Abstract
In mammals, sequence-specific termination of DNA replication within the ribosomal RNA genes is catalyzed by a defined DNA–protein complex that includes transcription termination factor I (TTF-I). Here we show that TTF-I acts as a polar contrahelicase contrary to the intrinsic 3′→5′ helicase activity of SV40 large T antigen. The contrahelicase activity requires binding of TTF-I to its cognate recognition site and the presence of an auxiliary GC-rich sequence, which is able to form a specific secondary structure. Mutations in the GC-rich sequence lead to a loss of folding into correct secondary structure and abrogate contrahelicase activity. The finding suggests that a specific interaction between the Sal box-bound TTF-I and the GC-rich sequence is essential for the inhibition of T antigen helicase. Analyses of N-terminally truncated mutants of TTF-I showed inhibition of helicase by the same domain of TTF-I, which is also responsible for replication fork arrest.
INTRODUCTION
Replication of mammalian ribosomal RNA genes (rDNA) occurs while the genes are actively transcribed. Since replication forks migrate bidirectionally, one of the two replication forks in an active replicon proceeds through a transcribed rRNA gene in a direction opposite to transcription and, hence, would repeatedly collide with approaching RNA polymerases. It is apparent that head-on collisions between the replication and transcription machinery are prevented by the action of replication fork barriers (RFBs). Site-specific arrest of replication forks in the rDNA locus has been observed in a wide range of eukaryotic species including yeast, plants, frog, mouse and human (reviewed by Bastia and Mohanty, 1996; Hyrien, 2000; Rothstein et al., 2000). Although in some species the RFBs vary in their efficiency, in all cases investigated they limit replication fork progression to the same direction as rDNA transcription.
In a recent study, we demonstrated that an RFB is located at the 3′ end of the murine rRNA transcription unit (Gerber et al., 1997). Previous studies revealed that this sequence domain contains binding sites for proteins involved in the termination of mouse, rat and human rRNA transcription. In mice, the terminator of transcription is an 18-bp motif called the Sal box, which is repeated 10 times downstream of the 3′ end of the pre-rRNA coding region. The Sal box is recognized by transcription termination factor I (TTF-I). TTF-I binds to the Sal box and this binding stops the elongation reaction of RNA polymerase I (Grummt et al., 1985). Using an SV40-based cell-free replication system, we have shown that Sal box 2 and its flanking regions constitute a polar barrier arresting DNA replication 122 bp downstream of the RNA polymerase I transcription termination site (Gerber et al., 1997). Binding of TTF-I to Sal box 2 is a prerequisite for replication fork arrest, and more importantly, RFB activity occurs also in the absence of transcription (Gerber et al., 1997; Lopez-Estrano et al., 1998). The region preceding Sal box 2, where replication termination occurs, exhibits an unusual nucleotide sequence. The most remarkable feature is a 39-bp GC-stretch consisting of a cluster of 20 uninterrupted cytosine residues followed by a 19 bp GC-rich sequence and a cluster of 26 thymidine residues. Removal of the T-cluster decreases, but does not eliminate, RFB activity. In contrast, point mutations within the GC-stretch or shortening the cluster of C-residues totally abolishes replication fork arrest. Thus, the integrity of the flanking sequences is as important as that of the Sal box for mediating RFB activity.
In this study, we investigated which replication enzymes or factors are targeted for inhibition by the RFB. In prokaryotes, DNA helicases appear to interact specifically with the replicative helicases. DNA helicases are probably ahead of the replication fork, opening the DNA in front of the DNA polymerase. The terminator proteins in Escherichia coli and Bacillus subtilis have been shown to impede DNA chain elongation by inhibiting the DNA unwinding activity of DNA helicases (Bedrosian and Bastia, 1991). Similarly, the Epstein–Barr nuclear antigen 1 (EBNA-1) protein, which causes pausing of replication forks in the Epstein–Barr virus genome, counteracts the helicase function of SV40 T antigen (Ermakova et al., 1996). In addition, an inhibition of SV40 T antigen helicase by the Ter protein of E. coli was observed.
