Abstract
The GABAA-receptor-associated protein (GABARAP) is a member of a growing family of intracellular membrane trafficking and/or fusion proteins and has been implicated in plasma membrane targeting and/or recycling of GABAA receptors. GABARAP is localized on intracellular membranes such as the trans-Golgi network, binds to the γ 2 subunit of GABAA receptors and interacts with microtubules and the N-ethylmaleimide-sensitive factor. We report the X-ray crystal structure of mammalian GABARAP at 2.0 Å resolution. GABARAP consists of an N-terminal basic helical region, which has been implicated in tubulin binding, and a core structure with a conserved ubiquitin-like fold. Consistent with the high extent of sequence conservation among GABARAP homologues from plants to mammals, one face of the core structure is absolutely conserved while the opposite face shows considerable divergence. These features are in agreement with the conserved surface mediating protein–protein interactions shared by all members of the family, whereas the non-conserved surface region may play specific roles, such as docking to particular membrane receptors.
INTRODUCTION
GABAA receptors (GABAARs) are ligand-gated pentameric chloride channels composed of five classes of subunits encoded by 17 genes (Moss and Smart, 2001). GABAARs are inserted into the plasma membrane and clustered beneath inhibitory nerve terminals to ensure efficient binding of γ-aminobutyric acid (GABA), the most abundant inhibitory neurotransmitter in the central nervous system. Defects in GABAAR clustering are associated with cellular dysfunction and anxiety disorders in mice (Crestani et al., 1999).
The tubulin-binding protein gephyrin (Prior et al., 1992) was originally found to anchor mammalian inhibitory glycine receptors to the subsynaptic cytoskeleton (Kirsch et al., 1991) and is essential for localizing them at the synapse (Feng et al., 1998). Gephyrin also plays a pivotal role in the clustering of GABAARs (Essrich et al., 1998; Kneussel et al., 1999). A recently identified GABAAR-associated protein, GABARAP, has been shown to interact with both the cytoplasmic loop region of the GABAAR subunit γ2 (Wang and Olsen, 2000) and gephyrin (Kneussel et al., 2000). GABARAP has been suggested to contain a tubulin-binding motif within its N-terminal region (Wang et al., 1999) and regulates GABAAR channel kinetics in a microtubule-dependent fashion (Chen et al., 2000). Since GABARAP is localized in intracellular compartments, including the Golgi network, but not enriched at gephyrin-positive synapses, it has been proposed that it plays a role in receptor targeting and/or degradation processes (Kneussel et al., 2000). This is consistent with its interaction and co-localization with the N-ethylmaleimide-sensitive factor (NSF) (Kittler et al., 2001), an ATPase and chaperone for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins that plays an important role in intracellular membrane trafficking and fusion events (Jahn and Sudhof, 1999).
Evidence of a role for GABARAP in membrane protein trafficking first emerged from its high sequence similarity to GATE-16 (Golgi-associated ATPase enhancer) (Legesse Miller et al., 1998), a protein that stimulates NSF and interacts with the SNARE protein GOS-28 (Sagiv et al., 2000). The structure of GATE-16 has recently been solved and shown to possess an ubiquitin-like fold (Paz et al., 2000). Secondly, Apg8p, also known as Aut7p and Cvt5p (Lang et al., 1998 and references therein), is a yeast orthologue of GATE-16/GABARAP and is essential for autophagocytosis, the degradation of cytoplasmic components by the lysosomal/vacuolar system. A third homologue, MAP-LC3 (microtubule-associated protein light chain 3), is part of the MAP-1 complex, which co-polymerizes with tubulin and has also been found on autophagosomal membranes (Kabeya et al., 2000). Together, these findings suggest that GABARAP may also be involved in intracellular membrane trafficking and/or fusion events.
