Abstract
Microglia, the resident immune cells of the central nervous system (CNS), have received significant attention due to their critical roles in maintaining brain homeostasis and mediating cerebral immune responses. Understanding the origin of microglia has been a subject of great interest, and emerging evidence suggests that microglia consist of multiple subpopulations with unique molecular and functional characteristics. These subpopulations of microglia may exhibit specialized roles in response to different environmental cues as in disease conditions. The newfound understanding of microglial heterogeneity has significant implications for elucidating their roles in both physiological and pathological conditions. In the context of disease, microglia have been studied rigorously as they play a very important role in neuroinflammation. Dysregulated microglial activation and function contribute to chronic inflammation. Further exploration of microglial heterogeneity and their interactions with other cell types in the CNS will undoubtedly pave the way to novel therapeutic strategies targeting microglia‐mediated pathologies. In this review, we discuss the latest advances in the field of microglia research, focusing specifically on the origin and subpopulations of microglia, the populations of microglia types in the brains of patients with neurodegenerative diseases, and how microglia are regulated in the healthy CNS.
Keywords: distribution, microglial origin, neurodegenerative diseases, regulation, subtypes
Microglia, the CNS's resident immune cells, display significant heterogeneity, with distinct subpopulations responding to various environmental cues in both health and disease. This diversity has vital implications for understanding their roles, especially in neuroinflammation. Investigating microglial origin, subtypes in neurodegenerative diseases, and healthy CNS regulation offer promising therapeutic avenues for addressing microglia‐related pathologies.
Abbreviations
- AD
Alzheimer's disease
- ALS
amyotrophic lateral sclerosis
- APOE
apolipoprotein
- APP
amyloid precursor protein
- ASD
autism spectrum disorder
- Aβ‐plaques
amyloid beta plaques
- BDNF
brain‐derived neurotrophic factor
- CCL4
C‐C Motif chemokine ligand 4
- CCR9
CC Chemokine receptor type 9
- CD40
cluster of Differentiation 4
- CNS
central nervous system
- CSF1
colony stimulating factor 1
- CX3CR1
CX3C receptor 1
- CyTOF
cytometry by time of flight
- DAMs
disease‐associated microglia
- EAE
experimental autoimmune encephalomyelitis
- EGA
estimated gestational age
- EGFP
enhanced green fluorescent protein
- ETS
E‐twenty six
- GABA
gamma‐aminobutyric acid
- H3K27me3
H3 lysine 27 trimethylation
- IBA1
ionized calcium‐binding adaptor molecule 1
- IL10
interleukin 10
- IL4
interleukin 4
- Irf8.
interferon regulatory factor 8
- KSPG
Keratan sulfate proteoglycan microglia
- MGnD
neurodegenerative microglia
- MS
multiple sclerosis
- NGF
Nerve Growth Factor
- P2RY12
purinergic receptor P2Y, G‐protein coupled 12
- PD
Parkinson's disease
- PGE2.
prostaglandin E2
- PRC2
polycomb repressive complex 2
- PRR
pattern recognition receptor
- RMS
rostral migratory stream
- SVZ
subventricular zone
- TNF‐α
tumor necrosis factor‐alpha
- TREM2
triggering receptor expressed on myeloid cells 2
The central nervous system (CNS), the most intricate human organ system, comprises billions of neurons coordinating the intricate interplay of thoughts, emotions, and physiological functions. However, the complex network requires constant surveillance and protection to maintain its delicate balance. So, in the brain, microglia serve as the guardians, playing a vital role in immune defense and maintaining cerebral homeostasis. As part of the innate immune system, microglia continuously monitor the CNS for potential internal and external threats [1, 2, 3]. In addition to their crucial role in defending the CNS against pathogens, microglia also contribute significantly to controlling neuronal proliferation, synapse formation, and elimination as well as debris clearance [4, 5, 6]. Moreover, they actively participate in remodeling neuronal circuits in postnatal mice [7]. Given the diverse functions, it is not surprising, given their diverse functions, that microglia represent a heterogeneous myeloid cell population, which is indispensable for the CNS in health and disease [8].
Considering the significance of microglia, it is important to explore their origin, their various subpopulations, and their precise mode of action. Microglia, as essential neuroglial cells, account for 5–20% of the entire glial population in mice while in humans, they constitute 0.5–16.6% of the total population [9].
In this article, we review the recent findings in the field of microglia research with a focus on their heterogeneity. The identification of distinct microglial subtypes and their unique functional characteristics have revolutionized our understanding of their roles in brain development, homeostasis, and disease processes. Understanding the modulation of these different microglial subpopulations will be key to develop precise therapeutic interventions targeting CNS innate immune mechanisms.
Origin of microglia
Over the past 160 years, multiple hypotheses about the origin of microglia have emerged. W. Ford Robertson was the first to introduce the term “mesoglia”, a phagocytic element derived from the mesoderm, distinct from neurons and other CNS cells. In 1856, Virchow coined the term “neuroglia” (originally “Nervenkitt”), referring to these cells as “nerve‐glue,” which was later translated to “neuroglia” [10]. Santiago Ramon y Cajal, a prominent scientist in the field of neuroscience, renamed the cells the “third element of the nervous system.” A student of his, Pio del Río‐Hortega, continued working on these cells and made significant contributions to the understanding of the “third element of the nervous system.” Del Río‐Hortega studied the third element of the nervous system using silver carbonate impregnation staining. He redefined the concept of the “third element” based on its morphology and function, which he named “microglia cells.” Microglia cells are characterized as a small population of phagocytic and migratory immune cells in the CNS, distinguishing them from neurons, astrocytes, and oligodendroglia, which are of neuroectodermal origin [11].
In murine models, the precise origin of microglia has sparked controversies. It was believed that microglia are present during early development, suggesting that they originated from embryonic progenitors. Del Río‐Hortega proposed an additional possibility, that microglia could derive from meningeal macrophages. Another hypothesis during that era was that microglia could originate from blood monocytes [10, 11]. Ashwell and colleagues initially observed the presence of amoeboid microglia cells at E11.0 in the fetal mouse cerebellum and later in the rat forebrain [12, 13]. Subsequently, Sorokin and his colleagues detected macrophage precursors and macrophage‐like cells in the embryonic mesenchyme and blood vessels in rats starting from E10.5, highlighting the brain as the first organ to be colonized [14].
The origin of microglia was studied using genetic mapping, which revealed that microglia originate from yolk sac primitive macrophages [15, 16, 17, 18, 19]. During embryonic development in mice, between embryonic days E8.0 and E10.0, there is blood vessel formation and remodeling [20]. Around E7.0, precursor cells expressing vascular endothelial growth factor migrate from the primitive streak to the proximal yolk sac, where they form blood islands. These blood islands house the multilineage c‐kit+ erythromyeloid yolk sac precursor cells that give rise to microglia [21, 22]. The yolk sac precursor cells mature from A1 (CD45+ c‐kitlo CX3CR1− F4/80−) to A2 (CD45+ c‐kit− CX3CR1+ F4/80hi) amoeboid macrophages in the blood islands and cephalic mesenchyme. Eventually, they acquire the phenotype of mature macrophages in the neuroepithelium by E10.5 [23]. Fate mapping studies using genetic targeting of hematopoietic precursors expressing the runt‐related transcription factor Runx1 between E6.5 and E10.5 have shown that yolk sac macrophages are specified between E7.0 and E7.5 [15]. An interesting study demonstrated that normal yolk sac hematopoiesis from E9.5 to 10.5 in Ncx‐1 knockout mice causes the absence of brain microglia progenitors. This supports the notion that brain recruitment of yolk sac progenitors depends on a functional circulatory system [15]. Microglia enter the developing brain through the leptomeninges and lateral ventricles at E9.5 and then distribute within the cortical walls from both directions. The speed of migration, proliferation rates, and maturation of microglia vary based on the region and developmental stage [15, 24, 25]. In early postnatal weeks of mouse development, there is an increase in the number of microglia cells. Subsequently, there is a gradual decrease in their number, reaching approximately 50% of the peak density. From Week 5 to 6, microglia density stabilizes [26].
