Abstract
Expression of the PrfA-controlled virulence gene hly (encoding the pore-forming cytolysin listeriolysin) is under negative regulation by readily metabolized carbon sources in Listeria monocytogenes. However, the hyperhemolytic strain NCTC 7973 exhibits deregulated hly expression in the presence of repressing sugars, raising the possibility that a defect in carbon source regulation is responsible for its anomalous behavior. We show here that the activity of a second glucose-repressed enzyme, α-glucosidase, is 10-fold higher in NCTC 7973 than in 10403S. Using hly-gus fusions, we show that the prfA allele from NCTC 7973 causes deregulated hly-gus expression in the presence of sugars in either the wild-type or the NCTC 7973 background, while the 10403S prfA allele restores carbon source regulation. However, the prfA genotype does not affect the regulation of α-glucosidase activity by repressing sugars. Of the two mutational differences in PrfA, only a Gly145Ser change is important for regulation of hly-gus. Therefore, NCTC 7973 and 10403S have genetic differences in at least two loci: one in prfA that affects carbon source regulation of virulence genes and another in an unidentified gene(s) that up-regulates α-glucosidase activity. We also show that the decrease in pH associated with utilization of sugars negatively regulates hly-gus expression, although sugars can affect hly-gus expression by another mechanism that is independent of pH.
Listeria monocytogenes is a gram-positive, facultative intracellular pathogen and is an important source of food-borne infection in humans (12). Several of the genes required for the pathogenesis of the bacterium are located on a 10-kb region of the chromosome. These genes include hly (encoding listeriolysin O, a thiol-activated cytolysin), plcA and plcB (encoding phosphatidylinositol-specific and phosphatidylcholine-specific lecithinases, respectively), actA (encoding a protein required for actin-based motility inside host cells), and mpl (encoding a metalloprotease involved in the maturation of the plcB gene product) (23, 34, 43). Expression of these genes is controlled by the virulence regulator PrfA. PrfA is a site-specific DNA-binding protein that recognizes the “PrfA box,” a 14-bp region of dyad symmetry in its target promoters (8, 14, 15, 28, 29). The prfA gene is also part of the virulence gene cluster and lies downstream of the plcA gene. It is expressed both as a monocistronic message from two promoters in the plcA-prfA intergenic region and as a bicistronic plcA-prfA message from the plcA promoter. This results in a positive feedback loop, since the expression of prfA is also up-regulated when it activates transcription from the upstream plcA promoter (7). PrfA has sequence similarity with cyclic AMP receptor protein (CRP), the global regulator of catabolite control in Escherichia coli, and significant structural and functional homology of PrfA with members of the CRP/Fnr family has been demonstrated by site-directed mutagenesis studies with the putative DNA-binding domain of PrfA (25, 41).
Several environmental signals regulate the expression of virulence genes in L. monocytogenes, including temperature (27), growth phase (29), composition of the medium (39), and the disaccharide cellobiose (32). It was reported recently that the effect of cellobiose on hly expression is not unique and that several utilizable sugars can down-regulate hly expression (30). Furthermore, while the presence of sugars resulted in over 50-fold down-regulation of hly, there was no detectable change in the level of PrfA protein itself. Therefore, it was hypothesized that regulation of hly expression by sugars is an aspect of global catabolite regulation rather than signature molecule-mediated signal transduction unique to cellobiose (30). Surprisingly, while the effect of utilizable sugars on hly expression was seen in three wild-type natural isolates, strain NCTC 7973 did not exhibit the same phenotype. In this strain, only cellobiose was found to repress virulence gene expression, leading to the suggestion that L. monocytogenes has at least two sugar-sensing pathways and that NCTC 7973 is a partially deregulated variant with a defect in some aspect of carbon source regulation. Ripio et al. showed recently that utilization of the carbohydrate glucose-1-phosphate in L. monocytogenes is coordinately regulated with other virulence factors and is dependent on PrfA (37). These results suggest the existence of important links between regulation of utilizable carbon sources and expression of virulence genes in L. monocytogenes and imply that coordinate regulation of these pathways may be a critical aspect of the pathogenicity and virulence of the bacterium.
While there has been much interest recently in sugar uptake mechanisms in L. monocytogenes (10, 33), catabolite regulation in this organism has never been studied. It is not known which genes are under catabolite regulation and what the mechanism of their regulation in Listeria may be. In this report, we establish that the metabolic enzyme α-glucosidase is under catabolite control in L. monocytogenes. We show that in NCTC 7973, a strain that exhibits high-level expression of hly, the activity of α-glucosidase also is over 10-fold higher than in the wild-type strain 10403S. However, the elevated activity of α-glucosidase and the sugar-insensitive expression of hly-gus in NCTC 7973 are due to distinct genetic defects. Furthermore, we show that the decrease in pH associated with utilization of sugars can itself down-regulate hly-gus expression. However, sugars affect hly-gus expression even if the pH is stringently controlled during growth, suggesting that the regulation of virulence genes by carbon sources involves both pH-dependent and pH-independent components.
MATERIALS AND METHODS
Bacterial strains.
The L. monocytogenes strains used in this work are listed in Table 1. E. coli DH5α mcr[φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 mcr] (Gibco BRL) was used for the construction of plasmids. This strain was grown in Luria-Bertani (LB) medium, and ampicillin was added at a concentration of 100 μg ml−1 for selection.