We have determined the crystal structure of rat GABARAP to 2.0 Å resolution. GABARAP contains an ubiquitin-like core with an additional N-terminal region, which may form a tubulin-binding surface distinct from previously described motifs. The highly conserved ubiquitin-like fold of GABARAP is discussed in the context of the growing family of GABARAP/GATE-16 homologues that appear to serve important functions in vesicle transport processes.
RESULTS AND DISCUSSION
Structure solution of GABARAP
Rat GABARAP was expressed in Escherichia coli as a maltose binding protein (MBP) fusion protein and, after cleaving off the MBP moiety with tobacco etch virus (TEV) protease, purified and crystallized. Crystals belonging to space group P212121 diffracted X-rays to 2.0 Å resolution (Tables I and II) and contain one monomer per asymmetric unit. The crystal structure of GABARAP was solved by molecular replacement using the GATE-16 structure (Paz et al., 2000) as a search model. The final model has been refined to a crystallographic R-factor of 23.5 (Rfree = 26.1) with good stereochemistry (Tables I and II) and includes residues 1–114, 57 water molecules, two Ni2+ ions and one Na+ ion. Only three C-terminal residues were disordered in the crystal.
Table I. X-ray data collection.
| Wavelength (Å) | 0.9333 |
| Resolution (Å) | 30.0–2.0 |
| Completeness (%) | 99.4 (95.5) |
| Total reflections | 56 243 |
| Unique reflections | 7790 |
| Redundancy | 3.8 (3.3) |
| Rmergea | 5.0 (43.9) |
| <I>/<σI> | 22.43 (2.11) |
Values in parentheses are for last shell limits, 2.07–2.0 Å.
aRmerge = ΣhklΣi│Ii(hkl) – <I(hkl)>│/ΣhklΣiIi(hkl).
Table II. Refinement statistics.
| Resolution range (Å) | 30–2.0 |
| Number of reflections | 7695 |
| Rfactor (%)a | 23.5 |
| Rfree (%) | 26.1 |
| Residues | 114 (out of 117) |
| Number of protein/solvent atoms | 960/57 |
| Average B-factor (Å2) | 42.4 |
| r.m.s.d. Δbond lengths (Å) | 0.006 |
| r.m.s.d. Δbond angles (°) | 1.146 |
| Ramachandran plot residues | |
| in most favoured and | |
| additionally allowed regions (%)b | 100 |
aRfactor = Σhkl││Fobs│ – k│Fcalc││/Σhkl│Fobs│.
bAs defined in PROCHECK (Laskowsky et al., 1993).
General architecture and structural similarity
GABARAP contains an N-terminal mostly basic helical region (α-helices 1 and 2) and a core composed of a mixed β-sheet. The core is classified as a β-Grasp fold by the SCOP server (Murzin et al., 1995) and is shared by the ubiquitin-like superfamily (Orengo et al., 1994). The core domain contains two parallel β-strands surrounded by one anti-parallel β-strand on each side. α-helix 3 is inserted between β-strands 2 and 3, while α-helix 4 is located between β-strands 3 and 4, packing towards the same side of the central β-sheet (Figure 1A).