In human fetuses, microglia‐like cells can be detected as early as 3 weeks of the estimated gestational age (EGA) [27]. By Week 4.5, amoeboid microglial cells migrate into the cerebral wall via the pial surface, ventricle, and choroid plexus [28, 29]. In the white matter, subplate, and cortical plate layers, radial and tangential migration was observed and then at 12–13 gestational weeks, the second wave of microglia was observed via the vasculature [29, 30]. Around Week 9, colonization of the spinal cord begins, and by Week 16, there is a significant influx and distribution of microglia throughout the entire CNS. It takes approximately 22 weeks for microglia to adopt a ramified form and to develop widely distributed processes. Importantly, well‐differentiated microglia are detected at 35 weeks of pregnancy [28, 30, 31, 32, 33]. These studies provide strong evidence that microglia originate from embryonic hematopoietic precursors that populate the CNS before birth and prior to bone marrow hematopoiesis. In zebrafish, yolk sac macrophages initially invade the entire cephalic mesenchyme and subsequently infiltrate epithelial tissues, including the brain. Additionally, other macrophages enter the blood circulation, indicating that colonization occurs independently of the blood circulation [34, 35]. Summarizing the current knowledge suggests the origin of microglia is from yolk sac primitive macrophages, and the colonization takes place before the formation of neuroectoderm‐derived cell types, such as astrocytes and oligodendrocytes. Microglia remain in the brain throughout life and self‐renew [15, 16, 17, 18, 19, 22, 25, 30, 36].
Microglia subpopulations
In the CNS, microglia are non‐uniformly distributed, as observed by Lawson and colleagues using polyclonal antiserum targeting the F4/80 marker. Their study revealed that the telencephalon region has the highest density of microglia, followed by the diencephalon, mesencephalon, and rhombencephalon, which contain fewer microglia. Furthermore, the gray matter is more densely populated with microglia compared with the white matter [37]. These studies have proposed the existence of different microglial subtypes, each associated with a distinct molecular signature [38]. Understanding these potential subtypes will provide insights into the differential responses of microglia to intrinsic and extrinsic stimuli i.e.; microglia near Aβ‐plaques show a neurodegenerative profile controlled by TREM2‐APOE, and targeting APOE can revert them to a healthy state, reducing apoptotic neuron phagocytosis [39, 40, 41, 42, 43, 44].
What makes a cell subtype? Traditionally, a cell type is defined based on its host tissue, morphology, lineage, function, and molecular composition [45]. From a historical perspective, del Río‐Hortega already defined a microglial subtype known as “satellite microglia,” which are located in close proximity to neuronal cell bodies [46]. This early observation highlights the concept of microglial subtypes based on their distinct anatomical localization and association with specific cellular components or morphological structures. It is important to continue investigating and characterizing microglial subtypes to gain a deeper understanding of their functional diversity and associated molecular signatures. Microglia distribution in the CNS is not only variable concerning localization but also varies in morphology based on their association with different cellular components such as neuronal cell bodies, dendrites, axons, myelinated axons, and blood vessels. This variability is also reflected at the transcriptional level [47, 48]. Several studies have mounted evidence that microglia isolated from unchallenged adult murine brain exhibit variability in gene expression patterns depending on the brain regions which they were isolated from. Various markers such as CD40, CD11b, CD45, CD80, CD86, F4/80, Triggering Receptor Expressed on Myeloid Cells 2b (TREM2b), CX3CR1, and CCR9 show variable expression levels in microglia based on brain area and transcriptome analysis using preselected panels [49, 50]. Furthermore, recent research by Jordão and colleagues revealed the diversity of CNS‐associated macrophages in three subsets expressing high levels of Mrc1, Ms4at, Pf4, Stab1, Cbr2, CD163, and Fcrls. These subsets are associated with different CNS compartments, including the leptomeninges, choroid plexus, and perivascular space [51]. The microglia in the vicinity of these different types of neurons, as well as other glial cells such as astrocytes, oligodendrocytes, and progenitor cells, show distinct gene expression profiles under steady‐state conditions. Several subtypes of microglia have been identified based on their unique genomic, morphological, and function specialization.
These subtypes include satellite microglia, keratan sulfate proteoglycan‐microglia (KSPG)‐microglia, microglia supporting neurogenesis, Hox8b+ microglia, CD11c+ microglia, dark microglia, and TREM2‐positive‐microglia (Table 1). Satellite microglia interact with the axon initial segment in the healthy brain and lose the interaction upon injury. The markers required to identify satellite microglia are IBA1, CD11b, and CX3CR1; these microglia are frequently detected in the cortex and hippocampus [52, 53, 54]. KSPG‐microglia appear upon different insults, for example around motoneurons in Amyotrophic Lateral Sclerosis (ALS), and can be identified by the microglia marker IBA1, CR3, and CD11b. KSPG‐microglia are mainly present in the olfactory bulb, hippocampus, and brainstem [55, 56, 57]. Microglia‐supporting neurogenesis are essential for neuroblast survival and migration in the subventricular zone (SVZ)/rostral migratory stream (RMS), and their characteristics include the expression of IBA1−, isolectin B4 −, CD68−, P2RY12low, pSTAT6+ cells as well as their ability to produce IL4 and IL10. Morphologically, these microglia are less ramified than microglia in neighboring brain cells and in the olfactory, subventricular zone, and rostral migratory stream, they can be identified by the expression of CX3CR1‐EGFP [58, 59, 60]. Hox8b+ microglia have a critical role in the functioning of the corticosteroid neuronal circuits. A deficiency of Hox8b+ microglia affects corticosteroid neuronal circuits negatively and leads to impaired grooming, anxiety, and altered social behaviors. Markers to identify Hox8b+ microglia are IBA1 and CD11b. These microglia are present in the olfactory bulb and cortex of the brain [61, 62, 63]. CD11c‐positive microglia promote myelination and neurogenesis in the neonatal brain and can be identified with markers IBA1, CD11c, CD45low, CX3CR1, and CCR2null. CD11c+ microglia cells are present in the corpus callosum and cerebellum [64]. Dark microglia, which interact with blood vessels and synapses, appear dark when detected by electron microscopy and can be identified by IBA1low, CX3CR1‐GFPlow, CD11b, TREM2, and 4D4. Dark microglia can be found in the cortex, hippocampus, amygdala, and hypothalamus [65, 66]. These subtypes of microglia demonstrate diversity in their localization and functional roles. The role of TREM2 microglia in Alzheimer's disease (AD) is essential for neuroprotection. However, it is important to note that not all microglia express the TREM2 receptor. TREM2‐positive microglia are known for their survival and proliferation, and they tend to cluster around Aβ‐plaques in AD [67]. In murine models, TREM2 expression in microglia varies across different brain regions. The highest levels of TREM2 expression are found in the cingulate cortex and lateral entorhinal cortex, while much lower levels are observed in regions such as the hypothalamus and habenula. Interestingly, some regions, such as the circumventricular organs, completely lack TREM2 expression [68]. These regional differences in TREM2 expression are also observed in humans. Microarray data from 101 individuals revealed significant variations in TREM2 expression between different brain regions, particularly in the white matter and cerebellum. This suggests the presence of specific subtypes of microglia with varying roles in different brain regions, which may be relevant to the progression of AD and other neurological disorders [69]. Of note, the described pattern may vary and be dynamic over the entire lifetime and in particular during inflammatory challenges and activation.