TABLE 1.
L. monocytogenes strains used in this work
| Strain | Genotype or description | Source or reference |
|---|---|---|
| 10403S | Laboratory strain, serotype 1 | 35 |
| NCTC 7973 | Laboratory strain, serotype 1/2a | 30 |
| LO28 | Laboratory strain, serotype 1/2c | 45 |
| EGD | Laboratory strain, serotype 1/2a | 26 |
| AML73 | 10403S hly-gus-neo | 31 |
| JB77 | NCTC 7973 hly-gus-neo | This work |
| JB136 | AML73 prfA::prfA10403S | This work |
| JB137 | JB77 prfA::prfA10403S | This work |
| JB138 | AML73 prfA::prfA7973 | This work |
| JB139 | JB77 prfA::prfA7973 | This work |
Cultivation of bacteria.
L. monocytogenes strains were grown on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.) or LB medium. Overnight cultures were grown in LB medium buffered with 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0) at 37°C with aeration for 12 to 15 h. Unless otherwise specified, overnight cultures were diluted 1:25 into fresh LB medium buffered with 100 mM MOPS (pH 7.4 for β-glucuronidase assay and pH 7.0 for α-glucosidase assay). Where indicated, filter-sterilized sugar supplements were added to cultures at a final concentration of 25 mM. Growth of cultures was monitored at a wavelength of 600 nm on a Shimadzu UV 1201 spectrophotometer. pH measurements were made on a digital ionalyzer (model 601A; Orion Research). For experiments measuring the effect of pH, LB medium was buffered with 100 mM MOPS (pH 7.4 to 6.5) or 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.0).
DNA sequencing.
To sequence the entire coding region of the prfA gene, a 1,039-bp DNA fragment was PCR amplified from each of the four L. monocytogenes strains by using primers PrfA1R (5′ AACCTCGGTACCATATACTAACTCT 3′) and PrfA4L (5′ GTACGCGTTCTAGAAAATGCTTCT 3′). DNA sequencing was carried out by the method of Sanger et al. (40) with the fmol thermal cycling sequencing system (Promega). Custom-synthesized oligonucleotides were purchased from the University of Georgia Molecular Genetics Facility or from DNAgency, Aston, Pa. Sequencing reactions were carried out on both strands of the PCR product amplified from strains 10403S and NCTC 7973 and on one strand of the product from strains EGD and LO28 (except at the 3′ end of the prfA coding sequence, where both strands were sequenced).
In vitro manipulations of DNA.
Restriction analyses and cloning were done by standard techniques as described before (1). Enzymatic reagents were purchased from New England Biolabs or Boehringer Mannheim and used as specified by the manufacturer. Amplification of DNA by PCR was carried out as previously described (19).
Construction of plasmids and strains for the prfA allele exchange.
The E. coli-L. monocytogenes shuttle vector pCON1 was used for cloning and strain construction. pCON1 was constructed by ligating EcoRI-ScaI-digested pUC18 with pKSV5-T (2) that had been digested with EcoRI and ScaI. pCON1 has a temperature-sensitive origin of replication, pE194ts, and a chloramphenicol resistance (Cmr) gene for selection in gram-positive bacteria. A 1,039-bp prfA fragment was amplified from either 10403S or NCTC 7973 chromosomal DNA with the primers PrfA1R and PrfA4L. There is an EcoRI site in the prfA structural gene (GenBank accession no. X61210), and the 3′ primer PrfA4L has an XbaI cloning site engineered into it. Therefore, digestion of the amplified product with EcoRI and XbaI resulted in a 790-bp prfA fragment lacking the promoter region and the first 19 nucleotides of the prfA coding region. This prfA fragment was ligated into EcoRI-XbaI-digested pCON1 and transformed into E. coli with ampicillin selection, resulting in plasmids pCON1-ΔprfA-10403S and pCON1-ΔprfA-7973. The plasmids were transformed into the conjugation donor strain E. coli S17-1 (44) and conjugated into either AML73 (10403S hly-gus-neo) or JB77 (NCTC 7973 hly-gus-neo) as described previously (2), except that cells were washed once in BHI medium and mating spots were incubated overnight at 30°C. To force chromosomal integration of the vectors, transconjugants were propagated in BHI medium with chloramphenicol (5 μg ml−1) selection for 3 h at 30°C and then shifted to 41°C for another 3 h. Appropriate dilutions were plated out on BHI agar containing chloramphenicol at 5 μg ml−1 and incubated for 2 to 3 days at 41°C. Integration of the vector into the chromosome and the two nucleotide changes in the prfA sequence were confirmed by PCR amplification of the gene and sequencing of the PCR products.
