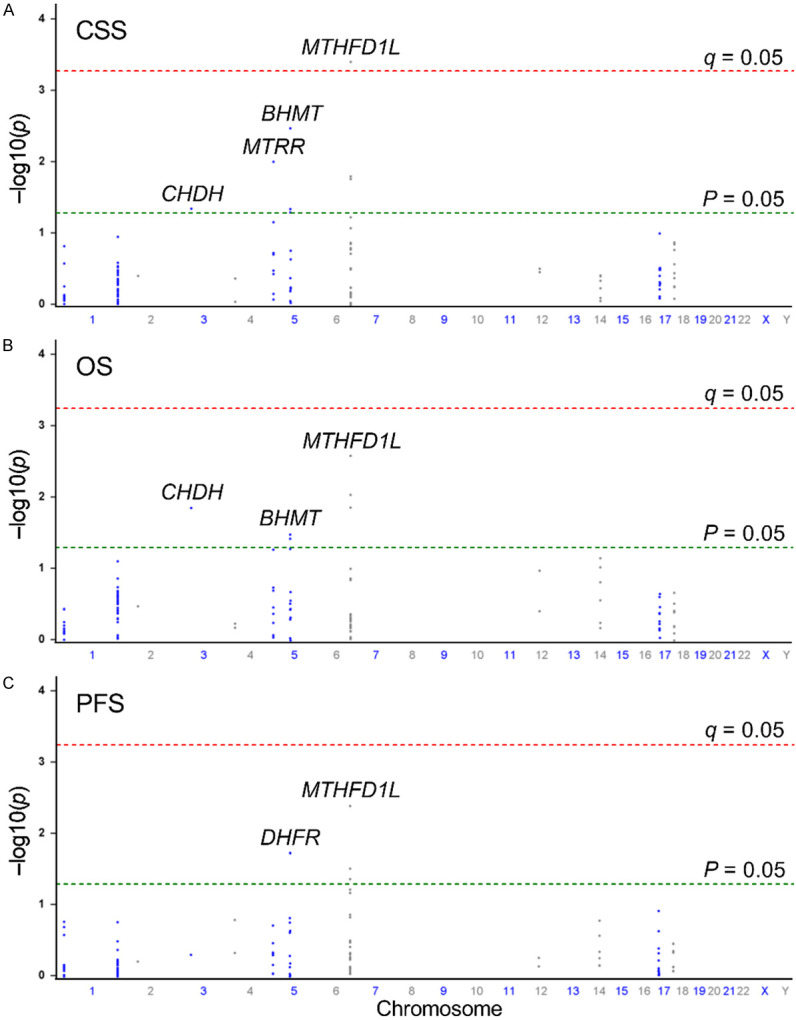
Manhattan plots illustrating the associations (-log10(P) values; Y-axis) between 130 single-nucleotide polymorphisms (SNPs; X-axis by chromosome positions) within 14 genes involved in the one-carbon metabolism pathway and (A) cancer-specific survival, (B) overall survival, and (C) progression-free survival in patients with prostate cancer undergoing androgen-deprivation therapy. Genes with significant associations (P < 0.05) are labeled. Red horizontal line indicates the significance threshold (q = 0.05), while green horizontal line represents the nominal significance threshold (P = 0.05).
Table 1.
The association of MTHFD1L rs2073190 with and cancer-specific, overall, and progression-free survival in prostate cancer patients receiving androgen deprivation therapy
| Genotype | Frequency | CSS | HR (95% CI) | P | q | HR (95% CI)a | P a | ||
|
| |||||||||
| TT/TG/GG | 406/193/30 | 185/105/23 | 1.38 (1.15-1.64) | 0.000385 | 0.033 | 1.44 (1.19-1.75) | 0.000184 | ||
|
| |||||||||
| OS | HR (95% CI) | P | HR (95% CI)a | P a | PFS | HR (95% CI) | P | HR (95% CI)a | P a |
|
| |||||||||
| 256/132/24 | 1.24 (1.06-1.45) | 0.009 | 1.27 (1.07-1.51) | 0.006 | 324/163/30 | 1.23 (1.07-1.42) | 0.004 | 1.29 (1.11-1.50) | 0.001 |
CSS, cancer-specific survival; OS, overall survival; PFS, progression-free survival; ADT, androgen deprivation therapy; HR, hazard ratio; CI, confidence interval.
