Abstract
We used retrovirus insertion-mediated random mutagenesis and tumor necrosis factor (TNF) selection to generate TNF-resistant lines from L929 cells. The metaxin gene, which encodes a protein located on the outer membrane of mitochondria, was identified to be the gene disrupted in one of the resistant lines. The requirement of metaxin in TNF-induced cell death of L929 was confirmed by the restoration of TNF sensitivity after ectopic reconstitution of metaxin expression. Analysis of the cell death induced by other stimuli revealed that metaxin deficiency-mediated death resistance was selective to certain stimuli. Studies using deletion mutants of metaxin showed that mitochondrial association of metaxin is required for the function of metaxin. Over-expression of truncated metaxin lacking the mitochondria anchoring sequence mimicked metaxin deficiency in wild-type cells. Interfering with metaxin prevented TNF-induced necrotic cell death in L929 cells and apoptosis in MCF-7 cells. Our work has thus defined a novel component in the death pathway used by TNF and some other death stimuli.
INTRODUCTION
Tumor necrosis factor α (TNF) is a proinflammatory cytokine originally identified and purified as a factor that leads to rapid hemorrhagic necrosis of an established tumor (Beutler and Cerami, 1988). TNF can induce cell death in numerous different tumor cell lines (Fiers et al., 1999). Interestingly, the morphological characteristics differ markedly among TNF-treated cell lines: KYM, MCF-7, and PC60 tumor lines die apoptotically in response to TNF treatment (Fiers et al., 1999), whereas L929 and WEHI 164 clone 13 cells die with necrotic phenotypes (Fiers et al., 1999). The typical symptoms of apoptosis such as nuclear condensation and DNA fragmentation are barely detectable in L929 cells after exposure to TNF, and cell death is accompanied by the loss of plasma membrane integrity (Fiers et al., 1999). Moreover, the caspase inhibitor zVAD does not block TNF-induced L929 cell death, but in fact enhances TNF-induced cell killing (Vercammen et al., 1998a).
Both apoptosis and necrosis are initiated by TNF receptor I (TNF-RI) clustering (Aggarwal et al., 1987; Ashkenazi and Dixit, 1998; Kroemer et al., 1998; Fiers et al., 1999). The clustered TNF-RI recruits the death domain (DD)-containing adaptor protein TRADD to its intracellular death domains. One function of TRADD is to bind to FADD, which in turn stimulates caspase-8 autoactivation in the apoptosis pathway. Active caspase-8 either acts via cytochrome c (cyt c) release or by the direct activation of effector caspases to execute apoptosis. Trimerization of the DD of TNF-RI also recruits TRADD in L929 (Fiers et al., 1999). It is known that neither the known pro-apoptotic caspases including caspase-8, nor cyt c release are involved in TNF-induced L929 cell death (Fiers et al., 1999). TNF stimulates reactive oxygen species (ROS) generation in mitochondria and the necrotic death of L929 cells is dependent on mitochondrial ROS generation (Fiers et al., 1999). The intracellular molecules linked to the TNF-induced necrosis are largely unknown.
We designed a functional gene identification procedure and applied it to L929 cells to search for the molecules required for TNF-induced necrotic cell death. As a result, we identified metaxin to be required for TNF-induced death of L929. Metaxin is a protein serendipitously identified in an attempt to establish a mouse model for Gaucher’s disease by introducing an A to G mutation into the glucocerebrosidase gene (GC) using homologous recombination (Armstrong et al., 1997). Because the metaxin gene is located near the GC gene, it was disrupted by this approach which led to the death of mice during early gestation. Bornstein and colleagues have demonstrated a mitochondrial localization and potential protein transport function of metaxin (Armstrong et al., 1997). Here we demonstrate that metaxin deficiency results in TNF resistance of L929 cells. Interfering with metaxin not only inhibited TNF-induced necrosis in L929 cells, but also TNF-induced apoptosis in MCF-7 cells.