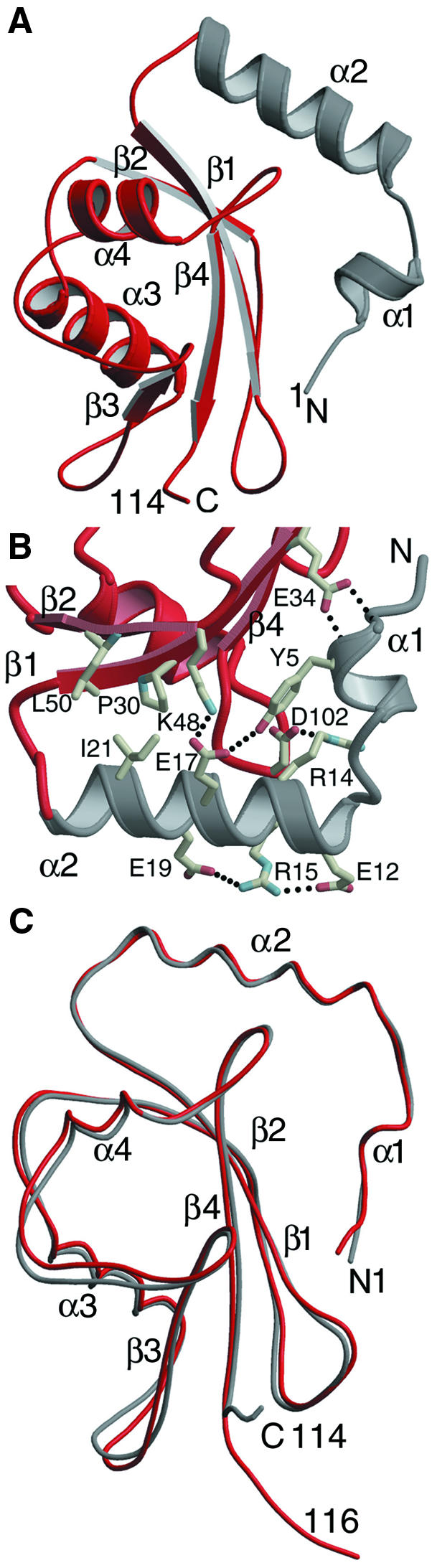
Fig. 1. (A) Ribbon diagram of GABARAP. The N-terminal region is shown in grey and the core structure displaying a conserved ubiquitin-like fold is shown in red. Secondary structure elements are labelled. (B) Close-up view of the interaction between the N-terminal region and the core structure. Hydrogen bonds are shown as dashed lines. (C) Superposition of the Cα atoms of GABARAP (grey) (residues 1–114) and corresponding residues of GATE-16 (molecule A; Paz et al., 2000) (red) shows an overall r.m.s.d. of 1.56 Å. This figure was generated using the programs Molscript (Kraulis, 1991) and lsqkab (CCP4, 1994).
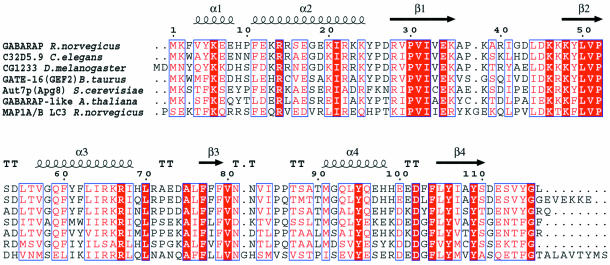
The small N-terminal region is composed of α-helices 1 and 2, which include residues 1–26. These amino acids are tightly packed against the ubiquitin-like folded core structure, and the buried surface between the N-terminal region and the core comprises 777 Å2. The interaction is stabilized by a hydrogen bond between the amide backbone at Tyr5 and Glu34 and by salt bridges between Arg14 and Asp102 and between Glu17 and Lys48 (Figure 1B). In addition, Glu17 hydrogen bonds with the hydroxyl group of Tyr5. The interaction between the N-terminal region and the core is also characterized by a small conserved hydrophobic region formed by Ile21, Pro30 and Leu50 (Figure 1B). All of the residues involved are highly conserved among the GABARAP-like molecules (Figure 2), with the exception of Tyr5, which is sometimes replaced by Trp (Figures 1B and 2).
Fig. 2. Sequence alignment of rat GABARAP (GenBank accession No. AF161586), bovine GATE-16 (AF020262, 57% identity), Drosophila melanogaster CG12334 (AAF55459.1, 78% identity), Caenorhabditis elegans C32D5 (AAC46797.1, 84% identity), Arabidopsis thaliana GABARAP-like (AAF08574.1, 42% identity) and Saccharomyces cerevisiae Aut7/Apg8 (CAA84899.1, 55% identity). Rat MAP-1A/B LC3 (U05784, 32% identity) is shown as a member of the MAP family, which is more distantly related. Secondary structure elements are shown for GABARAP above the sequence. The C-terminal end of the MAP1 LC3 sequence (ALKATATGREPCL) is not shown.