In the SV40 origin-dependent in vitro replication system (Li and Kelly, 1984), the SV40 large T antigen is the most prominent DNA helicase for the unwinding of duplex DNA (Stahl et al., 1986). Therefore, T antigen is a probable candidate for the interaction among the RFB and replication proteins. Our results demonstrate that the helicase activity of T antigen is indeed impaired by TTF-I in a polar manner when bound to a template that is serving as helicase substrate. The contrahelicase to be active requires the presence of the Sal box sequence and the binding of TTF-I, and in addition, the GC-rich sequences that are flanking the Sal box 2.
RESULTS
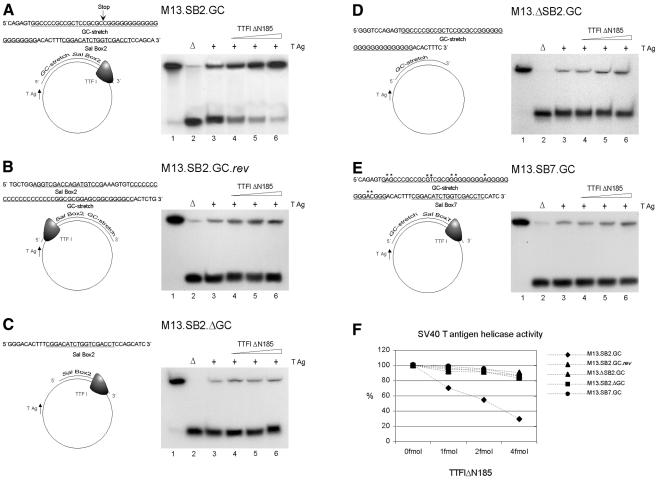
We investigated whether DNA-bound TTF-I would impair the helicase activity of SV40 large T antigen. In most previous studies, an N-terminally truncated form of murine TTF-I (TTFΔN185) was used rather than the intact protein. These studies revealed that the truncated protein bound specifically to the ‘Sal box’ target sequence in vitro, whereas the full-length protein did not (Sander et al., 1996). Therefore, we used TTFΔN185 for our studies. We performed assays for contrahelicase activity using partial duplex substrates that contained the Sal box 2 and the flanking GC-stretch of the RFB within the double-stranded region. The substrates were constructed in two different orientations: M13.SB2.GC for the terminating template (Figure 1A) and M13.SB2.GC.rev for the non-terminating template (Figure 1B) (Gerber et al., 1997). The helicase assay makes use of the fact that SV40 large T antigen translocates along a single-stranded template in a 3′→5′ direction and unwinds DNA hybrids when it encounters a duplex region. Addition of 10 pmol of T antigen caused extensive DNA unwinding resulting in the release of 60–80% of the annealed oligonucleotides (Figure 1A and B, lane 3). T antigen-dependent unwinding reaction on M13.SB2.GC was impaired by TTF-I in a dose-dependent manner (Figure 1A, lanes 4–6).
Fig. 1. Inhibition of SV40 large T antigen helicase by TTF-I at the Sal box 2 region. Left panels (A–E), partial duplex substrates used in the unwinding reactions. The oligonucleotides used to construct the five partial duplex substrates are shown at the top of the corresponding substrates. All fragments were 32P-radiolabeled by Klenow reaction. Right panels (A–E), autoradiographs of displacement analyses carried out by electrophoresis in non-denaturing 12% polyacrylamide gels; annealed partial duplex substrates are represented by the upper bands and displaced oligonucleotides the lower bands, respectively. For each reaction, 5 fmol of helicase substrate were used. Lane 1, substrate without SV40 T antigen; lane 2, heat denatured substrate; lanes 3–6 contained 10 pmol T antigen each and lanes 4–6 increasing amounts of TTFΔN185 (1, 2 and 3 fmol, respectively). (A) Contrahelicase activity on partial duplex substrate M13.SB2.GC, which is in the blocking orientation for SV40 T antigen helicase. (B) Contrahelicase activity on substrate M13.SB2.GC.rev, which is in the non-blocking orientation for SV40 T antigen helicase. (C) Contrahelicase activity on substrate M13.SB2.ΔGC lacking the GC-stretch in the terminating orientation. (D) Contrahelicase activity on substrate M13.ΔSB2.GC lacking Sal box 2. (E) Contrahelicase activity on substrate M13.SB7.GC containing Sal box 7 and its flanking GC-stretch. (F) Quantitative analyses of the displacement reactions using a PhosphorImager (Molecular Dynamics). The percentage of DNA unwinding by SV40 T antigen is given in the presence or absence of TTFΔN185. Extent of release of labeled oligonucleotides by SV40 T antigen in the absence of TTFΔN185 was set as 100%.
