Abstract
Translational regulation provides an efficient means to control the localization and production of proteins. The headcase (hdc) mRNA in Drosophila generates two overlapping proteins as a result of translational readthrough of an internal UAA stop codon. This readthrough event is necessary for the function of hdc as a branching inhibitor during tracheal development. By ectopic expression of different Hdc proteins in the trachea, we show that the long Hdc form alone, can function as a potent branching inhibitor whose activity is proportional to its amount. The suppression of termination in the hdc mRNA is not stop-codon dependent, suggesting that the readthrough does not involve codon specific suppressors. We have identified an 80 nucleotide sequence immediately downstream of the UAA, which is necessary and sufficient to confer termination readthrough in a heterologous mRNA. We present a novel mechanism of eukaryotic translational termination suppression that may regulate the amount of functional Hdc.
INTRODUCTION
During Drosophila tracheal development the growing tubules form an elaborate network as a result of primary, secondary and terminal branching events followed by branch fusion (Samakovlis et al., 1996a,b). headcase (hdc) is expressed in a subset of the tracheal fusion cells and inhibits terminal branching of neighbouring tracheal cells. From a single hdc trancsript two peptides are produced as a result of suppression of an internal UAA translation termination codon located in the hdc transcript. In the embryo, the 70 kDa termination product accounts for the majority of the Hdc protein and is four times more abundant than the longer 120 kDa readthrough product. The readthrough event is required for hdc function in the trachea, because expression of a construct containing the full-length cDNA in the tracheal fusion cells can rescue the hdc mutant phenotype, but a deletion construct, lacking the 3′ coding region of the long isoform, does not provide rescuing activity. In addition, ectopic expression of the full-length construct in all tracheal cells inhibits terminal cell sprouting but the deletion construct does not (Steneberg et al., 1998). In this study we show that the long Hdc protein is the main determinant for the tracheal function of the gene and that its branching inhibitory activity is proportional to its amount. We identify the sequences in the hdc mRNA necessary for stop codon readthrough and propose that a new translational readthrough mechanism may regulate the amount of functional Hdc protein.
RESULTS AND DISCUSSION
Hdc readthrough protein is the major branching inhibitor
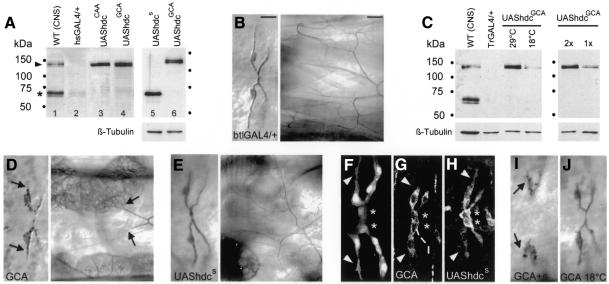
To investigate whether the two Hdc proteins produced operate together or whether the readthrough product alone is responsible for gene function, we made constructs where we replaced the hdc internal UAA stop codon with the codons for either alanine (GCA, UAShdcGCA) or glutamine (CAA, UAShdcCAA). Expression of these constructs in third instar larval salivary glands produced a single protein with a similar size as the endogenous Hdc readthrough protein (Figure 1A). We addressed the function of these Hdc proteins in ectopic expression experiments since we have shown previously that misexpression of full-length hdc cDNA, producing both proteins in all tracheal cells, resulted in suppression of terminal branching (Steneberg et al., 1998). Generalized tracheal expression of UAShdcGCA (UAShdcCAA showed almost identical phenotypes) using the btlGAL4 driver resulted in a dramatic suppression of terminal branching in the dorsal branches of late stage 16 embryos. The average length of the terminal branches, measured as lumen extensions past the terminal cell nucleus, was decreased by 87% (n = 423) compared to control embryos deriving from the driver strain. Continued expression of this construct in third instar larvae showed that several of these branches failed to generate tracheoles (Figure 1B and D). This phenotype was evident in the terminal cells of all tracheal branches in both embryos and larvae (data not shown). In wild-type embryos the terminal cells in the dorsal branches extend long cytoplasmic processes towards target tissues (Figure 1F). These extensions become severely reduced upon generalized tracheal misexpression of UAShdcGCA, suggesting that the failure in lumen growth is a consequence of the failure of the terminal cells to extend cytoplasmic processes (Figure 1G). Thus, the role of hdc in suppressing terminal branching in the trachea can be accounted for by the long product.