The general structural features of GABARAP are shared by GATE-16 (Paz et al., 2000), and Cα atoms of both structures (including residues 1–114, respectively) can be superimposed with an overall root-mean-square deviation (r.m.s.d.) of 1.56 Å. The structures show slight divergence at their respective C-termini and in the loop regions connecting β-strand 3 and α-helix 4, as well as α-helix 4 itself (Figure 1C). The r.m.s.d. values between the N-terminal parts (residues 1–25) of GABARAP and GATE-16 are as low as 0.48 Å, probably due to the high number of conserved interactions (Figure 1C). Despite the high backbone conservation in α-helix 2, there are also clear differences in solvent-exposed non-conserved side chains (Figures 3C), some of which line up and are stabilized through salt bridges (Figure 1B).
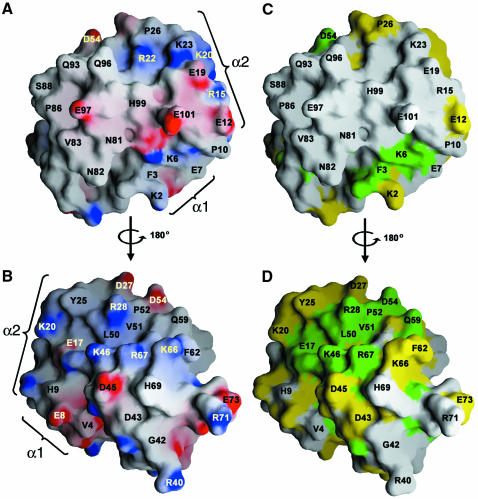
Fig. 3. Surface representations of GABARAP. (A) Electrostatic potential map of GABARAP surfaces (B, rotated by 180°). Regions of electrostatic potential less than 20 kBT are shown in red and those greater than +20 kBT are shown in blue (kB, Boltzmann constant; T, absolute temperature). (C and D) Representation of GABARAP in which identical (green) and homologous (yellow) residues are mapped to the protein surface using the sequence alignment shown in Figure 2. Orientations are identical to those shown in (A) and (B) and show that surface residues are conserved on one side of the molecule (C) but not on the other (D). Note the charged surface around α-helix 2 (region 1) (A) and a cleft lined by basic residues on the opposite side of the molecule (region 2) (B), which are highly conserved. Positions of α-helices 1 and 2 are marked. This figure was generated using the program GRASP (Nicholls et al., 1991).
Sequence conservation
GABARAP-like molecules are highly conserved among species ranging from mammals to yeast (Figure 2). Mammalian GABARAP sequences (human, rat, mouse) display 100% conservation and there are at least five GABARAP-like homologues in humans alone (Wang and Olsen, 2000; Xin et al., 2001). Their sequences can be roughly divided into two groups; the first includes GATE-16 and GABARAP-like molecules showing 60–85% identity (Figure 2). A second group, which is more closely related to MAPS, such as MAP1-LC3 (Mann and Hammarback, 1994), displays only ∼30% identity (Figure 2). GABARAP, MAP-1B and Apg8 bind tubulin, suggesting that tubulin binding may be a common function of all family members (Mann and Hammarback, 1994; Lang et al., 1998; Wang et al., 1999). Whether the interaction of GABARAP and GATE-16 with NSF (Sagiv et al., 2000; Kittler et al., 2001) is shared by the other homologues remains to be determined.