Construction of hly-gus fusions.
hly-gus fusion strains in either the NCTC 7973 or 10403S background were constructed with the vector pCON1-HGNH (31). pCON1-HGNH has a gusA gene (encoding β-glucuronidase) from E. coli (22) and a neomycin resistance (Nmr) cassette (20), flanked by a 489-bp hly fragment (5′ end) and a 397-bp hly-mpl fragment (3′ end) on a pCON1 vector backbone. Shifting of L. monocytogenes strains carrying the plasmid to the nonpermissive temperature (41°C) with chloramphenicol selection resulted in the chromosomal integration of pCON1-HGNH by homologous recombination between the 5′ hly fragment and the wild-type hly allele on the chromosome. These clones were Nmr and Cmr and were nonhemolytic on blood agar plates. Overnight cultures of these merodiploid strains were diluted 1:800 in BHI with only neomycin selection at the permissive temperature (30°C) to select for cells with spontaneous excision of the plasmid. To bring about vector curing after excision of the integrated construct, 1 ml of the saturated culture was diluted into 100 ml of BHI with 5 μg of neomycin ml−1 and incubated with aeration at the nonpermissive temperature to stationary phase. Appropriate dilutions were plated onto BHI plates with 5 μg of neomycin ml−1 at 41°C. Individual colonies were then patched onto neomycin, chloramphenicol, or blood agar plates. Excision of the plasmid via homologous recombination on the 3′ end of the chromosomal hly-mpl genes resulted in clones that had chromosomal hly-gus fusions and were Nmr, Cms, and Hly−. These clones were tested for β-glucuronidase activity on plates containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-GlcA) and confirmed by Southern blot hybridization.
Preparation of cell lysates.
Samples (10 ml) were collected by centrifugation and washed once with an equal volume of the assay buffer (50 mM potassium phosphate buffer for α-glucosidase and 50 mM sodium phosphate buffer for β-glucuronidase). The cells were resuspended in 2 ml of the same buffer and lysed by sonication on ice three times for 30 s each. The suspension was clarified by centrifugation, and up to 0.05 ml of the supernatant was used for assaying enzyme activity. Protein concentrations in cell lysates were determined by the method of Bradford (6), using a Bio-Rad protein assay, with bovine serum albumin as the standard.
Enzyme assays.
α-Glucosidase activity was assayed by using p-nitrophenylglucoside as a substrate essentially as described previously (9), except that the increase in absorbance was monitored at 405 nm on a Shimadzu UV-1201 spectrophotometer. β-Glucuronidase activity was assayed as described previously (21), except that the increase in p-nitrophenol absorbance was monitored at 405 nm.
RESULTS AND DISCUSSION
Utilization of sugars and catabolite regulation in NCTC 7973.
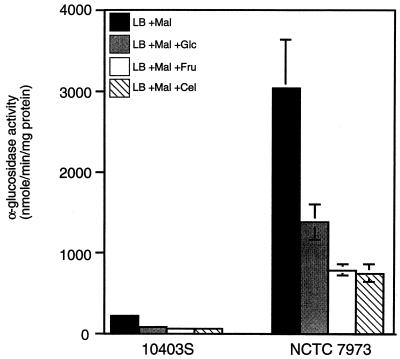
NCTC 7973 is a partially deregulated mutant in which utilizable sugars other than cellobiose do not affect virulence gene expression (30). To test the possibility that NCTC 7973 is generally defective in catabolite regulation, we examined the activity of a metabolic enzyme, α-glucosidase, which is subject to catabolite regulation in other gram-positive bacteria (13, 46). Specific activity of the enzyme in exponentially growing cells was measured. In 10403S, α-glucosidase activity was inducible by maltose (data not shown), and addition of glucose, fructose, or cellobiose to the cultures resulted in approximately fourfold repression of α-glucosidase (Fig. 1). Surprisingly, the specific activity of α-glucosidase was over 10-fold higher in NCTC 7973 than in 10403S, suggesting that a major difference between these two strains might be related to central pathways of catabolite repression. Addition of either glucose, fructose, or cellobiose resulted in a two- to fourfold repression of α-glucosidase activity in NCTC 7973, although the levels still remained severalfold higher than in the wild-type (10403S) control.
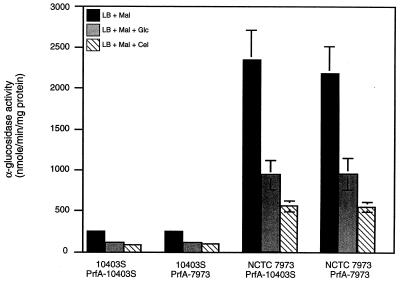
FIG. 1.
Catabolite regulation of α-glucosidase in strains 10403S and NCTC 7973. Cells were grown at 37°C in LB medium buffered with 100 mM MOPS (pH 7.0) and supplemented with the indicated sugars at 25 mM. The specific activity of α-glucosidase in exponentially growing cells was measured as described in Materials and Methods and is expressed as nanomoles of product formed minute−1 milligram of protein−1. Mal, maltose; Glc, glucose; Fru, fructose; Cel, cellobiose. Each sample was analyzed in triplicate, and the data represent the means and standard errors of the means for three independent experiments.