Table 1.
Microglia subpopulation in the CNS.
| Microglial subpopulation | Markers | Specific brain region |
|---|---|---|
| Satellite microglia [46] | IBA1, CD11b, and CX3CR1 | Cortex and hippocampus |
| Keratan sulfate proteoglycan‐microglia (KSPG)‐microglia [55, 56] | IBA1, CR3, and CD11b | Olfactory bulb, hippocampus, and brainstem |
| Microglia supporting neurogenesis [58] | IBA1−, isolectin B4 −, CD68−, P2RY12low, pSTAT6+ | Olfactory, subventricular zone, and rostral migratory stream |
| Hox8b+ microglia [61] | IBA1 and CD11b | Olfactory bulb and cortex of the brain |
| CD11c+ microglia [64] | IBA1, CD11c, CD45low, CX3CR1, and CCR2null | Corpus callosum and cerebellum |
| Dark microglia [65] | IBA1low, CX3CR1‐GFPlow, CD11b, TREM2, and 4D4 | The cortex, hippocampus, amygdala, and hypothalamus |
| TREM2‐positive‐microglia [67] | TREM2 | Cingulate cortex and lateral entorhinal cortex |
In addition to a subpopulation of microglia defined by differential gene expressions, there are differences in the microglia population between males and females [70, 71, 72, 73]. Microglial density in 13‐week‐old male mice is higher than in females, while the opposite holds true for 3‐week‐old mice, showcasing significant age‐ and gender‐related variation [74]. Hormones such as estradiol also play a major role in the gender‐dependent variation of microglia numbers [75, 76]. During early postnatal development, male mice have more microglia in the cortex, hippocampus, and amygdala compared with females. This is linked to increased expression of CC‐chemokine ligand (CCL) 20 and CCL4 due to testosterone, the primary masculinizing hormone, being aromatized to estradiol in the mouse brain. Adult female mice exhibit thicker microglia with longer processes in the hippocampus, amygdala, and cortex compared with male mice [76, 77].
Microglia regulation
Microglia are crucial components of the neuroglial network in the healthy CNS. In the “homeostatic” state, microglia exhibit small cell bodies with ramified processes [78]. In this state, microglia do not overlap with the processes of neighboring cells, and each microglia cell actively surveys its immediate vicinity. While the soma of microglia remains stable, the processes constantly elongate and retract, allowing them to constantly explore the tissue environment widely. Upon stimulation by pathogen exposure, microglia rapidly retract their processes and become mobile effector cells [2]. The activation of microglia is triggered by various immune receptors, both endogenous and exogenous, that are present on their surface and collectively described as pattern recognition receptors (PRR) [79, 80, 81]. Examples of these PPRs include Toll‐like receptors (TLRs), scavenger receptors, CD36, and CD47, as well as numerous cytokine and chemokine receptors. Further surface receptors are involved in the regulation of microglial homeostasis, their interaction with neighboring cells, and immune reactivity (Table 2):
Table 2.
Factors regulating microglia.
| Microglial regulation | Function |
|---|---|
| CD200 [82] | Helps to maintain resting state |
| CX3CR1 [85] | Regulate microglia recruitment to the site of neuroinflammation |
| CD47 [84] | Neuronal protein sends “do not eat me” signals to microglia via CD172a/Sirp alpha interaction |
| PRC2 [91] | PRC2 enzyme catalyzes H3K27me3 modification |
| TREM2 [93, 94] | Role in phagocytosis of debris and reducing proinflammatory cytokines |
| CSF1 [95] | Regulate the survival of myeloid lineage |
| Runx1, ETS, PU.1, Irf8, Hoxb8 [15, 22] | Regulating differentiation processes during the embryonic development |
| C‐myb [97] | Essential for microglia health, regulates proliferation, and survival in the CNS |
| Ionotropic receptors [98, 99] | Calcium influx and the release of pro‐inflammatory molecules |
| Metabotropic receptors [98, 99] | Activate intracellular signaling cascades that contribute to microglial activation and inflammation |
| Nerve Growth Factor [100] | Regulate microglial activation and survival |
| Prostaglandins E2 [101] | Modulate microglial activation and pro‐inflammatory responses |
| Gamma‐aminobutyric acid (GABA) [105] | Modulate microglial activation and inflammatory responses |
| Glucocorticoids [106] | Suppresses microglia activation and inflammation via glucocorticoid receptor binding |
| Estrogen [106] | Modulates microglia activation, migration, and phagocytic activity |
| Brain‐derived Neurotropic factor (BDNF) [107] | Modulate microglial function by influencing their activation, proliferation, and release of pro‐inflammatory cytokines |
| Norepinephrine [108, 109] | Controls the release of inflammatory factors like interleukin 6 (IL‐6), interleukin 1β (IL‐1β), and tumor necrosis factor α (TNF‐α) |
| Histamine and serotonin [111] | Increases of calcium in the microglia |
CD200 is a crucial molecule expressed on the surface of neurons, astrocytes, and oligodendrocytes. It serves as a receptor on microglia and macrophages, helping to maintain their resting state [82, 83, 84]. CX3CR1 is another essential molecule found on the surface of monocytes, macrophages, dendritic cells, and natural killer cells. Its ligand, fractalkine or neurotactin (CX3CL1), is present on neurons and interacts with microglia via CX3CR1. This interaction plays an important role in regulating microglia functions within the CNS [5, 85, 86, 87, 88, 89]. Loss of this interaction, as seen in animal models of PD and other neurodegenerative disorders, can lead to increased neuronal cell death [85]. CD47, expressed ubiquitously on neurons, transmits “do not eat me” signals to microglia through its interaction with CD172a/Sirp alpha [84, 90]. Ayata and colleagues showed that cerebellar microglia have a unique ability for clearance, while microglia in the striatum exhibit a homeostatic surveillance phenotype [90, 91]. It was also demonstrated that the suppression of clearance genes in striatal microglia is mediated by PRC2, an enzyme complex that catalyzes the repressive chromatin modification histone H3 lysine 27 trimethylation (H3K27me3). Removal of PRC2 in microglia results in enhanced clearance function in both the striatum and cerebral cortex even in the absence of dying neurons [91]. TREM2 is primarily known for its involvement in the phagocytosis of cellular debris and the downregulation of pro‐inflammatory cytokines [92]. Studies showed that the deletion of TREM2 showed identical phenotypes such as enhanced inflammatory cytokine production [93, 94]. Colony Stimulating Factor 1 (CSF1) plays a crucial role in regulating the survival of myeloid lineage cells in general [95]. Colony Stimulating Factor 1 binds to its receptor, CSF1R, and the absence of CSF1R results in a deficiency of several subsets of mononuclear phagocytes. Notably, in mice lacking CSF1R, microglia, are entirely absent [15, 96].