Adjustment for age, stage, Gleason score at diagnosis, PSA level at ADT initiation, PSA nadir, and time to PSA nadir.
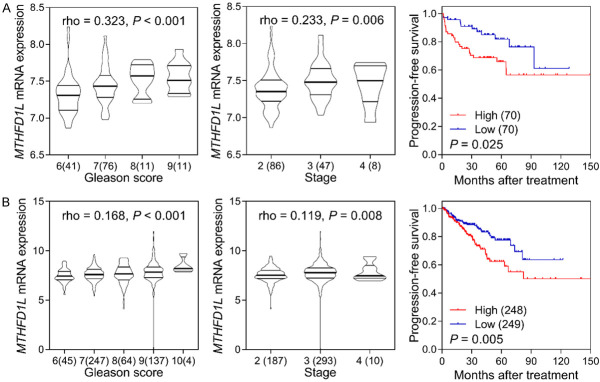
We next investigated the clinical significance of MTHFD1L in prostate cancer. Through a pooled analysis of 13 studies encompassing 1024 prostate cancer and 212 adjacent normal samples, we observed increased MTHFD1L expression in prostate cancer compared with those in normal tissues (standardized mean difference = 0.40, 95% CI = 0.05-0.75, P = 0.02; Figure 2). Furthermore, we found a strong correlation between high MTHFD1L expression and an elevated Gleason score as well as advanced tumor stage, as evidenced by the GSE21032 and TCGA PRAD datasets (P ≤ 0.008, Figure 3). Importantly, patients with higher MTHFD1L expression experienced significantly shorter PFS than those with lower expression levels (P ≤ 0.025, Figure 3).
Figure 2.
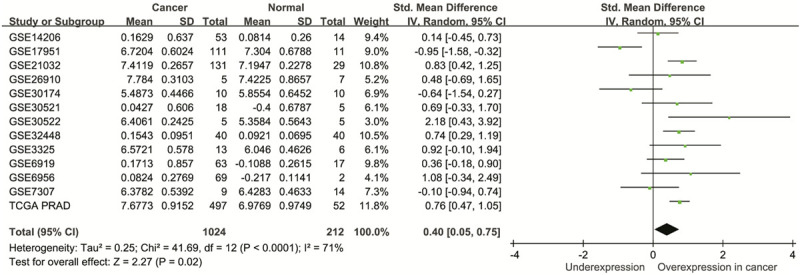
Pooled analysis of 13 studies reveals increased MTHFD1L expression in prostate cancer compared with normal tissues. TCGA PRAD, The Cancer Genome Atlas prostate adenocarcinoma; SD, standard deviation; IV, inverse variance; CI, confidence interval; Std, standardized; df, degrees of freedom.
Figure 3.
Clinical significance of MTHFD1L in prostate cancer. Expression of MTHFD1L was significantly upregulated in higher Gleason score and advanced stage tumor, and high MTHFD1L expression level was associated with poor progression-free survival in both GSE21032 (A) and The Cancer Genome Atlas prostate adenocarcinoma (B) datasets.