Fig. 1. The Hdc readthrough protein is a dose-dependent branching inhibitor. (A and C) Western blot of salivary gland extracts probed with the Hdc monoclonal antibody that recognizes both termination (asterisk) and readthrough (arrowhead) products. In both blots CNS extracts serve as wild-type controls (lane 1). (A) In larvae of the hsGAL4 driver strain a small amount of endogenous product is detected (lane 2) in salivary glands. The UAShdcCAA (lane 3) and the UAShdcGCA constructs produce only the long product (lanes 4 and 6) whereas theUAShdcs generates only the short protein under the control of the same driver (lane 5). Long and short proteins are produced at similar levels as shown by the tubulin loading control in lanes 5 and 6. (C) The amount of HdcGCA is higher at 29°C compared with 18°C due to the increased activity of the GAL4 driver similarly protein levels are higher in strains carrying two copies of the UAShdcGCA (2×) compared with one (1×). Tubulin was used as a loading control. (B, D–J) Dorsal view (anterior to the left) showing the tracheal dorsal branches in late stage 16 embryos (B–E, left panels and F–J) and in third instar larvae (B–E, right panels). (B, D, E, I and J) Embryos were stained to visualize tracheal lumen and terminal cells, in (G and H) with Hdc monoclonal antibody to visualize cell extensions and Hdc protein produced by the transgenes activated by the BtlGAL4 driver. Asterisks in (F), (G) and (H) indicate the fusion cells, where endogenous hdc is normally expressed. In (F), 1-eve-1 embryos were stained for β-galactosidase showing wild-type terminal cell extensions. Ectopic pan-tracheal expression of UAShdcGCA, producing the long Hdc protein, severely reduced terminal lumen (arrows in D), cellular extensions (arrowheads in G) and formation of tracheoles in larvae (arrows in D) compared to controls (B and arrowheads in F). Ectopic tracheal expression of UAShdcs, producing the short Hdc protein (lane 5 in A), caused less reduction in lumen length (E) and cell extension in embryos (H) and no inhibition of tracheoles in larvae (E). Ectopic tracheal coexpression of the two constructs (I) enhanced suppression of lumen extensions (arrows in I). The effect of UAShdcGCA on terminal lumen extension is milder at 18°C (J) compared to its effect at 29°C. Bars in (B) correspond to 10 µm for embryos and 50 µm for larvae.
To test whether the short product may also contribute in hdc function we generated a deletion construct (UAShdcs), where the sequence immediately downstream of the internal UAA stop codon was deleted. In salivary glands, expression of this construct generated a product of similar size as the endogenous short Hdc termination protein (Figure 1A). Ectopic expression of UAShdcs with the same driver also reduced terminal branch lumen length but to a much lower extent, 32% (n = 118), compared with the UAShdcGCA. Terminal cell extensions were also shorter than in wild-type embryos but the formation of tracheoles in third instar larvae expressing UAShdcs in the trachea was not affected (Figure 1E and H). These results suggest that both Hdc proteins can inhibit terminal branching but the longer protein is a much stronger inhibitor since the amount of protein produced by the two constructs both in the trachea and in other tissues is similar (Figure 1A and in situ staining data not shown).
To test whether the expression of the two proteins in the same cells may have a cooperative effect on their activity, we combined UAShdcGCA and UAShdcs and expressed them together with the btlGAL4 driver. In the progeny of these crosses lumen extensions in all terminal cells of the dorsal branches were severely reduced or absent (Figure 1I), and in sporadic severe cases some dorsal branches were missing (data not shown). The tracheal phenotype in embryos expressing both constructs is qualitatively similar to the phenotype produced by the expression of the long product alone, but the reduction of lumen length was >95% (n = 86), suggesting that the presence of both products may have an additive effect. We therefore tested whether the inhibitory function of hdc may be sensitive to the relative amounts of the Hdc product in two ways. We first expressed the long product in the trachea from two copies of UAShdcGCA, thus producing approximately double the amount of the long protein (Figure 1C). These embryos showed slightly stronger tracheal phenotypes (91% reduction of lumen extensions, n = 112) than the ones carrying one copy of the construct and they were comparable to the embryos coexpressing the two different Hdc proteins. We also compared the tracheal phenotypes generated by misexpression of UAShdcGCA at 29 and 18°C. At 18°C the activity of GAL4 is reduced and consequently the UAShdcGCA construct produced less than half the amount of protein than at 29°C (Figure 1C). At 18°C the length of lumen extensions was reduced to 60% (n = 102) compared with the 87% length reduction seen at 29°C (Figure 1C and J). These results argue that the readthrough protein is the major functional product of hdc in the trachea and that the expression level of the long Hdc protein can regulate cell and lumen extensions.