Conservation of putative protein–protein interaction sites
Two prominently charged regions characterize the surface of GABARAP. Region 1 includes residues contributed by α-helices 1 and 2 (Figure 3A). Region 2, located ∼180° away from region 1, forms a potential binding pocket, which is lined mostly by basic residues comprising Lys46, Arg67 and Lys66 on one side and Lys20, Arg18 and Asp54 on the other (Figure 3B). Both regions form a continuous belt of basic charges around the molecule (Figure 3). Mapping of residues homologous within the aligned primary sequences of GABARAP-like molecules shows that these charged regions are also largely conserved amongst family members (Figure 3C and D). Notably, one side of GABARAP (Figure 3C) shows almost no sequence conservation, whereas the opposite face is highly conserved (Figure 3D), including the solvent-exposed Phe62 in α-helix 3, which is replaced by Trp or Tyr in other members of the family (Figure 2). It is tempting to speculate that the presence of conserved and variable surface domains reflects the dual functions of GABARAP-like molecules, e.g. common and specific protein–protein interactions relevant for vesicle transport processes.
Tubulin and γ 2 loop interaction
Deletion of the first 35 residues of GABARAP was reported to abrogate tubulin binding (Wang et al., 1999). Such a construct, however, misses not only the N-terminal helices 1 and 2, but also β-strand 1, which occupies a central position in the β-sheet of the ubiquitin-like core (Figure 1A). This is likely to interfere with a stable folded globular domain. In addition, however, a synthetic peptide corresponding to the N-terminal 22 residues (α-helices 1 and 2, Figure 1A) has been described to promote tubulin assembly (Wang and Olsen, 2000), implicating the N-terminal basic region in tubulin association (Figure 1B). The structure shows a number of solvent-exposed charged residues in the N-terminal region that could potentially be involved in tubulin binding, including Lys6, Arg15, Glu19, Lys20, Arg22 and Lys23 (Figures 1B and 3A). Although many proteins interact with tubulin, there is no known common tubulin-binding motif. Interestingly, a stathmin-like domain from the neuronal protein RB3 forms a helical segment when bound to the αβ tubulin heterodimer (Gigant et al., 2000; Wallon et al., 2000). Generally, interactions with tubulin are mostly ionic, including a high number of basic charges (Nogales, 2000). Notably, the N-terminal regions of all GABARAP-like sequences present in the databases carry an overall basic charge; the calculated pI of a consensus sequence for this region is >9.0 (GABARAP pI = 9.8), whereas the ubiquitin-like fold of the core has an acidic pI (GABARAP core pI = 6.0).
Since this paper was submitted, a crystal structure of GABARAP and NMR binding studies performed with synthetic peptides corresponding to flexible α and β tubulin tails were reported (Knight et al., 2001), which showed only rather weak binding and did not involve GABARAP α1 and 2. However, there is also no clear indication that GABARAP should interact with the tubulin sequences tested for binding. Therefore, further studies are necessary to determine the interaction of GABARAP with tubulin, which may reside within the N-terminal helical region. In addition, it remains to be shown whether tubulin binding is a common function of all family members.
Deletion analysis has mapped the GABAAR interaction, mediated by the γ2 loop, to the N-terminus of the ubiquitin-like core using GST fusion constructs of GABARAP comprising residues 36–117, 1–68 and 1–117 (Wang and Olsen, 2000). These constructs point to a functional role of residues 36–68, which may involve solvent-exposed charged residues derived from β-strand 2 and α-helix 3 (described above as region 2, Figures 2 and 3B). However, these deletion constructs have the same caveat as described above in that they may not fold into a stable globular protein as represented by the structure of GABARAP. NMR binding studies performed using minimal binding sequences corresponding to the GABAAR γ2 loop also showed only extremely weak binding (Knight et al., 2001). Again, a more detailed study, namely by point mutation analysis, is necessary to pinpoint the exact interaction between GABARAP and the GABAAR.