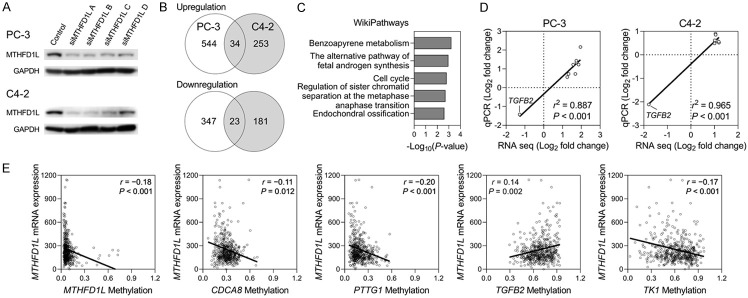
Based on the clinical data analyses, we established a correlation between MTHFD1L and the malignant progression of prostate cancer. To investigate the biological function of MTHFD1L, we employed a siRNA-based approach to reduce its expression in PC-3 and C4-2 human prostate cancer cell lines. Transfection of four gene-specific siRNAs (siMTHFD1L A-D) led to a significant downregulation of MTHFD1L protein expression in both PC-3 and C4-2 cells compared with that in the mock control (Figure 4A). To gain further insight into the underlying mechanisms, we conducted an RNA-seq transcriptome analysis. Venn diagram analysis revealed 57 significantly differentially expressed (SDE) genes that were commonly dysregulated upon MTHFD1L knockdown in both cell lines (Figure 4B). These 57 common SDE genes were annotated based on the WikiPathway database, which identified several pathways strongly associated with the genetic changes induced by MTHFD1L inhibition, including benzo[a]pyrene metabolism, androgen synthesis, cell cycle, sister chromatid separation, and endochondral ossification (Figure 4C). The enrichment findings hold particular relevance as cancer is characterized by uncontrolled cell proliferation resulting from dysregulation of the cell cycle machinery. This observation prompted us to investigate whether MTHFD1L differentially regulates cell cycle-related genes. To validate our results, we performed qRT-PCR analysis of nine cell cycle-related genes identified in the RNA-seq data (AURKA, CCNA2, CDC20, CDCA8, KIF20A, MYBL2, PTTG1, TGFB2, and TK1). The experimental results demonstrated a high level of consistency between the RNA-seq and qRT-PCR results, indicating that AURKA, CCNA2, CDC20, CDCA8, KIF20A, MYBL2, PTTG1, and TK1 were upregulated, while TGFB2 was downregulated in MTHFD1L knockdown PC-3 and C4-2 cells compared to their corresponding control cells (r2 ≥ 0.887, P < 0.001; Figure 4D). Given the pivotal role of one-carbon metabolism in DNA methylation, it is of interest to investigate whether MTHFD1L could impact the expression of the cell cycle-related genes identified in the RNA-seq by modulating DNA methylation. We employed the TCGA database to assess the correlation between MTHFD1L expression and the methylation status of these cell cycle-related genes (Figure 4E). Our analysis revealed a significant correlation between MTHFD1L expression and the methylation levels of specific cell cycle-related genes, including CDCA8, PTTG1, TGFB2, and TK1 (P ≤ 0.012). Notably, MTHFD1L expression displayed a negative correlation with the DNA methylation of MTHFD1L, CDCA8, PTTG1, and TK1 genes, while exhibiting a positive correlation with the DNA methylation of TGFB2. This observed pattern aligns with the gene expression results obtained from RNA-seq and qRT-PCR analyses, indicating that the expression of CDCA8, PTTG1, and TK1 is up-regulated, whereas TGFB2 is down-regulated in MTHFD1L-knockdown cells (Figure 4D). These findings suggest that MTHFD1L could, at least partially, influence the expression of cell cycle-related genes by modulating DNA methylation.
Figure 4.
Gene expression profiling reveals alterations in cell cycle-related genes upon silencing MTHFD1L expression. A. Silencing MTHFD1L expression using short interfering RNA (siRNA) decreased protein expression in the human prostate cancer PC-3 and C4-2 cell lines. The cells were transfected with control or MTHFD1L siRNAs (siMTHFD1L), and MTHFD1L protein expression in cells was examined via western blotting. B. Venn diagram analysis shows commonly significantly differentially expressed (SDE) genes upon MTHFD1L knockdown in PC-3 and C4-2 cells, displayed as the number of genes. C. WikiPathway analysis of 57 common SDE genes expressed in both PC-3 and C4-2 cells. D. Pearson correlation between fold changes in gene expression of cell cycle-related pathway genes measured by RNA-seq and qRT-PCR in PC-3 and C4-2 cells. E. Matched DNA methylation and gene expression data from The Cancer Genome Atlas were plotted to depict the correlation between MTHFD1L expression and the methylation status of the cell cycle-related genes.