Endogenous transcription factors, such as Runx1, ETS (E‐twenty six) family transcription factor PU.1 shown in Fig. 1, interferon regulatory factor 8 (Irf8), and Hoxb8, play crucial roles in regulating differentiation processes during the embryonic development [15, 22]. While c‐myb is not directly involved in microglia development, it plays a vital role in maintaining microglial homeostasis. C‐myb is an important regulator of cell proliferation and survival, and its function is essential for the normal functioning and maintenance of microglia in the healthy CNS [97]. ATP release in the CNS leads to various effects, including an inflammatory response, migration, and proliferation, ultimately resulting in a microglial activation [2]. This activation is mediated through the presence of purinergic receptors on the surface of microglia, including ionotropic receptors (P2X4, P2X7) and metabotropic receptors (P2Y1, P2Y2, and P2Y12) [98, 99]. Ionotropic purinergic receptors, such as P2X4 and P2X7, are involved in calcium influx and the release of pro‐inflammatory molecules. Metabotropic purinergic receptors, such as P2Y1, P2Y2, and P2Y12, activate intracellular signaling cascades contributing to microglial activation and inflammation. Nerve Growth Factor (NGF) can regulate microglial activation and survival. It has been shown to modulate microglial morphology and pro‐inflammatory responses [100]. Prostaglandins, including prostaglandin E2 (PGE2), can modulate microglial activation and pro‐inflammatory responses. They are synthesized by microglia through cyclooxygenase 1 and 2 and can act in an autocrine or paracrine manner [101].
Fig. 1.
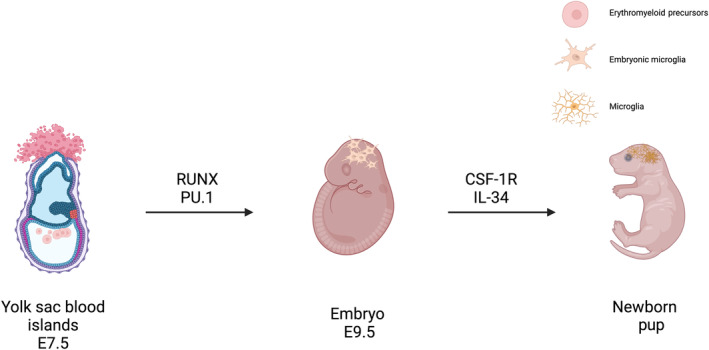
Origin of microglia in mice. Microglia stem from immature erythromyeloid precursors (EMPs) that depart the yolk sac blood island around E7.5, regulated by PU.1 and RUNX. By E9.5, these EMPs reach the neuroepithelium, giving rise to embryonic microglia, which mature into the fully developed form. Essential for mature microglia's growth are IL‐34 and CSF‐1, vital factors that support their proliferation.
Notably, microglia are well known to express receptors for neurotransmitters [102, 103, 104]. Gamma‐aminobutyric acid (GABA), an inhibitory neurotransmitter in the CNS, can also modulate microglial activation and inflammatory responses. Activation of GABA receptors on microglia has been shown to suppress their pro‐inflammatory phenotype, dampening the inflammatory response [105]. Glucocorticoids and estrogen have been shown to regulate microglia function. Glucocorticoids suppress microglia activation and pro‐inflammatory responses by binding to glucocorticoid receptors expressed on microglia. Estrogen modulates microglia activation, migration, and phagocytic activity [106]. Brain‐Derived Neurotrophic Factor (BDNF) can modulate microglial function by influencing their activation, proliferation, and release of pro‐inflammatory cytokines [107]. Norepinephrine regulates microglia and controls the release of inflammatory factors such as interleukin 6 (IL‐6), interleukin 1β (IL‐1β), and tumor necrosis factor α (TNF‐α). This regulation impacts neuroinflammation, neuropathic pain, anxiety, and depression [108, 109, 110]. Histamine and Serotonin induce the increase of calcium in the microglia [111]. Microglia and neurons communicate through receptors for various neurotransmitters, enabling neurons in specific brain regions to influence microglia heterogeneity based on their comcombined neurotransmitter profile. The communication between microglia and neurons via neurotransmitter receptors may be crucial for brain region‐specific differences, given variances in neuron‐derived transmitter profiles.
Microglia in disease
Emerging research has shown that microglia represent a highly heterogeneous population composed of distinct subpopulations with diverse functions (Table 1). Dysregulation of these subpopulations has been implicated in various neurological and neurodegenerative diseases. Here are some examples of microglial subpopulations involved in specific diseases: Activation of microglia in brain diseases mostly functions through the ligation of PRRs, including the Toll‐like receptor family, the scavenger receptors, CD47, CD36, and several others that cooperate to induce downstream immune signaling pathways [79, 80, 81]. Various cytokines, such as interleukin‐1 beta (IL‐1β) and tumor necrosis factor‐alpha (TNF‐α), can modulate microglial activation and immune responses. They can be released by microglia themselves or by other cells in the CNS [112]. Consequently, microglia adopt disease‐specific states (Table 3), which still must be defined in greater detail, as most of the existing literature is of cross‐sectional nature and may not fully account for highly dynamic changes along the respective disease trajectories. Nevertheless, AD is linked to amyloid‐beta plaques from increased amyloid precursor protein (APP) production or clearance issues and hyperphosphorylated tau protein buildup [113, 114, 115]. Disease‐associated microglia (DAMs) and Neurodegenerative microglia (MGnD) represent a subset of microglia with heightened expression of AD‐risk‐associated genes (ApoE, Trem2, and Clec7a) and functional activation of the TREM2‐APOE pathway [40, 42]. Disease‐associated microglia is associated with protective phagocytosis, and MGnD is a dysfunctional microglial phenotype [116]. While many genes differ in DAM and MGnD between mice and humans, they share some similarities. Notably, both human microglia and activated mouse microglia show increased APOE expression, along with a reduced TREM2 expression [42, 117, 118]. They play a role in amyloid‐beta plaque clearance and may have both protective and harmful effects on AD pathogenesis [40]. In AD, the CD33 transmembrane receptor is mainly expressed by microglia, which regulates innate immune responses [119, 120]. Parkinson's disease (PD, a movement disorder caused by the degeneration of dopaminergic neurons) is characterized by motor impairment and the presence of intraneuronal inclusions called Lewy bodies, which represent aggregates of misfolded alpha‐synuclein [121]. During the course of PD, reactive microglia also become activated by alpha‐synuclein and release pro‐inflammatory cytokines and oxidative stress‐inducing factors, contributing to PD‐associated neuroinflammation and likely causing dopaminergic neuron loss [122, 123]. Multiple Sclerosis (MS) is characterized by multifocal white matter lesions. Experimental Autoimmune Encephalomyelitis (EAE) serves as an animal model for studying inflammatory demyelination disease [124, 125, 126]. Both neuroinflammatory and neurodegenerative models develop double‐positive TNF‐alpha‐ and GM‐CSF‐producing cells, this subset abundance correlated best with the height of neuroinflammatory condition in the MS model [124]. Within EAE lesion sites, three distinct subtypes, namely daMG2, daMG3, and daMG4, have been identified. These subtypes exhibit variations in the expression of particular chemokines and cytokines, and they also exert differing effects on homeostatic markers such as P2RY12 and TMEM119 [51]. The expression of 5D4‐KSPG is elevated within a specific subgroup of microglia that are positive for IBA1/CD11b in the context of amyotrophic lateral sclerosis (ALS) and in a Wallerian degeneration mouse model for spinal cord injury [57, 127, 128]. In stroke, pro‐inflammatory microglia can become activated and exhibit a pro‐inflammatory phenotype. These microglia produce inflammatory cytokines and reactive oxygen species, contributing to secondary brain damage [129]. In autism spectrum disorder (ASD), microglia show impaired synaptic pruning. These microglia fail to efficiently eliminate excessive synapses during brain development, potentially leading to disrupted or dysfunctional neural circuits and subsequently altered connectivity [130]. In brain tumors, the analysis of activated microglia using CyTOF, which is a single‐cell‐based immune phenotyping technique that relies on time‐of‐flight mass cytometry, has revealed significant differential expressions of HLA‐DR, TREM2, and APOE. [131].