RESULTS AND DISCUSSION
Identification of metaxin as a required element in TNF-induced L929 cell death
Retroviral integration can generate null alleles, resulting in diminished endogenous gene expression. Since cells with mutations in genes required for TNF-induced killing may become TNF resistant and be selected by TNF treatment, we used a retrovirus-mediated mutagenesis to study the mechanism of TNF-induced L929 cell death. The retroviral vector we used is shown in Figure 1A. Open bars represent the flanking endogenous sequences from the insertion site. There are three potential gene products that can result from viral insertion: a truncated endogenous gene, GFP, and a neo gene product. GFP protein will be expressed irrespective of the viral integration site because the viral genome contains a complete expression cassette [with a 5′-LTR promoter and viral poly(A) signal sequence close to 3′-LTR]. Hence, GFP expression can be used to assess the efficiency of infection. The expression of two other gene products is dependent on the site of insertion. If integration occurs within an intron, the upstream splicing donor (SD) of the endogenous gene will splice with a vector-encoded splicing acceptor (SA) to generate a truncated endogenous gene. The SD downstream of the neo coding sequence will splice with endogenous SA to generate a fused mRNA capable of expressing full-length Neo protein. If integration occurs within an exon, no splicing will occur upstream of the vector, and the transcript of the endogenous gene will directly fuse with the viral gene to end at the vector poly(A) signal (pA), generating a truncated endogenous gene. The neo transcript will join exons downstream of the insertion site by fusion or splicing events, depending on the presence of a downstream SA. If the insertion occurs within a non-gene region, or within a gene but in a wrong orientation, the neo gene will lack a poly(A) signal and will not be expressed. Thus, G418 resistance can be used to select the cells that had a viral insertion in genes. We term such an event—functional insertion. As only 2% of the total genome encodes genes, a large number of non-functional insertions were eliminated by G418 selection.
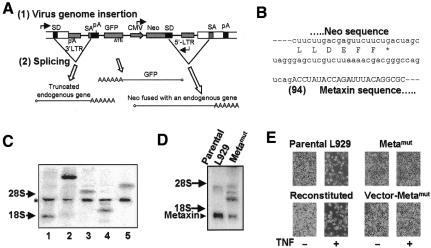
Fig. 1. Identification of metaxin as a gene required for TNF-induced L929 cell death. (A) Diagram of pDisrup1.0 retroviral vector structure and gene products resulting from viral insertion. The retroviral genome inserted within an intron of an endogenous gene was shown. Three transcripts are generated from the endogenous promoter, internal CMV promoter and retroviral 5′-LTR, respectively. Splicing occurs between SDs at the 3′-end of the endogenous exon and SA, as well as between retroviral SD 3′-end to neo gene and endogenous SA at the 5′-end of the downstream endogenous exon. pA, polyadenylation signal. See text for details including the description of exon-insertion. (B) mRNA sequence at the junction between neo and metaxin. Amino acid sequence at the C-terminus of Neo is shown beneath mRNA sequence. Splicing donor is shown in lower case. The number in parentheses shows the beginning of the disrupted metaxin gene relative to its start codon (as +1). (C) Northern blotting of five TNF-resistant clones using 32P-labeled double-strand neo probe. Lane 4 is the metaxin deficiency line. Positions of 28S and 18S ribosomal RNA are indicated by arrows. A common band derived from the viral genome by the 5′-LTR is marked by an asterisk (*). The bands of different size revealed by neo probe are fused mRNA of neo with the disrupted endogenous gene. (D) Northern blotting of metaxin mRNA in wild-type L929 cells and cells with disrupted metaxin gene (Metamut). The triangle shows the position of endogenous metaxin mRNA. (E) Disruption of the metaxin gene renders cells resistant to TNF and reconstitution of metaxin expression restores sensitivity of Metamut cells to TNF. Stable cell lines were generated out of Metamut cells by transfecting with metaxin expression vectors (reconstituted) or with empty vector (vector-Metamut). All three lines, together with parental wild-type L929 cells, were treated with 100 ng/ml TNF for 24 h and photographs were taken under phase contrast.