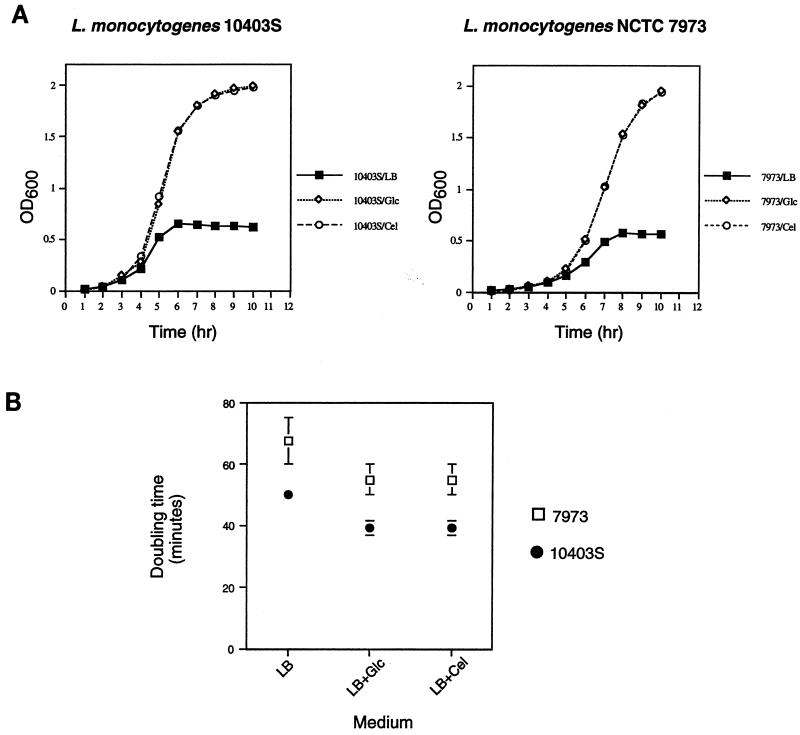
We also asked whether there was any difference in the utilization of glucose and cellobiose that might account for the differential effects of these sugars on hly expression in NCTC 7973 reported previously (30, 32). As indicated by effects on doubling time and final cell densities reached in the cultures, 10403S and NCTC 7973 could utilize glucose and cellobiose equally well (Fig. 2). However, NCTC 7973 had a longer lag phase and a growth rate approximately 34% lower than those of 10403S in the presence of both sugars. Thus, NCTC 7973 differs from 10403S both in general growth characteristics and in the specific activity of at least one glucose-repressed enzyme but exhibits no obvious defect in the ability to metabolize cellobiose.
FIG. 2.
(A) Growth of 10403S and NCTC 7973 in LB medium supplemented with various sugars. Overnight cultures were grown in LB medium buffered with 100 mM MOPS (pH 7.0) and were diluted 1:25 into fresh medium with or without either glucose or cellobiose at 25 mM. The results of one representative experiment are shown. Similar results were obtained in three independent experiments. OD600, optical density at 600 nm. (B) Doubling times of 10403S and NCTC 7973 in buffered LB medium with different carbon sources. Data represent means ± standard errors of the means for three independent experiments.
Sequence differences between PrfA proteins from NCTC 7973 and other natural isolates.
Other groups have noted the presence of mutational differences in the prfA alleles of various natural isolates, but the literature is contradictory and the functional significance of these mutations is unclear. With strain EGD as the wild-type reference, the deduced sequence of PrfA from NCTC 7973 was reported by two groups to contain two mutational substitutions, Gly145Ser and Cys229Tyr (4, 38). However, at least one other report suggested that there was only a single amino acid difference in PrfANCTC 7973, a Cys-to-Tyr change at position 229 (5). Furthermore, the sequence of PrfA from the wild-type strain EGD in the GenBank database (accession no. M55160) is different from that of PrfALO28 (accession no. X61210) at three amino acid residues at the carboxy terminus of the protein. The prfA gene from 10403S was not previously sequenced. To resolve the conflicting reports regarding the sequence of PrfA, we amplified and sequenced the prfA genes from the four strains used in a previous study (30). Figure 3 shows a partial alignment of the deduced amino acid sequences of PrfA from the four L. monocytogenes strains. We were able to confirm the nucleotide changes in codons 145 (GGT to AGT) and 229 (TGT to TAT) in prfANCTC 7973 that lead to Gly145Ser and Cys229Tyr substitutions in the PrfA protein as reported recently by Ripio et al. (38). However, we found the prfA sequences from LO28, EGD, and 10403S to be identical at the amino acid level, although there was a silent nucleotide change (T to C) at the third position of codon 127 in prfA10403S.
FIG. 3.
Alignment of the deduced carboxy-terminal amino acid sequences of PrfA proteins from four L. monocytogenes strains. The substitutions in the PrfA sequence from NCTC 7973 are indicated by dots above the alignment. The numbers to the left of the sequences correspond to the position of the first residue in the full-length protein. The helix-turn-helix motif is boxed (41). The three residues at the carboxy terminus of PrfAEGD that were found to be different from the published sequence (28) are indicated by asterisks. Identical residues are shown in white letters on a black background, while divergent residues are in black letters on a white background.
Effect of exchanging prfA alleles between 10403S and NCTC 7973 on hly-gus expression.