Table 3.
Microglia in disease.
| Microglia identity | Present in |
|---|---|
| Disease‐associated microglia (DAM) & Neurodegenerative microglia (MGnD) [40, 42] |
Alzheimer's disease AD model |
| Reactive microglia [122] |
Parkinson's disease Alzheimer's disease PD model |
| Double‐positive TNF‐alpha‐ and GM‐CSF‐producing cells [124] |
Multiple sclerosis MS model |
| daMG2, daMG3, and daMG4 [51] |
Expexrimental autoimmune encephalomyelitis EAE model |
| 5D4‐KSPG [57] |
Amyotrophic lateral sclerosis ALS model |
In summary, microglia are diverse and dynamic cells with important functions in the CNS and during CNS disorders. Understanding their origin, distribution, and subpopulations is crucial for unraveling their roles in both healthy and diseased conditions. Further research in this field will contribute to a deeper understanding of microglial biology and help to identify potential therapeutic interventions for CNS disorders.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
SD authored the manuscript, and MTH edited and enhanced it, contributing to its refinement.
Acknowledgements
This work was supported by funding from the Fonds National de la Recherche Luxembourg (FNR) within the PEARL program (FNR/16745220) to MTH, the EU Joint Programme—Neurodegenerative Disease Research (JPND/17063008) to MTH, the Gerhard und Ilse Schick Stiftung and the Luxembourg Centre for Systems Biomedicine (LCSB). The authors would like to thank Dr Arnaud Mary for his help to refine the manuscript.
Edited by Koji Yamanaka
References
- 1. Lehnardt S (2010) Innate immunity and neuroinflammation in the CNS: the role of microglia in toll‐like receptor‐mediated neuronal injury. Glia 58, 253–263. [DOI] [PubMed] [Google Scholar]
- 2. Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML and Gan W‐B (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8, 752–758. [DOI] [PubMed] [Google Scholar]
- 3. Nimmerjahn A, Kirchhoff F and Helmchen F (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- 4. Graeber MB (2010) Changing face of microglia. Science 330, 783–788. [DOI] [PubMed] [Google Scholar]
- 5. Hughes V (2012) Microglia: the constant gardeners. Nature 485, 570–572. [DOI] [PubMed] [Google Scholar]
- 6. Tremblay M‐È, Lowery RL and Majewska AK (2010) Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L et al. (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- 8. Perry VH, Nicoll JAR and Holmes C (2010) Microglia in neurodegenerative disease. Nat Rev Neurol 6, 193–201. [DOI] [PubMed] [Google Scholar]
- 9. Mittelbronn M, Dietz K, Schluesener HJ and Meyermann R (2001) Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol 101, 249–255. [DOI] [PubMed] [Google Scholar]
- 10. Rio‐Hortega P (1939) The microglia. Lancet 233, 1023–1026. [Google Scholar]
- 11. Rezaie P and Male D (2002) Mesoglia & microglia – a historical review of the concept of mononuclear phagocytes within the central nervous system. J Hist Neurosci 11, 325–374. [DOI] [PubMed] [Google Scholar]
- 12. Ashwell K (1990) Microglia and cell death in the developing mouse cerebellum. Dev Brain Res 55, 219–230. [DOI] [PubMed] [Google Scholar]
- 13. Ashwell KWS and Waite PME (1991) Cell death in the developing trigeminal nuclear complex of the rat. Dev Brain Res 63, 291–295. [DOI] [PubMed] [Google Scholar]
- 14. Sorokin SP, Hoyt RF Jr, Blunt DG and McNelly NA (1992) Macrophage development: II. Early ontogeny of macrophage populations in brain, liver, and lungs of rat embryos as revealed by a lectin marker. Anat Rec 232, 527–550. [DOI] [PubMed] [Google Scholar]
- 15. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER et al. (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, Leboeuf M, Low D, Oller G, Almeida F et al. (2012) Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac‐derived macrophages. J Exp Med 209, 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulz C, Perdiguero EG, Chorro L, Szabo‐Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW et al. (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. [DOI] [PubMed] [Google Scholar]
- 18. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F et al. (2015) Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 518, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC et al. (2015) C‐Myb+ Erythro‐myeloid progenitor‐derived fetal monocytes give rise to adult tissue‐resident macrophages. Immunity 42, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walls JR, Coultas L, Rossant J and Henkelman RM (2008) Three‐dimensional analysis of vascular development in the mouse embryo. PloS One 3, e2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones EAV (2011) The initiation of blood flow and flow induced events in early vascular development. Semin Cell Dev Biol 22, 1028–1035. [DOI] [PubMed] [Google Scholar]
- 22. Kierdorf K and Prinz M (2013) Factors regulating microglia activation. Front Cell Neurosci 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roumier A, Pascual O, Bechade C, Wakselman S, Poncer JC, Real E, Triller A and Bessis A (2008) Prenatal activation of microglia induces delayed impairment of glutamatergic synaptic function. PLoS One 3, e2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arnoux I, Hoshiko M, Mandavy L, Avignone E, Yamamoto N and Audinat E (2023) Adaptive phenotype of microglial cells during the normal postnatal development of the somatosensory “Barrel” cortex. Glia 61, 1582–1594. [DOI] [PubMed] [Google Scholar]
- 25. Swinnen N, Smolders S, Avila A, Notelaers K, Paesen R, Ameloot M, Brône B, Legendre P and Rigo J‐M (2013) Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia 61, 150–163. [DOI] [PubMed] [Google Scholar]
- 26. Nikodemova M, Kimyon RS, De I, Small AL, Collier LS and Watters JJ (2015) Microglial numbers attain adult levels after undergoing a rapid decrease in cell number in the third postnatal week. J Neuroimmunol 278, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hutchins KD, Dickson DW, Rashbaum WK and Lyman WD (1990) Localization of morphologically distinct microglial populations in the developing human fetal brain: implications for ontogeny. Dev Brain Res 55, 95–102. [DOI] [PubMed] [Google Scholar]
- 28. Rezaie P, Dean A, Male D and Ulfig N (2005) Microglia in the Cerebral Wall of the human telencephalon at second trimester. Cereb Cortex 15, 938–949. [DOI] [PubMed] [Google Scholar]
- 29. Monier A, Adle‐Biassette H, Delezoide A‐L, Evrard P, Gressens P and Verney C (2007) Entry and distribution of microglial cells in human embryonic and fetal cerebral cortex. J Neuropathol Exp Neurol 66, 372–382. [DOI] [PubMed] [Google Scholar]
- 30. Verney C, Monier A, Fallet‐Bianco C and Gressens P (2010) Early microglial colonization of the human forebrain and possible involvement in periventricular white‐matter injury of preterm infants. J Anat 217, 436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rezaie P and Male D (1999) Colonisation of the developing human brain and spinal cord by microglia: a review. Microsc Res Tech 45, 359–382. [DOI] [PubMed] [Google Scholar]