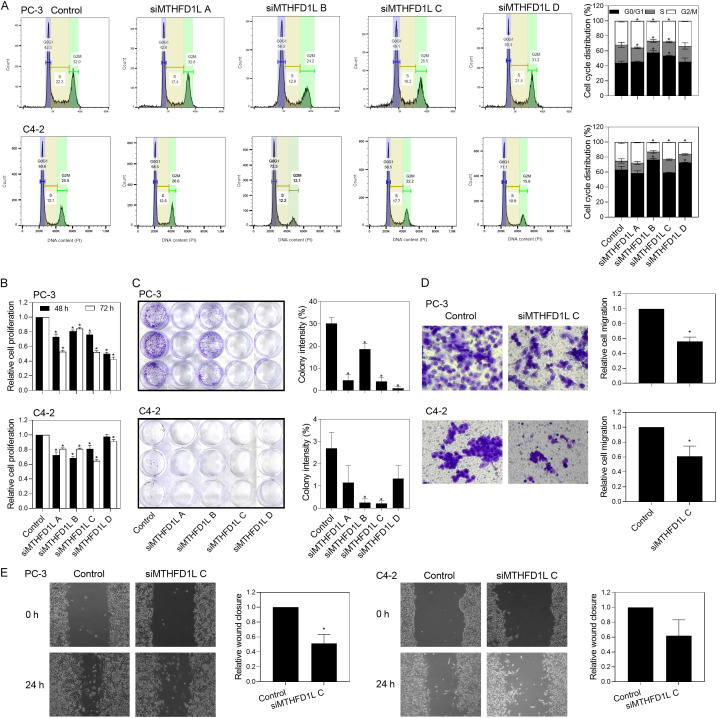
Additionally, when we monitored cell cycle progression in response to MTHFD1L knockdown by individual siRNAs, there was a higher frequency of G0/G1 elevation in PC-3 (siRNA B and C) and C4-2 (siRNA B and D) cells (Figure 5A). Although not all siRNAs exhibited the same arresting effect on the cell cycle, the majority reduced the rate of cell proliferation (Figure 5B). This inhibitory effect remained significant and consistent across all siRNAs when analyzing colony formation activities (Figure 5C). However, downregulation of MTHFD1L did not result in a noticeable change in the number of cells undergoing apoptosis (3.4% for PC-3 cells and 9.1% for C4-2 cells; data not shown) compared to cells treated with the mock control (3.9% for PC-3 cells and 6.4% for C4-2 cells), as assessed by annexin V and PI assay. Furthermore, the transwell cell migration assay revealed a significant decrease in migration ability upon MTHFD1L depletion, with a reduction of 48.0% in PC-3 cells and 41.9% in C4-2 cells (Figure 5D). The impact of MTHFD1L function loss on cell migration was further confirmed using the wound healing assay, yielding similar inhibitory results (Figure 5E). In summary, these findings provide evidence that MTHFD1L contributes to prostate cancer cell proliferation, migration, and colony formation, at least partially through regulation of the cell cycle.
Figure 5.
Silencing MTHFD1L expression reduces cell proliferation, colony formation, and cell migration by altering cell cycle-related pathways. A. Cell cycle distribution of control and MTHFD1L siRNAs (siMTHFD1L)-treated PC-3 and C4-2 cells was analyzed by flow cytometry. B. MTHFD1L knockdown decreases the proliferation of PC-3 and C4-2 cells. Cells transfected with control or siMTHFD1L were seeded in 96-well plates and allowed to proliferate for 48 or 72 h. Cell proliferation was then estimated using Cell Counting Kit-8 assay. C. MTHFD1L knockdown decreases the colony formation potential of PC-3 and C4-2 cells. Cells transfected with control or siMTHFD1L were seeded in 12-well plates and allowed to grow for 7 days. The colonies were fixed and counted using the ImageJ software. MTHFD1L knockdown reduces the migration ability of PC-3 and C4-2 cells. D. Control and MTHFD1L-knockdown cells were seeded into the insert chambers of transwell plates and allowed to migrate for 48 h. The number of migrated cells were counted by using ImageJ analysis of stained insert membranes. E. Control and MTHFD1L-knockdown cells were grown to confluency, scratched, and photographed 0 and 24 h. Relative wound closure was determined by measuring the recovery of wounded areas using the ImageJ software. Quantitative data are expressed as mean ± standard error of three or more repeated experiments, *P < 0.05.