Readthrough is stop-codon independent
To investigate the readthrough mechanism in the hdc mRNA we first tested its specificity for the UAA stop codon. Stop codon dependent readthrough events have been described in both in pro- and eukaryotes, where suppression of a UGA stop codon is mediated by the insertion of selenocystein (Farabaugh, 1997). Also in the Drosophila kelch mRNA the presence of UGA is required to generate the readthrough product presumably by incorporation of selenocystein (Robinson and Cooley, 1997).
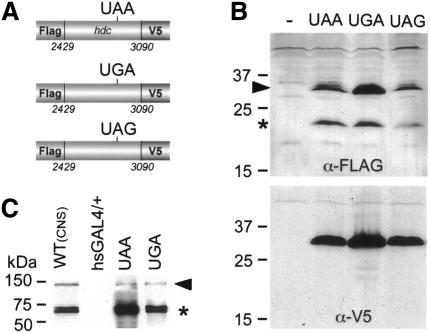
We first defined a crude region of ∼600 nucleotides in the hdc mRNA that was sufficient to promote translational readthrough of the UAA stop codon in cell culture experiments. Expected products from expression constructs were tagged so that identification of termination products, ending at the internal UAA, and readthrough products could be detected with two different antibodies (Figure 2A and Methods). The constructs were transiently transfected in Drosophila mbn2 cells and produced termination and readthrough products visualized by V5 and Flag antibodies on western blots of cell extracts (Figure 2B). To test whether the readthrough event is dependent on the specific stop codon we replaced the TAA stop codon with the TGA and TAG stop codons (Figure 2A). Expression of these constructs also resulted in readthrough of the respective stop codons arguing that the readthrough event does not depend on the stop codon itself. To confirm these findings we replaced the endogenous internal TAA stop codon in the full-length hdc cDNA with TGA and tested its products in larvae. Expression of this construct in salivary glands using the hsGAL4 driver (Brand and Perrimon, 1993) produced termination and readthrough products of similar sizes as the products generated by the expression of the full-length hdc cDNA and by the endogenous hdc gene (Figure 2C). Therefore, we conclude that readthrough in the hdc mRNA is not stop-codon dependent suggesting that specific tRNA suppressors are not required for termination suppression.
Fig. 2. All three stop codons are readthrough in the context of hdc sequences. (A) Schematic drawings of the constructs used to transfect mbn2 cells. The three constructs contain the same part of the hdc cDNA each including a different stop codon. They were expressed under the control of the actin promoter and were tagged at the 5′ end with a Flag epitope and at the 3′ end with the V5 epitope to visualize termination and readthrough products. (B) Western blot from cell extracts probed with FLAG (upper panel) or V5 (lower panel) antibodies. Expression of all three constructs produced two proteins, one termination product (asterisk) and one readthrough product (arrowhead). (C) UGA stop codon is also suppressed in larvae. Western blot probed with the Hdc antibody shows both termination and readthrough (arrow head) products in extracts from wild-type CNS and salivary glands of third instar larvae expressing the wild-type hdc cDNA (20 salivary glands), or a derivative with a stop codon substitution from TAA to TGA (six salivary glands).