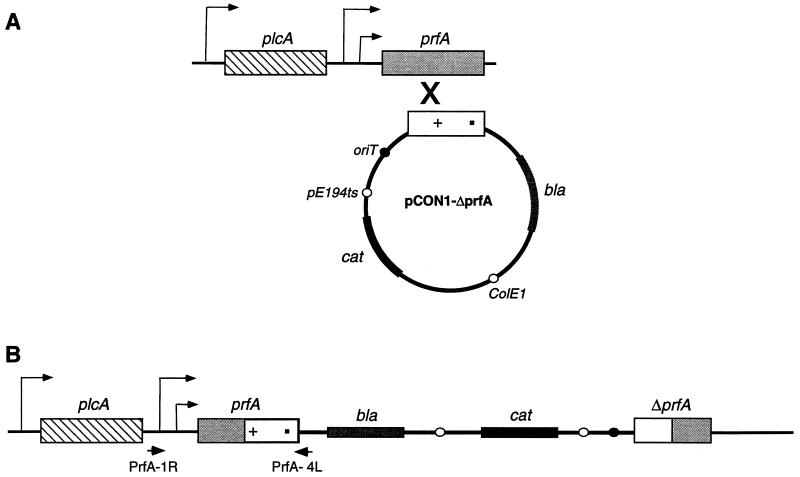
Ripio et al. reported recently that a Gly-to-Ser substitution at position 145 of PrfA leads to constitutive overexpression of virulence factors in the strain P14-A (belonging to serovar 4b) (38). Interestingly, this mutation in PrfA corresponds to a mutation in CRP that makes its activity independent of cyclic AMP in E. coli. Introduction of the prfA allele from P14-A on a multicopy vector into the prfA-deficient LO28 background resulted in overexpression of virulence genes. However, when the monocistronic wild-type prfA allele was introduced into the P14-A background, there was a decrease in expression levels of hly and plcB, which was attributed to competition between the mutant and wild-type forms of PrfA for their target promoters. Paradoxically, when the wild type bicistronic plcA-prfA region was introduced into P14-A, there was no significant reduction in the high levels of expression of virulence genes. Results based on propagation of regulatory factors on multicopy vectors can be misleading for many reasons. Therefore, it remained unclear whether the high levels of virulence gene expression in P14-A could be explained by the PrfA mutation alone or whether there were additional defects in the genetic background responsible for the phenotype. NCTC 7973 has a mutation in PrfA similar to that in P14-A, as well as aberrantly high activity of a glucose-repressed metabolic enzyme. Thus, we wanted to find out whether the sugar-insensitive expression of hly in NCTC 7973 was due to the prfA allele alone or to additional defects in the genetic background that affected several catabolite-controlled genes, including those of the virulence cluster. To differentiate between these possibilities, we exchanged the prfA alleles between the strains 10403S and NCTC 7973 in such a way that a single copy of either the wild-type or mutant allele of prfA would be under control of the natural promoter in either genetic background. To achieve this exchange of alleles, we cloned a promoterless, truncated copy of prfA from each of the two strains into the temperature-sensitive integrational vector pCON1 and introduced the vector by conjugative transfer into either 10403S or NCTC 7973 containing hly-gus transcriptional fusions. Integration of the vector into the chromosome placed either the wild-type or the mutant copy of prfA under control of the natural promoter (Fig. 4). The integration was confirmed by amplifying the prfA gene by using the primers PrfA1R and PrfA4L, and the mutations were confirmed by sequence analysis of the PCR product.
FIG. 4.
Strategy for construction of strains for prfA allele exchange by integrative replacement. (A) A promoterless, truncated copy of prfA from either 10403S or NCTC 7973 was cloned into the integrational vector pCON1 and conjugated into the host strains. (B) Shifting the strains to the nonpermissive temperature resulted in the integration of the temperature-sensitive vector into the chromosome at the prfA locus. The two nucleotide changes in the prfA sequence in codons 145 and 229 are represented as a cross and a dot, respectively. The integrated copy of prfA was amplified with the primers PrfA1R and PrfA4L, and the PCR product was sequenced to confirm the sequence changes. bla, β-lactamase gene; cat, chloramphenicol acetyltransferase gene; oriT, mobilization signal from plasmid RP4; pE194ts, replication functions derived from plasmid pE194ts; ColE1, replication functions derived from pUC18.
Introduction of the mutant copy of prfA into the 10403S background resulted in 10-fold higher expression of hly-gus (Fig. 5). The expression was also completely deregulated in the presence of glucose, while 25 mM cellobiose exerted a modest (twofold) effect. On the other hand, introduction of the wild-type copy of the prfA gene into the NCTC 7973 background decreased the level of hly-gus in LB medium alone and restored at least partial regulation by glucose and cellobiose. Significantly, we did not detect more than a twofold down-regulation of hly-gus by cellobiose in either NCTC 7973 or 10403S expressing prfANCTC 7973. Two previous papers had reported that cellobiose was unique in its ability to down-regulate virulence gene expression in NCTC 7973, raising the possibility that cellobiose has a signal transduction pathway distinct from that of other sugars (30, 32). However, Ripio et al. reported that cellobiose has almost no effect on virulence gene expression in P14-A (38), a result that was quite puzzling since P14-A, like NCTC 7973, has a Gly145Ser substitution in PrfA. While it is not clear why our results with cellobiose are different from those reported previously, we speculate that it could be due to subtle differences in growth conditions or to the elimination of the confounding effects of acidity on hly-gus expression by the stringent control of pH in our experiments. While it is certainly possible that independent sensing pathways exist for cellobiose or other sugars upstream of PrfA, it seems likely that modification of PrfA activity is the final common pathway for regulation by all sugars.
FIG. 5.
Effect of exchanging prfA alleles between strains 10403S and NCTC 7973 on the regulation of hly-gus expression by utilizable sugars. Cells were grown in LB medium buffered with 100 mM MOPS (pH 7.4) with or without either glucose or cellobiose at 25 mM. β-Glucuronidase activity was measured as described in Materials and Methods. Data represent means ± standard errors of the means for two independent experiments, each done in triplicate. Glc, glucose; Cel, cellobiose.