- 32. Esiri MM, al Izzi MS and Reading MC (1991) Macrophages, microglial cells, and HLA‐DR antigens in fetal and infant brain. J Clin Pathol 44, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rezaie P (2003) Microglia in the human nervous system during development. Neuroembryology 2, 18–31. [Google Scholar]
- 34. Herbomel P, Thisse B and Thisse C (1999) Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745. [DOI] [PubMed] [Google Scholar]
- 35. Herbomel P, Thisse B and Thisse C (2001) Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M‐CSF receptor‐dependent invasive process. Dev Biol 238, 274–288. [DOI] [PubMed] [Google Scholar]
- 36. Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR et al. (2011) Yap1 acts downstream of α‐catenin to control epidermal proliferation. Cell 144, 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawson LJ, Perry VH, Dri P and Gordon S (1990) Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170. [DOI] [PubMed] [Google Scholar]
- 38. Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM and McColl BW (2016) Microglial brain region−dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19, 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A et al. (2016) New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA 113, E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keren‐Shaul H, Spinrad A, Weiner A, Matcovitch‐Natan O, Dvir‐Szternfeld R, Ulland TK, David E, Baruch K, Lara‐Astaiso D, Toth B et al. (2017) A unique microglia type associated with restricting development of Alzheimer's disease. Cell 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 41. Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, Veras MM, Pereira TF, Leite REP, Möller T et al. (2017) Transcriptomic analysis of purified human cortical microglia reveals age‐associated changes. Nat Neurosci 20, 1162–1171. [DOI] [PubMed] [Google Scholar]
- 42. Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O'Loughlin E, Xu Y, Fanek Z et al. (2017) The TREM2‐APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakanishi A, Kaneko N, Takeda H, Sawasaki T, Morikawa S, Zhou W, Kurata M, Yamamoto T, Akbar SMF, Zako T et al. (2018) Amyloid β directly interacts with NLRP3 to initiate inflammasome activation: identification of an intrinsic NLRP3 ligand in a cell‐free system. Inflamm Regen 38, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G et al. (2019) Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature 566, 388–392. [DOI] [PubMed] [Google Scholar]
- 45. Stratoulias V, Venero JL, Tremblay ME and Joseph B (2019) Microglial subtypes: diversity within the microglial community. EMBO J 38, e101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rio‐Hortega P (1919) “Tercer elemento” de Los Centros Nerviosos. II. Intervencion de la microglia en los procesos patologicos (Cellulas en bastocito y cuerpos granulo‐adiposos). Bol Soc Esp Biol 9, 91–103. [Google Scholar]
- 47. Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F et al. (2014) Environment drives selection and function of enhancers controlling tissue‐specific macrophage identities. Cell 159, 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O'Connor C, Fitzpatrick C, Pasillas MP et al. (2017) An environment‐dependent transcriptional network specifies human microglia identity. Science 356, eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doorn KJ, Brevé JJP, Drukarch B, Boddeke HW, Huitinga I, Lucassen PJ and van Dam A‐M (2015) Brain region‐specific gene expression profiles in freshly isolated rat microglia. Front Cell Neurosci 9, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Haas AH, Boddeke HW and Biber K (2008) Region‐specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia 56, 888–894. [DOI] [PubMed] [Google Scholar]
- 51. Jordão MJC, Sankowski R, Brendecke SM, Sagar N, Locatelli G, Tai Y‐H, Tay TL, Schramm E, Armbruster S, Hagemeyer N et al. (2019) Single‐cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554. [DOI] [PubMed] [Google Scholar]
- 52. Baalman K, Marin MA, Ho TS‐Y, Godoy M, Cherian L, Robertson C and Rasband MN (2015) Axon initial segment–associated microglia. J Neurosci 35, 2283–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wogram E, Wendt S, Matyash M, Pivneva T, Draguhn A and Kettenmann H (2016) Satellite microglia show spontaneous electrical activity that is uncorrelated with activity of the attached neuron. Eur J Neurosci 43, 1523–1534. [DOI] [PubMed] [Google Scholar]
- 54. Ri‐o‐Hortega P (1919) “Tercer elemento” de Los Centros Nerviosos II. Intervenciion de la microglia en los procesos patologicos (Cellulas en bastocito y cuerpos granul‐o‐adiposos). Bol Soc Esp Biol 9, 91–103. [Google Scholar]
- 55. Bertolotto A, Agresti C, Castello A, Manzardo E and Riccio A (1998) 5D4 keratan sulfate epitope identifies a subset of ramified microglia in normal central nervous system parenchyma. J Neuroimmunol 85, 69–77. [DOI] [PubMed] [Google Scholar]
- 56. Bertolotto A, Caterson B, Canavese G, Migheli A and Schiffer D (1993) Monoclonal antibodies to keratan sulfate immunolocalize ramified microglia in paraffin and cryostat sections of rat brain. J Histochem Cytochem 41, 481–487. [DOI] [PubMed] [Google Scholar]
- 57. Hirano K, Ohgomori T, Kobayashi K, Tanaka F, Matsumoto T, Natori T, Matsuyama Y, Uchimura K, Sakamoto K, Takeuchi H et al. (2013) Ablation of Keratan sulfate accelerates early phase pathogenesis of ALS. PLoS One 8, e66969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shigemoto‐Mogami Y, Hoshikawa K, Goldman JE, Sekino Y and Sato K (2014) Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci 34, 2231–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xavier ALR, Kress BT, Goldman SA, Lacerda de Menezes JR and Nedergaard M (2015) A distinct population of microglia supports adult neurogenesis in the subventricular zone. J Neurosci 35, 11848–11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xavier AL, Lima FRS, Nedergaard M and Menezes JRL (2015) Ontogeny of CX3CR1‐EGFP expressing cells unveil microglia as an integral component of the postnatal subventricular zone. Front Cell Neurosci 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen S‐K, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G and Capecchi MR (2010) Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagarajan N, Jones BW, West PJ, Marc R and Capecchi MR (2018) Corticostriatal circuit defects in Hoxb8 mutant mice. Mol Psychiatry 23, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. De S, Van Deren D, Peden E, Hockin M, Boulet A, Titen S and Capecchi MR (2018) Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development 145, dev152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wlodarczyk A, Holtman IR, Krueger M, Yogev N, Bruttger J, Khorooshi R, Benmamar‐Badel A, de Boer‐Bergsma JJ, Martin NA, Karram K et al. (2017) A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J 36, 3292–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bisht K, Sharma KP, Lecours C, Gabriela Sánchez M, El Hajj H, Milior G, Olmos‐Alonso A, Gómez‐Nicola D, Luheshi G, Vallières L et al. (2016) Dark microglia: a new phenotype predominantly associated with pathological states. Glia 64, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hui CW, St‐Pierre A, El Hajj H, Remy Y, Hébert SS, Luheshi GN, Srivastava LK and Tremblay M‐È (2018) Prenatal immune challenge in mice leads to partly sex‐dependent behavioral, microglial, and molecular abnormalities associated with schizophrenia. Front Mol Neurosci 11, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yeh FL, Hansen DV and Sheng M (2017) TREM2, microglia, and neurodegenerative diseases. Trends Mol Med 23, 512–533. [DOI] [PubMed] [Google Scholar]