cis acting sequences regulate readthrough
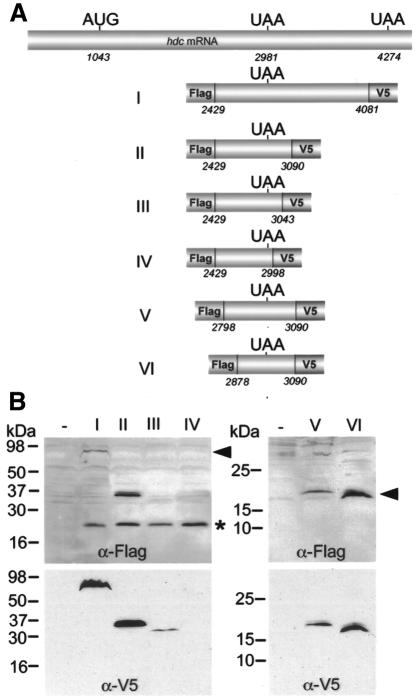
To address the role of sequences surrounding the UAA stop codon in termination readthrough we cloned different segments of the hdc cDNA, including the internal TAA codon, into expression vectors for transient transfection of mbn2 cells (Figure 3A). Subsequently, the tagged termination and readthrough products were detected on western blots from cell extracts with two different antibodies. Expression of construct I and II (Figure 3A) generated both termination and readthrough products of the expected sizes recognized by the Flag and V5 antibodies, indicating that the 109 nucleotides downstream of UAA are sufficient for termination suppression (Figure 3A and B, lanes I and II). We have also noticed that the ratio between the long and short products from these constructs was different, so that readthrough appears more efficient in the shorter construct (construct II), arguing that the region between 3090 and 4081 might negatively regulate termination suppression. Further 3′ deletions extending to 17 nucleotides downstream of the UAA stop codon totally abolished readthrough (Figure 3A and B, lane IV). The termination product from the same constructs was produced and recognized by the Flag antibody (Figure 3B, lane IV), indicating that the region between 2998 and 3090 is required for termination suppression. Expression of construct III, which deletes approximately half of this region, generated a readthrough product but at reduced levels compared to construct II (Figure 3A and B, lanes II and III), suggesting that progressive deletion of this region reduces the readthrough efficiency. 5′ deletions removing up to 103 nucleotides upstream of the stop codon did not affect the readthrough event (Figure 3B, lanes V and VI). Thus, we have identified a 109 nucleotide region located immediately downstream of the UAA stop codon required for termination suppression.
Fig. 3. Mapping of the hdc sequences required for readthrough. (A) Schematic drawing of the different deletion constructs that were used to transfect mbn2 cells. Constructs are tagged at both 5′ and 3′ ends with Flag and V5 epitope, respectively, to detect termination and readthrough products. (B) Termination (asterisk) and readthrough (arrowhead) products were visualized with the Flag antibody (upper panel) and the readthrough product with the V5 antibody (lower panel) on western blots of cell extracts. Construct II contains the minimal sequence required for readthrough. 3′ deletions of this segment reduced the amount of readthrough product (construct III) or completely eliminated it (construct IV). 5′ deletions (constructs V and VI) did not affect readthrough (right panel).
A predicted stem loop structure of 80 nucleotides is sufficient to confer readthrough
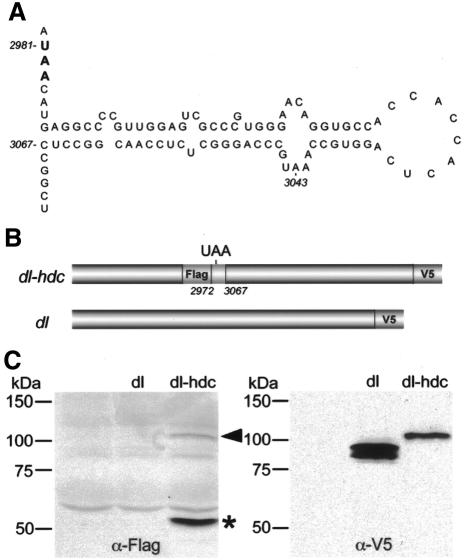
To investigate whether the identified region might promote readthrough by forming a secondary structure that interferes with the translation apparatus we performed secondary structure analysis of hdc sequence from nucleotides 2878 to 3090 (http://bioweb.pasteur.fr/seqanal). The programme predicted a putative hairpin loop engaging 80 nucleotides in the region. This putative loop is located three nucleotides downstream of the UAA stop codon (Figure 4A). To find out whether the region containing the putative hairpin loop was sufficient to confer readthrough in an unrelated mRNA, we cloned the hdc sequence from 2972 to 3067 nucleotides, containing the TAA stop codon, in to the dl cDNA (Figure 4B and Methods). Transfection of mbn2 cells with the hdc-dorsal construct generated termination and readthrough products of the predicted sizes recognized by the Flag and V5 antibodies, respectively (Figure 4C). The readthrough product was slightly larger than the product generated by the dorsal cDNA. The ability of the hdc-dorsal construct to perform readthrough indicates that the hdc sequence from nucleotides 2972 to 3067 in itself is sufficient to regulate suppression of translation termination not only in the hdc mRNA but also in a different context.