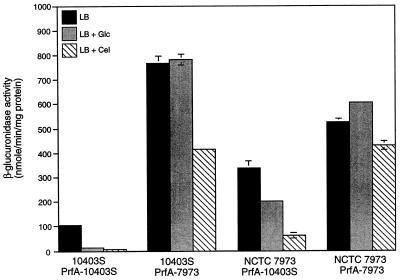
PrfA has been shown to be a structural and functional homolog of CRP, and so the possibility existed that the PrfA mutations, besides causing hly-gus overexpression, were also responsible for the up-regulation of α-glucosidase. An analogous situation exists in the opportunistic pathogen Pseudomonas aeruginosa, in which the expression of genes encoding exotoxin A and protease is regulated by the vfr gene product. Interestingly, the vfr gene product, which is also a homolog of CRP, not only controls virulence gene expression in P. aeruginosa but also can complement the β-galactosidase- and tryptophanase-deficient phenotypes of an E. coli crp deletion mutant (47). To determine whether the regulation of α-glucosidase in NCTC 7973 was related to its prfA genotype, we asked whether there was any effect of exchanging the prfA alleles on α-glucosidase activity in either the wild-type or the NCTC 7973 background. The levels and regulation of α-glucosidase activity were identical in 10403S containing either the wild-type or mutant prfA allele (Fig. 6). Similarly, the levels of the enzyme did not decrease upon introduction of the wild-type copy of prfA into NCTC 7973. We conclude from these data that NCTC 7973 has mutations in at least two loci: a defect in prfA that causes overexpression of hly and a second mutation that up-regulates α-glucosidase activity. While the nature of this second defect in NCTC 7973 is uncertain, we speculate that it could be similar to the ccrA1 mutation of Streptomyces coelicolor, which up-regulates the expression of several catabolite-controlled genes without affecting repression by carbon sources (18).
FIG. 6.
Effect of exchanging prfA alleles between 10403S and NCTC 7973 on the catabolite regulation of α-glucosidase. Cells were grown in LB medium buffered with 100 mM MOPS (pH 7.0) with or without either glucose or cellobiose at 25 mM. Specific activity of α-glucosidase from exponentially growing cells was measured as described in Materials and Methods. Data represent means ± standard errors of the means for three separate assays from each of two independent experiments. Mal, maltose; Glc, glucose; Cel, cellobiose.
During the construction of strains for the prfA allele exchange by integrative recombination, we obtained several clones that had the vector integrated in the chromosome of 10403S but which showed normal levels and patterns of regulation of hly-gus (data not shown). We reasoned that this phenotype could be due to a single recombinational event taking place between codons 145 and 229 in prfA, thus leaving the wild-type Gly residue at position 145 but resulting in a Cys-to-Tyr substitution at position 229 of PrfA. The result was confirmed by PCR amplification and sequence analysis of the prfA allele from this strain. This finding is consistent with the results of Ripio et al. with the strain P14-A, which contains only the Gly-to-Ser substitution at position 145 of PrfA (38).
Effect of extracellular pH on hly-gus expression.
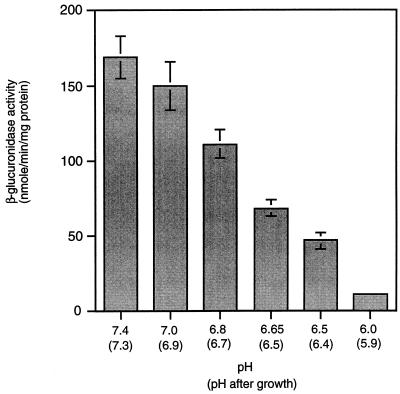
During the course of our experiments, we noticed that the final pH of the culture medium buffered with 100 mM MOPS was approximately 6.5 after growth in the presence of utilizable sugars. To test whether low extracellular pH could itself affect expression of hly-gus in the absence of sugars, we measured expression in LB medium buffered at various pH values ranging from 7.4 to 6.0. To our surprise, hly-gus expression was repressed approximately fourfold at pH 6.5 and eightfold at pH 6.0 relative to expression at pH 7.4 (Fig. 7). Over this range of pH, there was a significant decrease neither in the growth rates of cultures nor in the basal level of activity of α-glucosidase (data not shown). This result suggests that the regulation of virulence genes by extracellular pH is a specific regulatory effect and not the result of a general down-regulation of gene expression.
FIG. 7.
Effect of pH on the expression of hly-gus. Cells were grown in LB medium buffered with either 100 mM MOPS (pH 7.4 to 6.5) or MES (pH 6.0). Samples were collected at 1 h into stationary phase, and β-glucuronidase specific activity in cell lysates was measured. Data represent means ± standard errors of the means for three independent experiments, each done in triplicate.
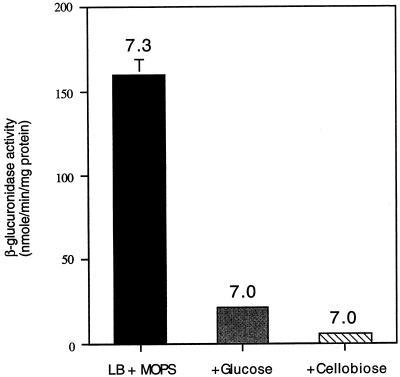
The effect of acidic extracellular pH on hly-gus expression raised the possibility that the effect of sugars on virulence genes observed by us and by others was due to the decrease in pH and not due to carbon source regulation. To test this possibility, we assayed hly-gus expression in medium buffered at pH 7.4 with 100 mM MOPS, so that the final pH of the cultures did not drop below 7.0 (Fig. 8). Under these conditions, the presence of either glucose or cellobiose at 25 mM still resulted in down-regulation of hly-gus, confirming that sugars could regulate virulence gene expression by a pH-independent mechanism.