- 68. Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS, Sutcliffe JG and Carson MJ (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells‐2 on adult murine microglia. J Neurochem 83, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Forabosco P, Ramasamy A, Trabzuni D, Walker R, Smith C, Bras J, Levine AP, Hardy J, Pocock JM, Guerreiro R et al. (2013) Insights into TREM2 biology by network analysis of human brain gene expression data. Neurobiol Aging 34, 2699–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mouton PR, Long JM, Lei D‐L, Howard V, Jucker M, Calhoun ME and Ingram DK (2002) Age and gender effects on microglia and astrocyte numbers in brains of mice. Brain Res 956, 30–35. [DOI] [PubMed] [Google Scholar]
- 71. Schwarz JM, Sholar PW and Bilbo SD (2012) Sex differences in microglial colonization of the developing rat brain. J Neurochem 120, 948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lenz KM, Nugent BM, Haliyur R and McCarthy MM (2013) Microglia are essential to masculinization of brain and behavior. J Neurosci 33, 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bollinger JL, Bergeon Burns CM and Wellman CL (2016) Differential effects of stress on microglial cell activation in male and female medial prefrontal cortex. Brain Behav Immun 52, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guneykaya D, Ivanov A, Hernandez DP, Haage V, Wojtas B, Meyer N, Maricos M, Jordan P, Buonfiglioli A, Gielniewski B et al. (2018) Transcriptional and translational differences of microglia from Male and female brains. Cell Rep 24, 2773–2783.e6. [DOI] [PubMed] [Google Scholar]
- 75. Villa A, Vegeto E, Poletti A and Maggi A (2016) Estrogens, neuroinflammation, and neurodegeneration. Endocr Rev 37, 372–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte‐Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G et al. (2018) Microbiome influences prenatal and adult microglia in a sex‐specific manner. Cell 172, 500–516.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wolf SA, Boddeke HWGM and Kettenmann H (2017) Microglia in physiology and disease. Annu Rev Physiol 79, 619–643. [DOI] [PubMed] [Google Scholar]
- 78. Cuadros MA and Navascués J (1998) The origin and differentiation of microglial cells during development. Prog Neurobiol 56, 173–189. [DOI] [PubMed] [Google Scholar]
- 79. Kawai T and Akira S (2010) The role of pattern‐recognition receptors in innate immunity: update on toll‐like receptors. Nat Immunol 11, 373–384. [DOI] [PubMed] [Google Scholar]
- 80. Kettenmann H, Hanisch U‐K, Noda M and Verkhratsky A (2011) Physiology of microglia. Physiol Rev 91, 461–553. [DOI] [PubMed] [Google Scholar]
- 81. Venegas C and Heneka MT (2017) Danger‐associated molecular patterns in Alzheimer's disease. J Leukoc Biol 101, 87–98. [DOI] [PubMed] [Google Scholar]
- 82. Barclay AN, Wright GJ, Brooke G and Brown MH (2002) CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol 23, 285–290. [DOI] [PubMed] [Google Scholar]
- 83. Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH et al. (2000) Down‐regulation of the macrophage lineage through interaction with OX2 (CD200). Science 290, 1768–1771. [DOI] [PubMed] [Google Scholar]
- 84. Biber K, Neumann H, Inoue K and Boddeke HWGM (2007) Neuronal ‘on’ and ‘off’ signals control microglia. Trends Neurosci 30, 596–602. [DOI] [PubMed] [Google Scholar]
- 85. Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A and Littman DR (2000) Analysis of Fractalkine receptor CX3CR1 function by targeted deletion and Green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim S‐Y, Lee H, Kim H‐J, Bang E, Lee S‐H, Lee D‐W, Woo D‐C, Choi C‐B, Hong KS, Lee C et al. (2011) In vivo and ex vivo evidence for ketamine‐induced hyperglutamatergic activity in the cerebral cortex of the rat: potential relevance to schizophrenia. NMR Biomed 24, 1235–1242. [DOI] [PubMed] [Google Scholar]
- 87. Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A and Schall TJ (1997) A new class of membrane‐bound chemokine with a CX3C motif. Nature 385, 640–644. [DOI] [PubMed] [Google Scholar]
- 88. Pan Y, Lloyd C, Zhou H, Dolich S, Deeds J, Gonzalo J‐A, Vath J, Gosselin M, Ma J, Dussault B et al. (1997) Neurotactin, a membrane‐anchored chemokine upregulated in brain inflammation. Nature 387, 611–617. [DOI] [PubMed] [Google Scholar]
- 89. Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, Streit WJ, Salafranca MN, Adhikari S, Thompson DA et al. (1998) Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1‐expressing microglia. Proc Natl Acad Sci USA 95, 10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. van Beek EM, Cochrane F, Barclay AN and van den Berg TK (2005) Signal regulatory proteins in the immune system. J Immunol 175, 7781–7787. [DOI] [PubMed] [Google Scholar]
- 91. Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YE, Ebert A, Pimenova AA, Ramirez BR, Chan AT et al. (2018) Epigenetic regulation of brain region‐specific microglia clearance activity. Nat Neurosci 21, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Colonna M (2003) TREMs in the immune system and beyond. Nat Rev Immunol 3, 445–453. [DOI] [PubMed] [Google Scholar]
- 93. Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH et al. (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160, 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M and Colonna M (2006) Cutting edge: TREM‐2 attenuates macrophage activation. J Immunol 177, 3520–3524. [DOI] [PubMed] [Google Scholar]
- 95. Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, Kitazawa M, Matusow B, Nguyen H, West BL et al. (2014) CSF1 receptor signaling is necessary for microglia viability, which unmasks a cell that rapidly repopulates the microglia‐depleted adult brain. Neuron 82, 380–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dai X‐M, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V and Stanley ER (2002) Targeted disruption of the mouse colony‐stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120. [DOI] [PubMed] [Google Scholar]
- 97. Michell‐Robinson MA, Touil H, Healy LM, Owen DR, Durafourt BA, Bar‐Or A, Antel JP and Moore CS (2015) Roles of microglia in brain development, tissue maintenance and repair. Brain 138, 1138–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Inoue K (2002) Microglial activation by purines and pyrimidines. Glia 40, 156–163. [DOI] [PubMed] [Google Scholar]