Fig. 4. Eighty nucleotides of the hdc sequence are sufficient to confer readthrough in a heterologous RNA. (A) Computer predicted hairpin loop structure of the region immediately downstream of the UAA stop codon including nucleotides 2987–3067. Note that UAA is not included in the loop. (B) Schematic drawing of dl-hdc fusion construct where the hdc sequence from 2972 to 3067 nucleotides has been inserted in the dl cDNA. The dl-hdc construct is tagged with Flag and V5 epitopes to detect termination and readthrough products whereas dl alone is only tagged with V5 epitope. (C) Western blots of mbn2 cell extracts showing expression of the dl-hdc fusion construct where termination (asterisk) and readthrough (arrowhead) products are detected with Flag and V5 antibodies, respectively.
Speculation
hdc is a unique example of a eukaryotic gene, where readthrough of an internal UAA stop codon results in the generation of the functional protein. The readthrough Hdc product acts as a potent branching inhibitor in the trachea and its relative amount in the cell can control the degree of cell and lumen extensions. It is tempting to speculate that its generation by translational termination suppression provides the tracheal cells with a precise control mechanism to regulate the length of cellular extensions and lumen growth. The molecular components that control termination suppression in the hdc mRNA remain to be identified but are unlikely to include specific suppressor tRNAs. Translational frameshifting is also an unlikely mechanism for generating functional Hdc, since both alternative ORFs in the mRNA are too short to account for the molecular weight of the protein. The 80 nucleotide long sequence downstream of UAA can form a secondary structure that may directly interfere with the ribosome, release factors and tRNAs to facilitate termination suppression and in-frame progression, perhaps in a similar way to the termination bypassing events described for viral RNAs (Farabaugh, 1997).
METHODS
Embryo fixation and staining. Embryo fixation and antibody staining was as described (Samakovlis et al., 1996b). The mAb2A12 against a tracheal lumenal antigen was diluted 1:5, the mAb2-161 against D-SRF (from M. Gilman, Ariad Corporation, Boston, MA) was diluted 1:1000, the mAbU33.7 against Headcase was diluted 1:1 and the rabbit anti-β galactosidase antiserum was diluted 1:1500.
Generation of hdc constructs and transgenic strains. To replace the hdc internal TAA stop codon at position 2981 of the published cDNA sequence with TGA or TAG or CAA or GCA by in vitro mutagenesis we used primers containing the TAA replacements and the QuickChangeTM Site-Directed Mutagenesis Kit from Stratagene. The resulting mutated fragments were cloned into the NotI–KpnI sites of the pUAST vector. The UAShdcs construct was generated as a PCR fragment using a 5′-primer starting at position 1042 containing the FLAG epitope: 5′-GCTCTAGAACCACAATGGATTACAAGGATGACGACGATAAGCGTCGCAACAGCAACATC-3′ and 3′-primer 5′-TTTCTAGACAACGGGGCCTCATGTTAT-3′ terminating at position 2998. The resulting PCR fragment was cloned into the pUAST vector using XbaI. All constructs were verified by sequencing and were injected into w1118 embryos to generate transgenic strains following standard procedures (Roberts, 1998).