FIG. 8.
Sugars regulate hly-gus expression by a mechanism that is independent of pH. Cells were grown in LB medium (with 100 mM MOPS, pH 7.4) with or without either glucose or cellobiose at 25 mM. Samples were collected at 1 h into stationary phase, and β-glucuronidase specific activity was measured. The starting pH of each culture was 7.4. The number above each bar represents the pH of the culture at the time samples were collected.
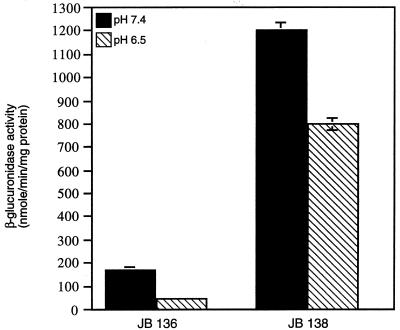
We then asked whether the influence of pH on hly-gus was also mediated through PrfA by comparing the expression of hly-gus at pH 6.5 and 7.4 in either 10403S or 10403S expressing prfANCTC 7973 (Fig. 9). The presence of the prfANCTC 7973 allele in a 10403S background resulted in partial deregulation of hly-gus at pH 6.5 (1.5-fold repression, compared to nearly 4-fold repression in the wild-type strain). This observation suggests that the effect of pH, like those of sugars and other environmental factors, is mediated by alteration of PrfA activity, possibly by changing the levels or activity of the putative PrfA-associated factor (Paf) (4, 36, 38, 42).
FIG. 9.
Effect of prfA10403S and prfANCTC 7973 alleles on pH regulation of hly-gus expression. Culture conditions were as described in the legend to Fig. 7. Results are expressed as nanomoles of p-nitrophenol formed minute−1 milligram of protein−1 and represent the means ± standard errors of the means for two independent experiments, each done in triplicate.
L. monocytogenes requires the hly gene product to escape from a vacuole in the mammalian cell (3, 11, 16, 24, 35). Listeriolysin has a pH optimum of 5.5 (17). It is therefore surprising that the expression of hly should be shut off at a pH which is optimal for its activity. We speculate that the expression of hly takes place at a pH of 7.4, a condition which the bacterium would encounter in the bloodstream, while low pH might be a signal for secretion of preformed listeriolysin, enabling the rapid escape of the bacterium from the vacuole into the cytoplasm of the host cell.
Conclusions.
We have demonstrated catabolite regulation of the metabolic enzyme α-glucosidase in L. monocytogenes and have shown that the specific activity of this enzyme is significantly higher in the hyperhemolytic mutant NCTC 7973 than in 10403S but is still subject to repression by glucose, fructose, and cellobiose. We have confirmed that the sequences of PrfA from three wild-type L. monocytogenes strains are identical but that there are two mutations in PrfANCTC 7973. By introducing a single copy of the mutant prfA allele from NCTC 7973 into the wild-type 10403S background, under control of its natural promoter, we have confirmed that the defect in prfA is responsible for the sugar-insensitive expression of hly-gus. Conversely, introduction of the prfA allele from 10403S into the NCTC 7973 background is sufficient to lower expression levels of hly-gus and restore at least partial regulation by sugars. However, there is no change in the levels of α-glucosidase activity when either the wild-type or mutant copy of prfA is introduced under control of the natural prfA promoter in the 10403S or the NCTC 7973 background. These results suggest that NCTC 7973 is anomalous not only with respect to the regulation of PrfA-controlled virulence genes but also in the expression of at least one other metabolic gene. Our results demonstrating the regulation of hly-gus by low extracellular pH suggest that the down-regulation of hly-gus expression in the presence of sugars may be due to a combination of effects produced by changes in pH or growth rate or by specific regulatory proteins. Therefore, it seems not only that several environmental factors regulate virulence gene expression but that any one factor may affect this regulation by several different mechanisms. Our results also underscore the importance of stringent control of extracellular pH during experiments measuring the effects of utilizable carbon sources on virulence genes. The fact that the Gly145Ser mutation in PrfA results in defective pH regulation suggests that as with several other environmental factors, the final common pathway of regulation of virulence genes by pH may be through modification of PrfA activity. Therefore, our results are consistent with the model of Ripio et al. (38), which proposes that regulation of virulence genes by environmental cues is achieved by alterations in the level or activity of a factor(s) required by PrfA for its activity.
ACKNOWLEDGMENTS
We are grateful to Dan Portnoy for the generous gift of bacterial strains and experimental protocols. We thank Andrea Milenbachs and Kai Hung for help with vector and strain construction and Tracey Foulger and David Brown for construction of pCON1. Thanks are also due to David Hodgson, Marlena Moors, Andrea Milenbachs, David Brown, Janet Hatt, and Paul Fawcett for many helpful discussions and suggestions during the preparation of the manuscript. We also thank Jan Westpheling and Tad Seyler for critically reading the manuscript.