- 99. Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan W‐B and Julius D (2006) The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9, 1512–1519. [DOI] [PubMed] [Google Scholar]
- 100. De Simone R, Ambrosini E, Carnevale D, Ajmone‐Cat MA and Minghetti L (2007) NGF promotes microglial migration through the activation of its high affinity receptor: modulation by TGF‐β. J Neuroimmunol 190, 53–60. [DOI] [PubMed] [Google Scholar]
- 101. Niraula A, Fasnacht RD, Ness KM, Frey JM, Cuschieri SA, Dorfman MD and Thaler JP (2023) Prostaglandin PGE2 receptor EP4 regulates microglial phagocytosis and increases susceptibility to diet‐induced obesity. Diabetes 72, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McMullan SM, Phanavanh B, Guo Li G and Barger SW (2012) Metabotropic glutamate receptors inhibit microglial glutamate release. ASN Neuro 4, e00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Noda M, Nakanishi H, Nabekura J and Akaike N (2000) AMPA–Kainate subtypes of glutamate receptor in rat cerebral microglia. J Neurosci 20, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sivakumar V, Ling E‐A, Lu J and Kaur C (2010) Role of glutamate and its receptors and insulin‐like growth factors in hypoxia induced periventricular white matter injury. Glia 58, 507–523. [DOI] [PubMed] [Google Scholar]
- 105. Bhandage AK and Barragan A (2021) GABAergic signaling by cells of the immune system: more the rule than the exception. Cell Mol Life Sci 78, 5667–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sierra A, Gottfried‐Blackmore A, Milner TA, McEwen BS and Bulloch K (2008) Steroid hormone receptor expression and function in microglia. Glia 56, 659–674. [DOI] [PubMed] [Google Scholar]
- 107. Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, Lafaille JJ, Hempstead BL, Littman DR and Gan W‐B (2013) Microglia promote learning‐dependent synapse formation through BDNF. Cell 155, 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Feinstein DL, Heneka MT, Gavrilyuk V, Russo CD, Weinberg G and Galea E (2002) Noradrenergic regulation of inflammatory gene expression in brain. Neurochem Int 41, 357–365. [DOI] [PubMed] [Google Scholar]
- 109. Hammerschmidt T, Kummer MP, Terwel D, Martinez A, Gorji A, Pape H‐C, Rommelfanger KS, Schroeder JP, Stoll M, Schultze J et al. (2013) Selective loss of noradrenaline exacerbates early cognitive dysfunction and synaptic deficits in APP/PS1 mice. Biol Psychiatry 73, 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Heneka MT, Nadrigny F, Regen T, Martinez‐Hernandez A, Dumitrescu‐Ozimek L, Terwel D, Jardanhazi‐Kurutz D, Walter J, Kirchhoff F, Hanisch UK et al. (2010) Locus ceruleus controls Alzheimer's disease pathology by modulating microglial functions through norepinephrine. Proc Natl Acad Sci USA 107, 6058–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pannell M, Szulzewsky F, Matyash V, Wolf SA and Kettenmann H (2014) The subpopulation of microglia sensitive to neurotransmitters/neurohormones is modulated by stimulation with LPS, interferon‐γ, and IL‐4. Glia 62, 667–679. [DOI] [PubMed] [Google Scholar]
- 112. Liu X and Quan N (2018) Microglia and CNS Interleukin‐1: beyond immunological concepts. Front Neurol 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE and Bateman RJ (2010) Decreased clearance of CNS β‐amyloid in Alzheimer's disease. Science 330, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sarlus H and Heneka MT (2017) Microglia in Alzheimer's disease. J Clin Invest 127, 3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Johnson GVW and Stoothoff WH (2004) Tau phosphorylation in neuronal cell function and dysfunction. J Cell Sci 117, 5721–5729. [DOI] [PubMed] [Google Scholar]
- 116. Wei Y and Li X (2022) Different phenotypes of microglia in animal models of Alzheimer disease. Immun Ageing 19, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang H (2021) Microglia heterogeneity in Alzheimer's disease: insights from single‐cell technologies, Frontiers in synaptic . Neuroscience 13, 773590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mathys H, Davila‐Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X et al. (2019) Single‐cell transcriptomic analysis of Alzheimer's disease. Nature 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gingrich MB and Traynelis SF (2000) Serine proteases and brain damage – is there a link? Trends Neurosci 23, 399–407. [DOI] [PubMed] [Google Scholar]
- 120. Jiang T, Yu J‐T, Hu N, Tan M‐S, Zhu X‐C and Tan L (2014) CD33 in Alzheimer's disease. Mol Neurobiol 49, 529–535. [DOI] [PubMed] [Google Scholar]
- 121. Spillantini MG, Crowther RA, Jakes R, Hasegawa M and Goedert M (1998) α‐Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc Natl Acad Sci USA 95, 6469–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. McGeer PL, Itagaki S, Boyes BE and McGeer EG (1988) Reactive microglia are positive for HLA‐DR in the substantia nigra of Parkinson's and Alzheimer's disease brains. Neurology 38, 1285. [DOI] [PubMed] [Google Scholar]
- 123. Hirsch L, Jette N, Frolkis A, Steeves T and Pringsheim T (2016) The incidence of Parkinson's disease: a systematic review and meta‐analysis. Neuroepidemiology 46, 292–300. [DOI] [PubMed] [Google Scholar]
- 124. Milo R and Kahana E (2010) Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev 9, A387–A394. [DOI] [PubMed] [Google Scholar]
- 125. Miron VE, Boyd A, Zhao J‐W, Yuen TJ, Ruckh JM, Shadrach JL, van Wijngaarden P, Wagers AJ, Williams A, Franklin RJM et al. (2013) M2 microglia/macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat Neurosci 16, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ajami B, Samusik N, Wieghofer P, Ho PP, Crotti A, Bjornson Z, Prinz M, Fantl WJ, Nolan GP and Steinman L (2018) Single‐cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat Neurosci 21, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Foyez T, Takeda‐Uchimura Y, Ishigaki S, Narentuya, Zhang Z, Sobue G, Kadomatsu K and Uchimura K (2015) Microglial Keratan sulfate epitope elicits in central nervous tissues of transgenic model mice and patients with amyotrophic lateral sclerosis. Am J Pathol 185, 3053–3065. [DOI] [PubMed] [Google Scholar]
- 128. Shinjo R, Imagama S, Ito Z, Ando K, Nishida Y, Ishiguro N and Kadomatsu K (2014) Keratan sulfate expression is associated with activation of a subpopulation of microglia/macrophages in Wallerian degeneration. Neurosci Lett 579, 80–85. [DOI] [PubMed] [Google Scholar]
- 129. Wang SB, Wang YY, Zhang QE, Wu SL, Ng CH, Ungvari GS, Chen L, Wang CX, Jia FJ and Xiang YT (2018) Cognitive behavioral therapy for post‐stroke depression: a meta‐analysis. J Affect Disord 235, 589–596. [DOI] [PubMed] [Google Scholar]
- 130. Schaefer GB (2016) Clinical genetic aspects of autism Spectrum disorders. Int J Mol Sci 17, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, Seredenina T, Muhs A, Scheiwe C, Shah MJ et al. (2019) Mapping microglia states in the human brain through the integration of high‐dimensional techniques. Nat Neurosci 22, 2098–2110. [DOI] [PubMed] [Google Scholar]