Importantly, in experiments using the substrate in the reversed orientation, M13.SB2.GC.rev, the helicase activity of T antigen was impaired to a lesser extent when compared with the M13.SB2.GC template upon addition of TTF-I (Figure 1B, lanes 4–6). These findings indicate that the contrahelicase activity of TTF-I is of polar nature. The low degree of inhibition observed with the reversed template M13.SB2.GC.rev did not exceed that of a control experiment using a template lacking the RFB sequences. In experiments with this template, the effect of increasing amounts of TTF-I on the T antigen unwinding activity in a non-DNA binding manner is measured. In the control template the duplex region was within the lac Z operator sequence, which is not a target site for the TTF-I protein. As expected, only a slight inhibitory effect was observed upon addition of increasing amounts of TTF-I (data not shown). Quantitative analysis of the data shown in Figure 1A and B clearly demonstrates that TTF-I has a polar contrahelicase activity (Figure 1F).
To investigate if the 39-bp stretch of guanosine and cytidine residues on the 3′ end of the Sal box 2 is also involved in mediating contrahelicase activity of the RFB, a template containing only the Sal box element, but lacking the GC-rich sequence, was constructed (M13.SB2.ΔGC). No significant contrahelicase activity was observed with this substrate (Figure 1C, lanes 4–6). Quantitative analysis of the data of Figure 1C demonstrates that elimination of the GC-stretch abrogates the contrahelicase activity of Sal box 2-bound TTF-I (Figure 1F). These results suggest that TTF-I bound to Sal box 2 acts in conjunction with the adjacent GC-rich sequence.
Experiments with a template containing the GC-stretch, but lacking Sal box 2 (M13.ΔSB2.GC), revealed that deletion of the TTF-I binding site abrogates the contrahelicase activity (Figure 1D, lanes 4–6 and F) demonstrating that the binding of TTF-I to Sal box 2 is indispensable for the contrahelicase activity of TTF-I.
Most interestingly, there is a second Sal box flanked by a GC-rich region in the decameric Sal box region, Sal box 7. These sequences indicate that the termination region may have evolved by duplication of a primordial pentameric Sal box unit. However, we demonstrated previously that Sal box 7 region is unable to stall the replication fork movement (Gerber et al., 1997). To demonstrate whether the Sal box 7 region is unable to support TTF-I contrahelicase activity, Sal box 7 and the flanking GC-rich sequence were inserted into M13mp18 (M13.SB7.GC). We found that the unwinding reaction of SV40 T antigen on M13.SB7.GC was unimpaired by TTF-I (Figure 1E, lanes 4–6 and F). Therefore, only the Sal box 2 region is able to inhibit the helicase activity of SV40 T antigen. This experiment also rules out the possibility that the loss of contrahelicase activity in the case of the construct M13.SB2.GC.rev is due to a shorter length of duplex DNA between the end encountered by T antigen and the binding site of TTF-I (Figure 1B), since in construct M13.SB7.GC the distance between the Sal box and the 5′ end is the same as in M13.SB2.GC.