Discussion
In this study, we conducted a comprehensive analysis of genetic variants in the one-carbon metabolism pathway and their impact on clinical outcomes in men with advanced prostate cancer undergoing ADT. Our findings revealed a significant association between MTHFD1L rs2073190 and CSS, OS, and PFS, after adjusting for relevant covariates. Notably, our pooled analysis of 13 independent gene expression studies demonstrated significantly higher levels of MTHFD1L mRNA expression in prostate tumor tissue specimens than in adjacent noncancerous tissue samples. Moreover, elevated transcript levels of MTHFD1L correlated with higher Gleason scores, advanced disease stage, and unfavorable prognosis in patients with prostate cancer. Subsequent investigations revealed that silencing the MTHFD1L gene led to decreased cell proliferation and colony formation in the two prostate cancer cell lines, suggesting a potential role of MTHFD1L in promoting prostate cancer pathogenesis.
Recent studies have revealed significant associations between genetic variants in the one-carbon metabolism pathway and clinical outcomes in patients with early-stage non-small cell lung cancer [22]. However, the effect of these genetic variants on prostate cancer remains unclear. In our study, the risk SNP rs2073190 was located in the intron region of MTHFD1L. Functional annotations from the HaploReg database indicated that rs2073190 coincides with the potential promoter and enhancer regions of MTHFD1L and alters the putative transcription factor-binding sites for forkhead box P1 and histone deacetylase 2, suggesting a regulatory role for rs2073190. We performed expression quantitative trait loci analysis to explore the relationship between rs2073190 and MTHFD1L expression. The rs2073190 risk allele G showed a suggestive correlation with elevated MTHFD1L mRNA expression levels in monocytes (P = 0.063) [23] and lymphoblastoid cells in the 1000 Genomes Project (P = 0.244) [24]. Given that rs2073190 was not genotyped in the GTEx Project [25], we identified a proxy untyped SNP, rs12199747, which exhibited high linkage disequilibrium (D’ = 1.0) with rs2073190. This proxy SNP displayed similar expression patterns in prostate tissue samples, with the risk allele correlating with increased MTHFD1L expression levels compared to the reference allele (P = 0.049).
Recent studies have highlighted the importance of one-carbon units derived from the mitochondria in fueling cytoplasmic reactions, where MTHFD1L serves as a key enzyme responsible for the final step of the reaction in the mitochondrial chamber, generating formate as a critical source of one-carbon donor molecules [26]. Elevated MTHFD1L expression has been associated with adverse clinical outcomes, increased cell proliferation, and enhanced cell migration in various cancer types, including colorectal [27], liver [28], bladder [29], thyroid [30], and esophageal [31] cancers. Conversely, the knockdown of MTHFD1L expression inhibited cancer cell growth and induced apoptosis in these malignancies. Furthermore, downregulation of MTHFD1L or use of the antifolate drug methotrexate effectively reduced nicotinamide adenine dinucleotide phosphate levels, leading to reactive oxygen species (ROS) accumulation, ROS-associated cell cycle delay, and hindered cell growth in MHCC97L human hepatocellular carcinoma cells [28]. Mechanistic studies employing gene expression profiling have highlighted that ERK5 signaling is one of the most inhibited pathways in MTHFD1L-knockdown KYSE-150 human esophageal squamous cell carcinoma cells [31]. Notably, treatment with the highly selective ERK5 inhibitor (BIX 02189) rescued the proliferation and metastasis induced by MTHFD1L overexpression in esophageal cancer cells. Additionally, in TPC-1 human papillary thyroid carcinoma cells, MTHFD1L knockdown hindered proliferation and induced apoptosis, with transcriptomic analysis indicating the involvement of the Notch2 and CCND1 signaling pathways [30]. CCND1, which encodes cyclin D1, plays a critical role in cell cycle regulation by controlling the G1 to S phase transition [32]. Collectively, our findings align with the existing evidence implicating MTHFD1L in cancer cell proliferation through its potential role in cell cycle regulation. Nonetheless, further comprehensive biological and functional studies are imperative to determine the precise role of SNPs/genes in prostate cancer progression.
Despite these noteworthy findings, several limitations of this study should be acknowledged. First, the sample size was relatively small, which might have hindered the detection of modest risks. Further, the study population exclusively comprised Taiwanese individuals, warranting further validation in diverse ethnic cohorts. Furthermore, despite our efforts to correct for multiple tests using q values, the possibility of false positives cannot be entirely excluded. Finally, although we present in silico functional evidence elucidating the potential impact of rs2073190 on MTHFD1L transcriptional regulation, additional investigations are necessary to unravel the underlying mechanisms involved.