Role in ubiquitination-like processes
Autophagocytosis in yeast requires the formation of autophagosomes, vesicles whose transport and fusion with the vacuole depend on SNAREs, NSF and Apg8/Aut7, an orthologue of mammalian GABARAP, which also binds to microtubules (Lang et al., 1998; Huang et al., 2000). Apg8 is modified by Apg4 protease, which leaves a C-terminal glycine residue for reversible covalent conjugation of phosphatidyl-ethanolamine (PE) to anchor Apg8 transiently to the membrane prior to autophagosome formation (Ichimura et al., 2000; Kirisako et al., 2000; Kim et al., 2001). Cleavage of Apg8–PE by Apg4 then releases Apg8 from membrane structures (Kirisako et al., 2000). Interestingly, formation of Apg8–PE involves an ubiquitination-like enzyme system, including activation of the ubiquitin-like protein Apg8 by the E1-like enzyme Apg7 and the E2 enzyme Apg3 (Tanida et al., 1999).
Recently, a human orthologue of Apg7 was found to act as an E1 enzyme in a putative ubiquitination-like mechanism, activating GATE-16, GABARAP and MAP-LC3, all three of which can be co-immunoprecipitated with Apg7 (Tanida et al., 2001). This suggests that GABARAP and its mammalian homologues may undergo similar processing and modification reactions. The glycine residue modified in Apg8 corresponds to GABARAP Gly116, which is conserved in all members of the family (Figure 2). However, Gly116 is part of the flexible C-terminal end and is disordered in the crystal. PE conjugation could thus take place opposite to the putative tubulin-binding motif (Figure 1) without interfering with other potential protein–protein interactions mediated through the conserved (Figure 3C) and variable protein surfaces of GABARAP-like molecules (Figure 3D). Further analysis of the dynamics of these interactions should help to define the precise functions of GABARAP in vesicle-mediated GABAAR sorting and trafficking.
METHODS
Expression and crystallization. The rat GABARAP cDNA encoding full-length GABARAP (residues 1–117; see Kneussel et al., 2000) was cloned by standard PCR methods into expression vector pMAL-c2g (New England Biolabs), engineered to contain a TEV protease cleavage site. The sequence of the clone was confirmed by DNA sequencing and the MBP–GABARAP fusion protein was expressed in E. coli cells BL21 codon+ (Stratagene). The MBP–GABARAP fusion protein was purified in a buffer containing 50 mM Tris pH 7.2 and 100 mM NaCl from cell lysates using an amylose column (New England Biolabs). Subsequently, TEV protease cleavage at a ratio of 1:500 (w/w) in a buffer containing 50 mM Tris pH 7.2, 50 mM NaCl, 1 mM DTT and 0.5 mM EDTA was performed at 4°C overnight GABARAP and cleaved MBP were separated on an SP Sepharose column and then purified further on a Superdex 75 (Amersham Pharmacia) size exclusion chromatography column in a buffer containing 20 mM Tris pH 7.2 and 100 mM NaCl. Fractions containing GABARAP were concentrated to 25 mg/ml.
Crystals of GABARAP were grown in drops of a mixture of 1 µl of protein solution and 1 µl of reservoir solution, containing 0.1 mM Tris pH 8.5, 30% (w/v) PEG 2000 monomethyl ether (MME) and 5 mM NiCl2. Cryo treatment was performed by a quick transfer of the crystals into a solution containing 30% (w/v) PEG 2000 MME, 0.1 M Tris pH 8.5 and 15% (w/v) glycerol, followed by flash-cooling in a gaseous nitrogen stream. Data to 2.0 Å resolution were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) beamline ID14 EH2 at a wavelength of 0.933 Å, with a 1° oscillation range. X-ray data were processed using the program packages DENZO, SCALEPACK (Otwinowski and Minor, 1997) and TRUNCATE (CCP4, 1994). The crystals belong to space group P212121 and contain one molecule per asymmetric unit, with unit cell dimensions of a = 29.72, b = 56.04 and c = 64.98 Å.