We infected ∼106 cells and obtained ∼104 G418-resistant clones. Ten TNF-resistant clones were isolated from the G418 resistant clones. 3′-rapid amplification of cDNA ends (RACE) of the mRNA fused with neo was used to identify the disrupted gene in each clone. The metaxin gene was identified as having been disrupted in one of the TNF-resistant lines. The junction sequence of neo–metaxin fusion mRNA is shown in Figure 1B. The pDisrup insertion was mapped to the 5′-portion of the metaxin gene (94 bp from the starting codon) and thus caused a truncation. We analyzed the metaxin-disrupted line (termed Metamut) together with the other four mutation lines by northern blotting using randomly 32P-labeled double-strand DNA probe for the neo gene. As shown in Figure 1C, a common band was seen in all clones (indicated by an asterisk), which is most likely the transcript derived from the viral genome driven by the 5′-LTR. The fused neo mRNA driven by the CMV promoter from the opposite direction was of a different size, indicating that insertion of the viral genome was random. Only one fused gene was found in each of the cell lines including Metamut (lane 4). It is crucial to the efficiency of this protocol that multiple functional insertions in a single cell be avoided, since these events will cause difficulty in determining which disrupted gene is responsible for the functional alteration.
Northern blotting with a metaxin probe showed that the mRNA level of metaxin was significantly reduced in Metamut cells (Figure 1D), suggesting that a functional allele of the metaxin gene was indeed disrupted. In support of this, we found an mRNA with a larger mass detected in the metaxin line with the metaxin probe, indicating a probable metaxin-vector fusion product.
The cell viability after TNF treatment of wild-type parental L929, Metamut, Metamut cells transfected with metaxin expression vector (termed reconstituted), and Metamut cells transfected with empty vector (vector-Metamut), is shown in Figure 1E. Parental L929 cells were very sensitive to TNF-induced cell killing, while Metamut cells were resistant to TNF-induced cell death. Expression of metaxin in Metamut cells regained the TNF sensitivity to almost the same level as wild-type L929, while vector control had no effect. Thus metaxin is required for TNF-induced L929 cell death.
The TNF resistance of the metaxin-deficient line
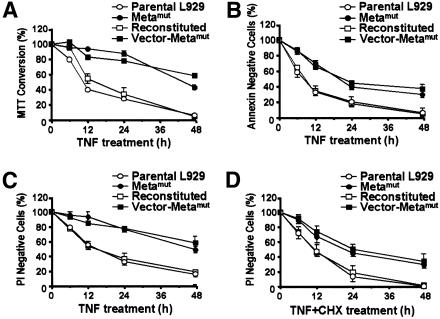
To further evaluate TNF resistance in the Metamut line, we analyzed the TNF sensitivity of parental, Metamut, reconstituted, and vector-Metamut lines using several different criteria. Conversion of the yellow 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to the dark-blue formazan measures the activity of various dehydrogenases and quantifies the reductive capacity of the cell. This assay has been widely applied to the measurement of the cytolytic response to TNF. We used the MTT assay to compare TNF-induced cell death in the four cell lines (Figure 2A). Metamut and vector-Metamut lines had a smaller reduction in their capacity to convert MTT compared to the parental and reconstituted lines, supporting the conclusion that metaxin mutation promotes resistance to TNF-induced cell death.
Fig. 2. TNF resistance in Metamut cells was demonstrated by different assays. Wild-type parental L929 cells (open circle), Metamut cells (filled circle), reconstituted cells (open square), and vector control cells (filled square) were treated with TNF or TNF + cycloheximide (CHX, 0.3 µg/ml) for different periods of time. Cell viability was measured by MTT assay (A), annexin staining (B), or PI staining (C and D).