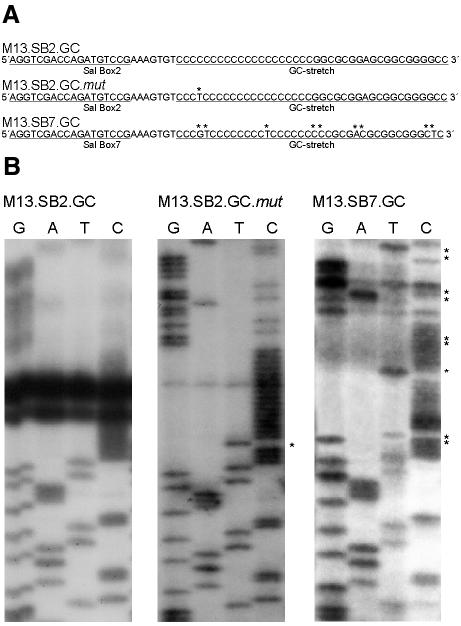
Comparison of sequences reveals that the difference between the two GC-stretches adjacent to Sal boxes 2 and 7 is that three cytosine residues within the cytosine stretch of the Sal box 2 cluster are replaced by one guanosine and by two thymidine residues, respectively, in the Sal box 7 region (Figure 2A). Furthermore, six more changes in the base composition are observed in the GC-rich sequence of the Sal box 7 region compared with the Sal box 2 region. To evaluate whether the GC-stretch of the Sal box 2 region may form secondary structures that, simultaneously with Sal box-bound TTF-I, cause replication termination as well as contrahelicase activity, we sequenced the purine-rich strand. Figure 2B shows that there are prominent stopping points for DNA synthesis, as reflected by strong bands at the same position in all four sequencing ladders when M13.SB2.GC is used as a template. However, no DNA polymerase stops were observed in sequencing reactions with M13.SB7.GC as template, suggesting that this template, in contrast to M13.SB2.GC, does not contain pronounced secondary structures of high thermodynamic stability. This assumption was substantiated by the fact that introduction of a random C→T point mutation into the 20-bp cytosine sequence of M13.SB2.GC (Figure 2A) abolished the interruption of the sequencing reaction (Figure 2B, M13.SB2.GC.mut). Furthermore, mutations made anywhere within the cytosine stretch also abrogated RFB activity (Gerber et al., 1997). Thus, formation of a strong secondary structure in the wild-type region flanking the Sal box 2 is a prerequisite for both contrahelicase and RFB activity. These findings suggest a functional cooperativity between these two activities.
Fig. 2. The influence of secondary structures on DNA polymerization. (A) Comparison of the sequences of M13.SB2.GC, M13.SB2.GC.mut and M13.SB7.GC. (B) DNA polymerization with different single-stranded DNA templates. G, A, T and C are ddGTP, ddATP, ddTTP and ddCTP termination reactions, respectively. Asterisks indicate the sequence differences in comparison to M13.SB2.GC.
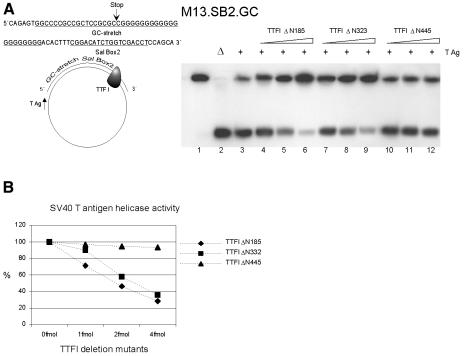
Having established that TTF-I is involved in contrahelicase activity, we examined which TTF-I protein domain mediates inhibition of the unwinding activity of SV40 T antigen. Therefore, the contrahelicase activity of three N-terminally truncated derivatives of TTF-I (TTFΔN185, TTFΔN323 and TTFΔN445) was compared (Figure 3). All three TTF-I mutants exhibit the same DNA binding activity, but show remarkable differences in their ability to terminate RNA polymerase I transcription as well as DNA replication (Sander et al., 1996; Gerber et al., 1997). Mutants TTFΔN185 and TTFΔN323 terminate both transcription and replication nearly as efficiently as full-length TTF-I. TTFΔN445, however, fail to support transcription termination as well as replication fork arrest. These data indicate that the DNA binding domain of TTF-I alone is not sufficient to arrest the replication fork. These data also suggest that a protein domain of TTF-I, which includes amino acids 323 to 445, is essential for both transcription termination and RFB activity. Figure 3 shows that the addition of increasing amounts of either TTFΔN185 or TTFΔN323 inhibited the strand-displacement reaction of T antigen (lanes 4–9), indicating that these TTF-I mutants possessed an intrinsic contrahelicase activity. In contrast, TTFΔN445 had no contrahelicase activity over a wide range of TTF-I–substrate ratios (Figure 3, lanes 10–12).