For the cell culture experiments we cloned the hdc fragments between positions 2429–4081, 2429–3090, 2429–3043 and 2429–2998 by PCR using full-length hdc cDNA as template. We used a common 5′-primer (2429) containing a KpnI site, a Kozak translation initiation sequence and a Flag epitope, 5′-TTAGGTTACCACCACAATGGATTACAAGGATGACGACGATAAGAAGATTGAGGACGAGGGCAACC-3′ together with either of the following 3′-primers containing an EcoRI or XbaI site: 5′-GGAATTCGCTGTTGTCCTGACTGATGC-3′ (4081), 5′-GCTCTAGATGCGATGGTGTTGCTGCTG-3′ (3090), 5′-TTTCTAGATTTGG-CACCTGAGTGGTG-3′ (3043) and 5′-TTTCTAGACAACGGGGCCTCATGTTAT-3′ (2998). For constructs 2878–3090 and 2798–3090 we used the following 5′-primers: 5′-TTAGGTTACCACCACAATGGATTACAAGGATGACGACGATAAGGGAGTGACCTGTCCGTATTG-3′ (for 2878) and 5′-TTAGGTTACCACCACAATGGATTA-CAAGGATGACGACGATAAGGAGCGGTTCAAGTGCAATAA-3′ (for 2798) together with the 3′-primer 3090. PCR products were cloned in-frame with the V5 epitope of the pAc5.1/V5-HisA expression vector (Invitrogen) containing the actin promoter. The constructs containing the TGA and TAG stop codon replacements were derived from PCR fragments using primers 2429 and 3090 cloned into pAc5.1/V5-HisA. For the dl-hdc fusion construct we cloned the hdc sequence 2972–3067 generated by PCR using 5′-primer (containing Flag epitope and SacII site) 5′-TCCCCGCGGATTACAAGGATGACGACGATAAGACGCTCGCATAACATGAGG-3′ (2972) and 3′-primer 5′-TCCCCGCGGAGGCCGTTGGAGAGC-3′ (3067). The PCR product was cut with SacII and cloned into pAc5.1/V5-HisA containing the full-length dl cDNA. All constructs were verified by sequencing.
GAL4 strains and UAS constructs for ectopic expression. The GAL4 drivers used and UAShdc were as described previously (Steneberg et al., 1998). BtlGAL4 was used for expression in the trachea whereas TrGAL4 and hsGAL4 was used for expression in salivary glands of third instar larvae. Embryos carrying one copy of the driver and one copy of the UAS strain were collected for 8 h, aged for 10 h at 29 or 18°C, and then processed as described previously (Steneberg et al., 1998). Several strains of each of the UAShdcGCA, UAShdcCAA and UAShdcTGA constructs were established and tested giving similar results. For the UAShdcs construct we only obtained and analysed one transgenic strain.
Cell culture, protein extractions and western blots. Mbn2 cells were grown in Drosophila Schneider Medium supplemented with 10% fetal calf serum. They were transfected using 2.5 M CaCl2 and harvested after 48 h. Protein extracts and western blots from dissected third instar CNS and salivary glands were processed as reported previously (Sambrook et al., 1989). The anti-Hdc antibody was used at 1:3 dilution (Weaver and White, 1995). Western analyses using the V5 and M5 antibodies were done in parallel with samples from the same extract. The anti-V5 (Invitrogen) and the anti-Flag (M5 from Sigma) were used at 1:5000 and 1:400, respectively. Signals were visualized using Amersham-ECL reagents.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. White for the Hdc mAb. We also thank G. Björk, D. Hultmark and our colleagues at UCMP for useful discussions and comments on the manuscript. This work was supported by grants from NFR and SSF to C.S.
REFERENCES
- Brand A.H. and Perrimon, N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. (1997) Programmed Alternative Reading of the Genetic Code. R.G.Landes Company and Chapman & Hall, Austin, TX, USA.
- Roberts D.B. (1998) Drosophila, A Practical Approach. Oxford University Press, New York, NY.
- Robinson D.N. and Cooley, L. (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development, 124, 1405–1417. [DOI] [PubMed] [Google Scholar]
- Samakovlis C., Hacohen, N., Manning, G., Sutherland, D., Guillemin, K. and Krasnow, M.A. (1996a) Branching morphogenesis of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development, 122, 1395–1407. [DOI] [PubMed] [Google Scholar]
- Samakovlis C., Manning, G., Steneberg, P., Hacohen, N., Cantera, R. and Krasnow, M.A. (1996b) Genetic control of epithelial tube fusion during Drosophila tracheal development. Development, 122, 3531–3536. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- Steneberg P., Englund, C., Kronhamn, J., Weaver, T.A. and Samakovlis, C. (1998) Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the Drosophila trachea. Genes Dev., 12, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver T. and White, R. (1995) headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development, 121, 4149–4160. [DOI] [PubMed] [Google Scholar]