Translocation of phosphatidylserine (PS) from the inner part of the plasma membrane to the outer layer is a common early event in apoptosis. Annexin staining of PS was also observed in TNF-treated L929 and may represent a change in TNF-induced necrosis of L929 (Fiers et al., 1999). Uptake of propidium iodide (PI) is an indicator of a loss in cell-membrane integrity which is widely used as a measurement of cell death. We used annexin and PI staining to analyze the death profile of the four lines in response to TNF treatment. As shown in Figure 2B and C, metaxin deficiency reduced TNF-induced annexin V and PI staining. Ectopical expression of metaxin in Metamut cells restored TNF sensitivity.
To address whether new protein synthesis is involved in metaxin mutation-mediated TNF resistance, we treated cells with TNF in the presence of the translation inhibitor cycloheximide (CHX). CHX enhanced cell killing by TNF in both normal and metaxin-deficient cells (Figure 2D and C). As observed without CHX, a higher survival rate was seen in metaxin-deficient cells, suggesting that metaxin mutation-mediated death resistance is independent from new protein synthesis.
Metamut line is selectively resistant to some death stimuli
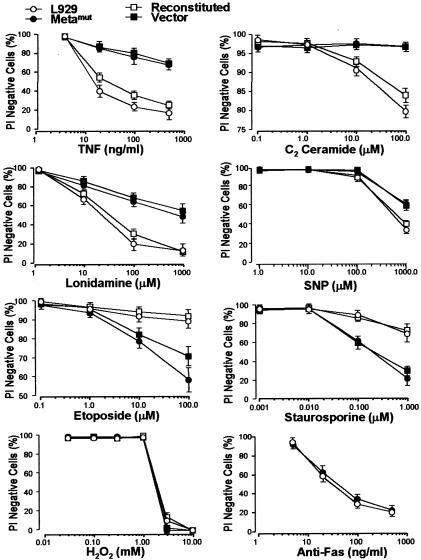
To establish whether Metamut is selectively resistant to different death triggers, we examined its sensitivity to several death stimuli. As shown in Figure 3, Metamut was resistant to TNF-, ceramide-, lonidamine- and sodium nitroprusside (SNP)-induced cell death. In contrast, Metamut was equally sensitive to H2O2-induced cell killing and was even more sensitive to staurosporine and etoposide. Both parental and Metamut lines were insensitive to dexamethasone, serum starvation, UV- and γ-irradiation (data not shown). Expression of Fas renders L929 cells sensitive to Fas ligation-induced cell killing (Vercammen et al., 1998b). We transfected fas cDNA into both parental and Metamut cells and selected the clones that express Fas as described (Vercammen et al., 1998b). Anti-Fas antibody treatment caused very quick cell death within 3 h, which was not influenced by metaxin mutation (Figure 3, bottom right panel). The different sensitivities of Metamut cells to different death stimuli supported the idea that there are multiple ways of cell death (Fiers et al., 1999) and demonstrated that metaxin mutation-mediated TNF resistance is not due to a general promotion of cell survival, but an impairment of the death pathway used by TNF.
Fig. 3. The sensitivity to cell death of metaxin-deficient cells was decreased to certain death stimuli but increased to others. Wild-type parental L929 cells (open cirle), Metamut cells (closed cirle), reconstituted cells (open square), and vector control cells (filled square) were treated with different concentrations of TNF, C2 ceramide, SNP, lonidamine, H2O2, etoposide or staurosporine for 24, 24, 48, 4, 8, 24 and 24 h, respectively. Parental and Metamut cells transfected with fas expression plasmid were treated with anti-Fas antibody (CH-11) for 3 h. Cell viability was measured by PI staining.