Fig. 3. Contrahelicase activity of TTF-I deletion mutants. (A) The substrate M13.SB2.GC was incubated with 1, 2 and 3 fmol of TTF-I mutant proteins, respectively, as described in the legend to Figure 1. Lanes 4–6, TTFIΔN185; lanes 7–9, TTFIΔN323; lanes 10–12, TTFIΔN445. (B) Quantitative analyses of the displacement reactions using a PhosphorImager (Molecular Dynamics). The percentage of DNA unwinding by SV40 T antigen is given in the presence or absence of various TTF-I mutant proteins. Extent of release of labeled oligonucleotides by SV40 T antigen in the absence of TTF-I was set as 100%.
DISCUSSION
We have shown previously that a polar barrier to replication fork movement exists in the spacer sequences of the murine rDNA locus. This RFB is constituted by sequences encompassing Sal box 2 and the flanking regions. Binding of the TTF-I protein to Sal box 2 is a prerequisite for replication fork arrest. If TTF-I bound to the Sal box was functionally homologous to prokaryotic replication terminator proteins, then it should inhibit the intrinsic helicase activity of SV40 large T antigen, the most prominent unwinding enzyme in SV40 DNA replication.
In fact, we have demonstrated here that the helicase activity of T antigen is inhibited upon binding of TTF-I to its target site, indicating that TTF-I mediates RFB activity by the impediment of unwinding of duplex DNA. T antigen translocates 3′→5′ on template DNA (Goetz et al., 1988; Wiekowski et al., 1988) and this movement is inhibited by the RFB in a polar fashion. Deletion of the GC-stretch abrogates the contrahelicase activity implying that Sal box 2 acts in conjunction with this adjacent sequence. This explains why in previous studies we were unable to observe an impediment of the unwinding reaction when the GC-stretch flanking Sal box 2 was not included in the partial duplex substrate (Gerber et al., 1997).
Furthermore, we found that the GC-rich region flanking Sal box 2 cannot be replaced by the GC-stretch flanking Sal box 7. The most obvious difference between these sequences is that only the GC-rich element flanking Sal box 2 contains an uninterrupted 20-bp cytosine sequence. Sequencing the single-stranded helicase substrates demonstrated that only the wild-type sequence of the GC-rich element flanking Sal box 2 contains strong stop positions for the sequencing reaction, but neither the mutated GC-stretch nor the GC-rich sequence that flank the Sal box 7 shows a similar effect. Termination of the sequencing reaction of single-stranded purine sequences can be expected, since it is well known that such sequences allow the formation of triplex structures with daughter DNA chains being involved (Baran et al., 1991; Dayn et al., 1992). Point mutations interrupting the 20 contiguous cytosines could destroy such secondary structures. In addition, Hanvey et al. (1987) have demonstrated that G19C19 sequences adopt a non-B right-handed structure, and Kohwi and Kohwi-Shigematsu (1988) suggested that poly(dG–dC) sequences are folded into halves from the center of the sequence to form a tetra-strand-like structure. These structures can contain a triple helix consisting of poly(dG–dG–dC) strands in the presence of Mg2+ and a fourth strand not involved in triplex formation but closely associated with the triplex. These observations strongly suggest that the C-rich sequence flanking Sal box 2 forms a triple helix structure, which is indispensable for the contrahelicase activity of the RFB.
Besides its role in transcription and replication termination, TTF-I also functions as a contrahelicase. The same functional domains of the TTF-I protein appear to be involved in all these activities. Previously, it was shown that TTF-I has a modular structure containing at least two structurally and functionally distinct domains, one that interacts with DNA and a second that is required for transcription termination (Sander et al., 1996). Replication forks are also arrested by a domain of TTF-I that is critical for transcription termination (Gerber et al., 1997), and the same holds true for the inhibition of the T antigen helicase activity.
Since TTF-I terminates transcription and replication with the opposite polarity and also impedes the T antigen helicase activity only in the direction of replication termination, we suggest that the TTF-I–DNA complex is structurally asymmetrical, which causes the polarity of the contrahelicase activity of this complex. Since the same cis and trans elements, Sal box 2, GC-stretch and TTF-I, are involved in contrahelicase activity of TTF-I and in the build up of an active RFB, we propose that the mode of action of the murine RFB is accomplished through inhibition of the replicative helicase.