In conclusion, our comprehensive genetic and functional investigations provide compelling evidence that genetic variants within one-carbon metabolism pathway genes, particularly MTHFD1L, play a role in driving prostate cancer progression by influencing the cell cycle pathway. These findings suggest MTHFD1L as a novel prognostic biomarker or potential indicator for precise treatment, shedding light on the molecular mechanisms underlying the observed associations with prostate cancer survival.
Acknowledgements
This work was supported by the National Science and Technology Council of Taiwan (grant Nos: 110-2314-B-038-135-MY3, 111-2320-B-039-021-MY3, 111-2218-E-037-001, 111-2622-E-039-004, 112-2311-B-039-001, 112-2314-B-037-127, 112-2622-E-039-001, and 112-2218-E-037-001), the Kaohsiung Medical University (grant No: NHRIKMU-111-I002), and the China Medical University (grant Nos: CMU110-MF-59, CMU111-MF-09, and CMU112-MF-10). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Authors thank Chao-Shih Chen for data analysis, and the National Centre for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support. The results published here are based in part on data generated by the HaploReg, 1000 Genomes, and TCGA projects.
Disclosure of conflict of interest
None.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castrationresistant prostate cancer (CRPC) Transl Androl Urol. 2015;4:365–380. doi: 10.3978/j.issn.2223-4683.2015.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito S, Sakamoto S, Higuchi K, Sato K, Zhao X, Wakai K, Kanesaka M, Kamada S, Takeuchi N, Sazuka T, Imamura Y, Anzai N, Ichikawa T, Kawakami E. Machine-learning predicts time-series prognosis factors in metastatic prostate cancer patients treated with androgen deprivation therapy. Sci Rep. 2023;13:6325. doi: 10.1038/s41598-023-32987-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clare CE, Brassington AH, Kwong WY, Sinclair KD. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu Rev Anim Biosci. 2019;7:263–287. doi: 10.1146/annurev-animal-020518-115206. [DOI] [PubMed] [Google Scholar]
- 7.Ding K, Jiang J, Chen L, Xu X. Methylenetetrahydrofolate dehydrogenase 1 silencing expedites the apoptosis of non-small cell lung cancer cells via modulating DNA methylation. Med Sci Monit. 2018;24:7499–7507. doi: 10.12659/MSM.910265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levesque N, Christensen KE, Van Der Kraak L, Best AF, Deng L, Caldwell D, MacFarlane AJ, Beauchemin N, Rozen R. Murine MTHFD1-synthetase deficiency, a model for the human MTHFD1 R653Q polymorphism, decreases growth of colorectal tumors. Mol Carcinog. 2017;56:1030–1040. doi: 10.1002/mc.22568. [DOI] [PubMed] [Google Scholar]
- 9.Yu H, Wang H, Xu HR, Zhang YC, Yu XB, Wu MC, Jin GZ, Cong WM. Overexpression of MTHFD1 in hepatocellular carcinoma predicts poorer survival and recurrence. Future Oncol. 2019;15:1771–1780. doi: 10.2217/fon-2018-0606. [DOI] [PubMed] [Google Scholar]
- 10.Rao K, Zheng K, Zhao Q, He J, Zhou B, Hou G, Sha N, Wang W, Yan M, Zhou Y, Jin Y, Jiang Y, Xia Q. The negative effect of G1958A polymorphism on MTHFD1 protein stability and HCC growth. Cell Oncol (Dordr) 2023;46:735–744. doi: 10.1007/s13402-023-00780-2. [DOI] [PubMed] [Google Scholar]
- 11.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, Lan YH, Lu TL, Lee HZ, Chen LM, Ting WC, Hsieh CJ, Huang SP. Significant associations of prostate cancer susceptibility variants with survival in patients treated with androgendeprivation therapy. Int J Cancer. 2012;130:876–884. doi: 10.1002/ijc.26091. [DOI] [PubMed] [Google Scholar]