Mitochondrial association is essential for metaxin function
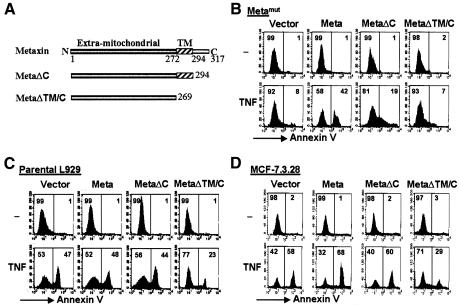
Metaxin is a protein of 317 amino acids (aa) with an extra-mitochondrial domain (aa 1–271), a transmembrane domain (aa 272–294) and an Aps/Glu rich C-terminal domain inside the outer membrane of mitochondria (Figure 4A) (Armstrong et al., 1997). Deletion of the C-terminal domain (ΔC) did not affect the mitochondrial location of metaxin, while further deletion of transmembrane domain (ΔTM/C) disrupts its mitochondrial localization (Armstrong et al., 1997). We examined whether the C-terminal intra-mitochondrial domain and the transmembrane domain are required for the function of metaxin in TNF-induced cell death. As shown in Figure 4B, full-length metaxin reconstituted TNF sensitivity of Metamut cells. While MetaΔC had a partial reconstitution effect, MetaΔTM/C did not restore the TNF sensitivity in the Metamut line. We also over-expressed wild-type and truncated metaxin mutants in parental L929 cells. metaxin and MetaΔC did not have a significant effect on TNF-induced parental L929 cell death (Figure 4C); however, transfection with MetaΔTM/C led to TNF resistance in the parental cells (Figure 4C). The dominant-negative effect by the over-expression of MetaΔTM/C lacking its mitochondria-targeting sequence is likely to be elicited by binding to metaxin-interacting proteins and subsequently disrupting the function of the complex. Since MetaΔTM/C can act as a dominant-negative mutant, we tested whether over-expression of MetaΔTM/C in MCF-7 and MCF-7.3.28 [a cell line generated from MCF-7 by transfection of caspase-3 cDNA (Janicke et al., 1998)] cells influenced TNF-induced apoptosis. Indeed, MetaΔTM/C expression inhibited TNF-induced MCF-7 and MCF-7.3.28 cell death (Figure 4D and data not shown). Thus, metaxin is required for TNF-induced apoptosis and necrosis.
Fig. 4. Mitochondrial anchor is required for metaxin function. (A) Schematic representation indicates that metaxin is 317 aa in length, and its N-terminal extramitochondrial region (aa 1–272), transmembrane domain (TM, aa 272–294), and C-terminal region are inside the outer membrane (aa 295–317). Also shown here are the two deletion mutants of metaxin: MetaΔTM/C and MetaΔC. (B) MetaΔTM/C mutant fails to restore the sensitivity of Metamut cells to TNF, as assayed by annexin staining. Metamut cells were stably transfected with the empty vector, metaxin, MetaΔTM/C, or MetaΔC, respectively, and were treated with or without TNF. Cells were then analyzed for annexin staining using flow cytometry as above. (C) MetaΔTM/C exerts a dominant-negative effect on parental L929 cells in TNF sensitivity. (D) MetaΔTM/C expression leads to resistance to TNF-induced cell death in MCF-7.3.28 cells.
Since free radical generation in mitochondria has an essential role in TNF-induced L929 cell death, we measured mitochondrial ROS levels in TNF-treated Metamut and parental lines. The ROS levels of Metamut and parental cells were similar at rest; ROS induction profiles in response to TNF stimulation were also comparable over a period of 24 h (data not shown). In addition, similar inhibition of TNF-induced cell death by the free radical scavenger butylated hydroxyanisole (BHA) was observed between the parental and Metamut cells; GSH/GSSG contents are also similar between these two lines (data not shown). Hence, metaxin should function at a position downstream of free radical generation.