Structure solution and refinement. The structure was solved by molecular replacement with the program MolRep5 (CCP4, 1994) using a modified version (Vriend, 1990) of the GATE-16 structure (Paz et al., 2000) as a search model. This resulted in a clear peak with a correlation coefficient of 62.3 and an R-factor of 41.9 before rigid body refinement, corresponding to the correct solution. Initial automatic model building and refinement were performed with the program ARP-wARP (Perrakis et al., 1999) and REFMAC (CCP4, 1994), and 5% of the reflections were selected at random and set aside for the R-free calculation. Model building was completed by tracing missing areas manually with the program TURBO (Roussel and Cambillau, 1989) in several steps alternated with cycles of automated refinement using data to 2.0 Å resolution. The refinement included conjugate gradient minimization and restrained B-factor refinement, where bulk solvent correction was applied, using the program CNS (Brunger et al., 1998). The solvent molecules, two Ni2+ ions whose polar interactions form crucial crystal contacts and one Na+ ion were included in the last stages of interactive model correction and automatic refinement. The final model was refined to an R-factor of 23.5 (Rfree = 26.1) with good stereochemistry (Tables I and II). All residues (100%) lie in the most favoured regions of the Ramachandran plot, and no residues are located in disallowed areas (PROCHECK) (Laskowsky et al., 1993) (Tables I and II).
Acknowledgments
ACKNOWLEDGEMENTS
We thank all members of the EMBL/ESRF Joint Structural Biology Group (Grenoble) for their support at the ESRF beamlines. V.N.B. is supported by a pre-doctoral fellowship from the ‘Fondation Loius-Jeantet de Médicine’. M.S. and A.B. are both supported by European Union Marie-Curie-fellowships. The atomic coordinates have been deposited in the RCSB Protein Data Bank (accession code 1KJT).
REFERENCES
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 157–163. [DOI] [PubMed] [Google Scholar]
- Chen L., Wang, H., Vicini, S. and Olsen, R.W. (2000) The γ-aminobutyric acid type A (GABAA) receptor-associated protein (GABARAP) promotes GABAA receptor clustering and modulates the channel kinetics. Proc. Natl Acad. Sci. USA, 97, 11557–11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crestani F. et al. (1999) Decreased GABAA-receptor clustering results in enhanced anxiety and a bias for threat cues. Nature Neurosci., 2, 833–839. [DOI] [PubMed] [Google Scholar]
- Essrich C., Lorez, M., Benson, J.A., Fritschy, J.M. and Luscher, B. (1998) Postsynaptic clustering of major GABAA receptor subtypes requires the γ 2 subunit and gephyrin. Nature Neurosci., 1, 563–571. [DOI] [PubMed] [Google Scholar]
- Feng G., Tintrup, H., Kirsch, J., Nichol, M.C., Kuhse, J., Betz, H. and Sanes, J.R. (1998) Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science, 282, 1321–1324. [DOI] [PubMed] [Google Scholar]
- Gigant B., Curmi, P.A., Martin-Barbey, C., Charbaut, E., Lachkar, S., Lebeau, L., Siavoshian, S., Sobel, A. and Knossow, M. (2000) The 4 Å X-ray structure of a tubulin:stathmin-like domain complex. Cell, 102, 809–816. [DOI] [PubMed] [Google Scholar]
- Huang W.P., Scott, S.V., Kim, J. and Klionsky, D.J. (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem., 275, 5845–5851. [DOI] [PubMed] [Google Scholar]
- Ichimura Y. et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature, 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Jahn R. and Sudhof, T.C. (1999) Membrane fusion and exocytosis. Annu. Rev. Biochem., 68, 863–911. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y. and Yoshimori, T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J., 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang, W.P. and Klionsky, D.J. (2001) Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol., 152, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T. et al. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol., 151, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch J., Langosch, D., Prior, P., Littauer, U.Z., Schmitt, B. and Betz, H. (1991) The 93-kDa glycine receptor-associated protein binds to tubulin. J. Biol. Chem., 266, 22242–22245. [PubMed] [Google Scholar]