- 12.Yu CC, Huang SP, Lee YC, Huang CY, Liu CC, Hour TC, Huang CN, You BJ, Chang TY, Huang CH, Bao BY. Molecular markers in sex hormone pathway genes associated with the efficacy of androgen-deprivation therapy for prostate cancer. PLoS One. 2013;8:e54627. doi: 10.1371/journal.pone.0054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang HH, Lee CH, Chen YT, Huang CY, Yu CC, Lin VC, Geng JH, Lu TL, Huang SP, Bao BY. Genetic analysis reveals the prognostic significance of the DNA mismatch repair gene MSH2 in advanced prostate cancer. Cancers (Basel) 2022;14:223. doi: 10.3390/cancers14010223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ke CC, Chen LC, Yu CC, Cheng WC, Huang CY, Lin VC, Lu TL, Huang SP, Bao BY. Genetic analysis reveals a significant contribution of CES1 to prostate cancer progression in Taiwanese men. Cancers (Basel) 2020;12:1346. doi: 10.3390/cancers12051346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12(Suppl 5):101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Choi C. Oncopression: gene expression compendium for cancer with matched normal tissues. Bioinformatics. 2017;33:2068–2070. doi: 10.1093/bioinformatics/btx121. [DOI] [PubMed] [Google Scholar]
- 19.You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, Davicioni E, Karnes RJ, Klein EA, Den RB, Ross AE, Schaeffer EM, Garraway IP, Kim J, Freeman MR. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res. 2016;76:4948–4958. doi: 10.1158/0008-5472.CAN-16-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do SK, Choi SH, Lee SY, Choi JE, Kang HG, Hong MJ, Kim JH, Baek SA, Lee JH, Lee WK, Do YW, Lee EB, Shin KM, Jeong JY, Lee YH, Seo H, Yoo SS, Lee J, Cha SI, Kim CH, Seok Y, Cho S, Jheon S, Park JY. Genetic variants in onecarbon metabolism pathway predict survival outcomes of early-stage non-small cell lung cancer. Oncology. 2020;98:897–904. doi: 10.1159/000509658. [DOI] [PubMed] [Google Scholar]
- 23.Quach H, Rotival M, Pothlichet J, Loh YE, Dannemann M, Zidane N, Laval G, Patin E, Harmant C, Lopez M, Deschamps M, Naffakh N, Duffy D, Coen A, Leroux-Roels G, Clement F, Boland A, Deleuze JF, Kelso J, Albert ML, Quintana-Murci L. Genetic adaptation and neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656. e617. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiura T, Nagano Y, Inoue T, Hirotani K. A novel mitochondrial C1-tetrahydrofolate synthetase is upregulated in human colon adenocarcinoma. Biochem Biophys Res Commun. 2004;315:204–211. doi: 10.1016/j.bbrc.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Lee D, Xu IM, Chiu DK, Lai RK, Tse AP, Lan Li L, Law CT, Tsang FH, Wei LL, Chan CY, Wong CM, Ng IO, Wong CC. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest. 2017;127:1856–1872. doi: 10.1172/JCI90253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eich ML, Rodriguez Pena MDC, Chandrashekar DS, Chaux A, Agarwal S, Gordetsky JB, Ferguson JE, Sonpavde GP, Netto GJ, Varambally S. Expression and role of methylenetetrahydrofolate dehydrogenase 1 like (MTHFD1L) in bladder cancer. Transl Oncol. 2019;12:1416–1424. doi: 10.1016/j.tranon.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi D, Yilihamu Y, Jiang C, Wang R, Lu X, Sang J, Su L. MTHFD1L knockdown diminished cells growth in papillary thyroid cancer. Tissue Cell. 2022;77:101869. doi: 10.1016/j.tice.2022.101869. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Yang Y, Cheng J, Luan S, Xiao X, Li X, Fang P, Gu Y, Shang Q, Zhang H, Chen L, Zeng X, Yuan Y. MTHFD1L confers a poor prognosis and malignant phenotype in esophageal squamous cell carcinoma by activating the ERK5 signaling pathway. Exp Cell Res. 2023;427:113584. doi: 10.1016/j.yexcr.2023.113584. [DOI] [PubMed] [Google Scholar]
- 32.Montalto FI, De Amicis F. Cyclin D1 in cancer: a molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells. 2020;9:2648. doi: 10.3390/cells9122648. [DOI] [PMC free article] [PubMed] [Google Scholar]