Cell death is controlled by both death and survival pathways. We have determined by using electro-mobility shift assay and reporter gene assay that TNF-induced NF-κB activation was comparable between the parental and Metamut cells (data not shown). The PI3K-Akt pathway is another well-studied survival pathway, but its role in TNF-induced cell death is unclear. We found that PI3K inhibitor LY294002 did not enhance, but rather inhibited TNF-induced L929 cell death (data not shown). The effect of LY294002 in both parental and metaxin-mutated cells was the same (data not shown). In addition, TNF did not induce any detectable change in Akt phosphorylation in either parental or metaxin-mutated cells. Thus, we did not have evidence to suggest that the survival pathway was affected by metaxin mutation at this moment.
An updated database search revealed that three proteins from Caenorhabditis elegans, Drosophila and fission yeast, shared sequence similarity with metaxin in the extra-mitochondrial domain, transmembrane domain, and Asp/Glu rich C-terminal residues. The hypothetical protein F39B2.11 from C. elegans (DDBJ/EMBL/GenBank accession No. T21992), CG9393 gene product from Drosophila (AAF54402), and a metaxin homolog from fission yeast (T40446), share 34, 30 and 27% identity, respectively, with metaxin in their extra-mitochondrial domain. The function of these proteins has not been studied. Metaxin also shares a low degree of sequence similarity with TOM37, a yeast protein which is located in the mitochondrial outer membrane and is a receptor subunit for general import pore (GIP) (Gratzer et al., 1995). Armstrong et al. (1997) found that anti-metaxin antibody, when preincubated with mitochondria, partially inhibited the uptake of pre-adrenodoxin into mitochondria. Thus, metaxin was proposed to be a component of a pre-protein import complex in the outer mitochondrial membrane. However, the high degree of identity (24%) only occurs between the N-terminal halves of TOM37 and metaxin; TOM37 has an N-terminal mitochondrial targeting sequence, while metaxin has a C-terminal targeting sequence (Armstrong et al., 1997). We found the TOM37-disrupted yeast strain obtained from Dr N. Pfanner was as sensitive as the parental strain to Bax-induced cell death (data not shown). Thus, metaxin may have a different function from that of TOM37. Supporting this, anti-metaxin 2 antibody did not have an effect on pre-adrenodoxin uptake (Armstrong et al., 1999). Despite the function of metaxin in pre-protein import still needing further investigation, the possibility that the TNF resistance in the Metamut line is due to inefficient protein transport into mitochondria cannot be completely excluded. However, we have found no difference between parental and metaxin-mutant cells in ATP levels, mitochondrial succinate dehydrogenase enzyme complex (complex II) activity, creatine kinase activity, mitochondrial quantity or the major protein amount and composition in the mitochondria (data not shown). The selective resistance of metaxin-mutant cells to certain death stimuli, but not to others (Figure 3), further supports the idea that metaxin deficiency-mediated death resistance does not result from a generalized deleterious effect of cellular physiology, but a specific defect of a cell death pathway used by TNF. The early embryonic lethality (<9 days) in mice lacking metaxin demonstrates a requirement of metaxin in development, and may be caused, at least in part, by an impaired process of cell death.
METHODS
Cell culture. L929 murine fibrosarcoma cells from ATCC were cultured in RPMI-1640 media as described (Vercammen et al., 1998a). Stable cell lines derived from L929 cells were established either by retrovirus infection or plasmid transfection by GenePorterTM transfection reagent (GTS Inc., San Diego, CA). Selection of transfected clones was performed by adding 1 mg/ml G418 or 10 µg/ml blasticidin S (Invitrogen, Carlsbad, CA) to the culture medium.
Plasmid construction. The pDisrup retroviral vector was constructed based upon MMLV retroviral vector pLNCX as backbone. The splicing donor and acceptor were designed according to human adenovirus type 2 major late mRNA intron sequence. Details of the plasmid as well as its construction are available upon request. metaxin and fas cDNA were subcloned into the expression vector pcDNA6 (Invitrogen) with blasticidin resistance. Metaxin truncation was made as described by Armstrong et al. (1997).