This work was supported by Public Health Service grant GM35495 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Barak I, Behari J, Olmedo G, Guzman P, Brown D P, Castro E, Walker D, Westpheling J, Youngman P. Structure and function of the Bacillus SpoIIE protein and its localization to sites of sporulation septum assembly. Mol Microbiol. 1996;19:1047–1060. doi: 10.1046/j.1365-2958.1996.433963.x. [DOI] [PubMed] [Google Scholar]
- 3.Bielecki J, Youngman P, Connelly P, Portnoy D A. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990;345:175–176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- 4.Bockmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol Microbiol. 1996;22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- 5.Bohne J, Kestler H, Uebele C, Sokolovic Z, Goebel W. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol Microbiol. 1996;20:1189–1198. doi: 10.1111/j.1365-2958.1996.tb02639.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Camilli A, Tilney L G, Portnoy D A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chasin L A, Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968;243:5165–5178. [PubMed] [Google Scholar]
- 10.Christensen D P, Hutkins R W. Glucose uptake by Listeria monocytogenes Scott A and inhibition by pediocin JD. Appl Environ Microbiol. 1994;60:3870–3873. doi: 10.1128/aem.60.10.3870-3873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cossart P, Vicente M F, Mengaud J, Baquero F, Perez-Diaz J C, Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher S H, Sonenshein A L. Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu Rev Microbiol. 1991;45:107–135. doi: 10.1146/annurev.mi.45.100191.000543. [DOI] [PubMed] [Google Scholar]
- 14.Freitag N E, Rong L, Portnoy D A. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect Immun. 1993;61:2537–2544. doi: 10.1128/iai.61.6.2537-2544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freitag N E, Youngman P, Portnoy D A. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J Bacteriol. 1992;174:1293–1298. doi: 10.1128/jb.174.4.1293-1298.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard J L, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geoffroy C, Gaillard J L, Alouf J E, Berche P. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect Immun. 1987;55:1641–1646. doi: 10.1128/iai.55.7.1641-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram C, Delic I, Westpheling J. ccrA1: a mutation in Streptomyces coelicolor that affects the control of catabolite repression. J Bacteriol. 1995;177:3579–3586. doi: 10.1128/jb.177.12.3579-3586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. [Google Scholar]
- 20.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jefferson R A, Burgess S M, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow M L, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn M, Goebel W. Molecular studies on the virulence of Listeria monocytogenes. Genet Eng. 1995;17:31–51. [PubMed] [Google Scholar]
- 24.Kuhn M, Kathariou S, Goebel W. Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect Immun. 1988;56:79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampidis R, Gross R, Sokolovic Z, Goebel W, Kreft J. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol Microbiol. 1994;13:141–151. doi: 10.1111/j.1365-2958.1994.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 26.Leimeister-Wachter M, Chakraborty T. Detection of listeriolysin, the thiol-dependent hemolysin in Listeria monocytogenes, Listeria ivanovii, and Listeria seeligeri. Infect Immun. 1989;57:2350–2357. doi: 10.1128/iai.57.8.2350-2357.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leimeister-Wachter M, Domann E, Chakraborty T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J Bacteriol. 1992;174:947–952. doi: 10.1128/jb.174.3.947-952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci USA. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland J A, Milon G, Cossart P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol. 1991;5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 30.Milenbachs A A, Brown D P, Moors M, Youngman P. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol Microbiol. 1997;23:1075–1085. doi: 10.1046/j.1365-2958.1997.2711634.x. [DOI] [PubMed] [Google Scholar]
- 31.Milenbachs, A. A., and P. Youngman. Unpublished data.
- 32.Park S F, Kroll R G. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol Microbiol. 1993;8:653–661. doi: 10.1111/j.1365-2958.1993.tb01609.x. [DOI] [PubMed] [Google Scholar]
- 33.Parker C, Hutkins R W. Listeria monocytogenes Scott A transports glucose by high-affinity and low-affinity glucose transport systems. Appl Environ Microbiol. 1997;63:543–546. doi: 10.1128/aem.63.2.543-546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portnoy D A, Jacks P S, Hinrichs D J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renzoni A, Klarsfeld A, Dramsi S, Cossart P. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun. 1997;65:1515–1518. doi: 10.1128/iai.65.4.1515-1518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripio M T, Brehm K, Lara M, Suarez M, Vazquez-Boland J A. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripio M T, Dominguez-Bernal G, Lara M, Suarez M, Vazquez-Boland J A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J Bacteriol. 1997;179:1533–1540. doi: 10.1128/jb.179.5.1533-1540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripio M T, Dominguez-Bernal G, Suarez M, Brehm K, Berche P, Vazquez-Boland J A. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res Microbiol. 1996;147:371–384. doi: 10.1016/0923-2508(96)84712-7. [DOI] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehan B, Klarsfeld A, Ebright R, Cossart P. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol Microbiol. 1996;20:785–797. doi: 10.1111/j.1365-2958.1996.tb02517.x. [DOI] [PubMed] [Google Scholar]
- 42.Sheehan B, Klarsfeld A, Msadek T, Cossart P. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J Bacteriol. 1995;177:6469–6476. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 44.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 45.Vazquez-Boland J A, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner E, Marcandier S, Egeter O, Deutscher J, Gotz F, Bruckner R. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J Bacteriol. 1995;177:6144–6152. doi: 10.1128/jb.177.21.6144-6152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West S E, Sample A K, Runyen-Janecky L J. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]