- Kittler J.T., Rostaing, P., Schiavo, G., Fritschy, J.M., Olsen, R., Triller, A. and Moss, S.J. (2001) The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol. Cell. Neurosci., 18, 13–25. [DOI] [PubMed] [Google Scholar]
- Kneussel M., Brandstatter, J.H., Laube, B., Stahl, S., Muller, U. and Betz, H. (1999) Loss of postsynaptic GABA(A) receptor clustering in gephyrin-deficient mice. J. Neurosci., 19, 9289–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneussel M., Haverkamp, S., Fuhrmann, J.C., Wang, H., Wassle, H., Olsen, R.W. and Betz, H. (2000) The γ-aminobutyric acid type A receptor (GABAAR)-associated protein GABARAP interacts with gephyrin but is not involved in receptor anchoring at the synapse. Proc. Natl Acad. Sci. USA, 97, 8594–8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight D., Harris, R., McAlister, M.S., Phelan, J.P., Geddes, S., Moss, S.J., Driscoll, P.C. and Keep, N. (2001) The X-ray crystal structure and putative ligand-derived peptide-binding properties of GABAA receptor associated protein (GABARAP). J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) Molscript: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lang T., Schaeffeler, E., Bernreuther, D., Bredschneider, M., Wolf, D.H. and Thumm, M. (1998) Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J., 17, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowsky R.A., McArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 276, 307–326. [Google Scholar]
- Legesse Miller A., Sagiv, Y., Porat, A. and Elazar, Z. (1998) Isolation and characterization of a novel low molecular weight protein involved in intra-Golgi traffic. J. Biol. Chem., 273, 3105–3109. [DOI] [PubMed] [Google Scholar]
- Mann S.S. and Hammarback, J.A. (1994) Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem., 269, 11492–11497. [PubMed] [Google Scholar]
- Moss S.J. and Smart, T.G. (2001) Constructing inhibitory synapses. Nature Rev. Neurosci., 2, 240–250. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner, S.E., Hubbard, T. and Chothia, C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nogales E. (2000) Structural insights into microtubule function. Annu. Rev. Biochem., 69, 277–302. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Jones, D.T. and Thornton, J.M. (1994) Protein superfamilies and domain superfolds. Nature, 372, 631–634. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Paz Y., Elazar, Z. and Fass, D. (2000) Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem., 275, 25445–25450. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris, R. and Lamzin, V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Prior P. et al. (1992) Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron, 8, 1161–1170. [DOI] [PubMed] [Google Scholar]
- Roussel A. and Cambillau, C. (1989) TURBO. In Silicon Graphics (ed.), Silicon Graphics Geometry Partner Directory (Fall 1989). Silicon Graphics, Mountain View, CA, pp. 77–78.
- Sagiv Y., Legesse-Miller, A., Porat, A. and Elazar, Z. (2000) GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J., 19, 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Mizushima, N., Kiyooka, M., Ohsumi, M., Ueno, T., Ohsumi, Y. and Kominami, E. (1999) Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell, 10, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake, E., Ueno, T. and Kominami, E. (2001) The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem., 276, 1701–1706. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Wallon G., Rappsilber, J., Mann, M. and Serrano, L. (2000) Model for stathmin/OP18 binding to tubulin. EMBO J., 19, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. and Olsen, R.W. (2000) Binding of the GABA(A) receptor-associated protein (GABARAP) to microtubules and microfilaments suggests involvement of the cytoskeleton in GABARAPGABA(A) receptor interaction. J. Neurochem., 75, 644–655. [DOI] [PubMed] [Google Scholar]
- Wang H., Bedford, F.K., Brandon, N.J., Moss, S.J. and Olsen, R.W. (1999) GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature, 397, 69–72. [DOI] [PubMed] [Google Scholar]
- Xin Y., Yu, L., Chen, Z., Zheng, L., Fu, Q., Jiang, J., Zhang, P., Gong, R. and Zhao, S. (2001) Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics, 74, 408–413. [DOI] [PubMed] [Google Scholar]