Northern blotting. The entire coding region of the G418-resistant gene was used as a probe in the detection of neo-fusion mRNA. metaxin cDNA was used for detecting metaxin mRNA levels in L929 and Metamut lines. The probes were labeled by Prime-It kit (Stratagene, La Jolla, CA) with [α-32P]dCTP.
3′-RACE. The portion of endogenous gene fused with neo gene was amplified by a 3′-RACE technique. Total RNA was isolated and reverse transcription was performed with an RT primer (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGC(T)17-3′). A nested PCR was performed with the RT product by separately using primers P1/Q1 (5′-ATGGGCTGACCGCTTCCT-3′/5′-CCAGTGAGCAGAGTGACG-3′) and P2/Q2 (5′-GACGAGTTCTTCTGACTAGCTAG-3′/5′-GAGGACTCGAGCTCAAGC-3′). P1 and P2 are located in the neo resistance gene while Q1 and Q2 are on the anchor sequence of QT. The PCR fragments were subcloned into the TA-cloning vector (Stratagene) and sequenced.
Cell death/survival assays. Mouse recombinant TNF was used with a concentration of 100 ng/ml unless otherwise indicated. Plasma membrane integrity was determined according to the exclusion of PI (Vercammen et al., 1998a). Annexin V-FITC kit (Roche Molecular Biochemicals) was used for measuring PS flipping according to the manufacturer’s protocol, and MTT assay was carried out as described previously (Vercammen et al., 1998a).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Alan G. Porter for MCF-7.3.28 cells and Dr Nikolaus Pfanner for yeast strains and Ms Janet V. Kuhns for excellent secretarial assistance. This is publication No. 13864-IMM from the Department of Immunology, The Scripps Research Institute, La Jolla, CA. This work was supported by grants from the National Institutes of Health and the California Cancer Research Program.
REFERENCES
- Aggarwal B.B., Aiyer, R.A., Pennica, D., Gray, P.W. and Goeddel, D.V. (1987) Human tumour necrosis factors: structure and receptor interactions. Ciba Found. Symp., 131, 39–51. [DOI] [PubMed] [Google Scholar]
- Armstrong L.C., Komiya, T., Bergman, B.E., Mihara, K. and Bornstein, P. (1997) Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem., 272, 6510–6518. [DOI] [PubMed] [Google Scholar]
- Armstrong L.C., Saenz, A.J. and Bornstein, P. (1999) Metaxin 1 interacts with metaxin 2, a novel related protein associated with the mammalian mitochondrial outer membrane. J. Cell Biochem., 74, 11–22. [PubMed] [Google Scholar]
- Ashkenazi A. and Dixit, V.M. (1998) Death receptors: signaling and modulation. Science, 281, 1305–1308. [DOI] [PubMed] [Google Scholar]
- Beutler B. and Cerami, A. (1988) Tumor necrosis, cachexia, shock, and inflammation: a common mediator. Annu. Rev. Biochem., 57, 505–518. [DOI] [PubMed] [Google Scholar]
- Fiers W., Beyaert, R., Declercq, W. and Vandenabeele, P. (1999) More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene, 18, 7719–7730. [DOI] [PubMed] [Google Scholar]
- Gratzer S. et al. (1995) Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol., 129, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke R.U., Sprengart, M.L., Wati, M.R. and Porter, A.G. (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem., 273, 9357–9360. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Dallaporta, B. and Resche-Rigon, M. (1998) The mitochondrial death/life regulator in apoptosis and necrosis. Annu. Rev. Physiol., 60, 619–642. [DOI] [PubMed] [Google Scholar]
- Vercammen D., Beyaert, R., Denecker, G., Goossens, V., Van Loo, G., Declercq, W., Grooten, J., Fiers, W. and Vandenabeele, P. (1998a) Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med., 187, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D., Brouckaert, G., Denecker, G., Van de Craen, M., Declercq, W., Fiers, W. and Vandenabeele, P. (1998b) Dual signaling of the Fas receptor: initiation of both apoptotic and necrotic cell death pathways. J. Exp. Med., 188, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]