METHODS
Helicase substrates. The single-stranded DNAs used in the helicase assays are derivatives of phage M13 and contain rDNA sequences (M13.SB2.GC and M13.SB2.GC.rev) from +636 to +712 in two different orientations. M13.SB2.ΔGC contains the sequence from +633 to +669 bp, M13.ΔSB2.GC from +658 to +714 bp and M13.SB7.GC from +1039 to +1114 bp. All the nucleotide numbering is made in accordance with the 3′ end of 28S rRNA. Substrates for helicase assays were prepared by incubating 50 pmol of oligonucleotides with 10 pmol of single-stranded DNA. Excess oligonucleotides were removed by passing through a PCR purification spin column (Amicon). Five hundred femtomoles of each substrate were 32P-labeled by Klenow reaction omitting one nucleotide.
Proteins. TTF-I and SV40 T antigen were expressed in insect cells and purified as described previously (Gerber et al., 1997).
DNA helicase assay. Reaction mixtures (20 µl) contained 50 mM Tris–HCl pH 7.5, 8 mM MgCl2, 6 mM ATP, 0.1 mg/ml bovine serum albumin, 10 pmol of T antigen and 5 fmol of the DNA substrate. Increasing amounts of TTF-I were added and the assays were incubated for 30 min at 37°C. TTF-I-containing reactions were preincubated for 20 min at 4°C, followed by addition of T antigen. The reaction was stopped by the addition of 0.1 volume of 3.3% SDS/50 mM EDTA, and the reaction products were analyzed by electrophoresis in 12% polyacrylamide gels.
Sequencing of single-stranded DNA. For each sequencing reaction, 1 µg of single-stranded DNA and 10 pmol of M13 primer from the T7 Sequencing Kit (Amersham Pharmacia Biotech) were used. Annealing and sequencing reactions were carried out according to the T7 Sequencing Kit Protocol.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. Gärtner and E. Kunkel for expert technical assistance. We also thank Drs A. Szalay and M. Wallisch for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- Baran N., Lapidot, A. and Manor, H. (1991) Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC) and d(GA) tracts. Proc. Natl Acad. Sci. USA, 88, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D. and Mohanty, B.K. (1996) Mechanisms for completing DNA replication. In DePamphilis, M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 177–215.
- Bedrosian C. and Bastia, D. (1991) Escherichia coli replication terminator protein impedes simian virus 40 (SV40) DNA replication fork movement and SV40 large tumor antigen helicase activity in vitro at a prokaryotic terminus sequence. Proc. Natl Acad. Sci. USA, 88, 2618–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayn A., Samadashwily, G.M. and Mirkin, S.M. (1992) Intramolecular DNA triplexes: unusual sequence requirements and influence on DNA polymerization. Proc. Natl Acad. Sci. USA, 89, 11406–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova O.V., Frappier, L. and Schildkraut, C.L. (1996) Role of the EBNA-1 protein in pausing of replication forks in the Epstein–Barr virus genome. J. Biol. Chem., 271, 33009–33017. [DOI] [PubMed] [Google Scholar]
- Gerber J.-K., Gögel, E., Berger, C., Wallisch, M., Müller, F., Grummt, I. and Grummt, F. (1997) Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell, 90, 559–567. [DOI] [PubMed] [Google Scholar]
- Goetz G.S., Dean, F.B., Hurwitz, J. and Matson, S.W. (1988) The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem., 263, 383–392. [PubMed] [Google Scholar]
- Grummt I., Maier, U., Öhrlein, A., Hassouna, N. and Bachellerie, J.-P. (1985) Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell, 43, 801–810. [DOI] [PubMed] [Google Scholar]
- Hanvey J.C., Klysik, J. and Wells, R.D. (1987) Influence of DNA sequence on formation of non-B right-handed helices in oligopurine:oligopyrimidine inserts in plasmids. J. Biol. Chem., 263, 7386–7396. [PubMed] [Google Scholar]
- Hyrien O. (2000) Mechanisms and consequences of replication fork arrest. Biochimie, 82, 5–17. [DOI] [PubMed] [Google Scholar]
- Kohwi Y. and Kohwi-Shigematsu, T. (1988) Magnesium ion-dependent triple-helix structure formed by homopurine–homopyrimidine sequences in supercoiled plasmid DNA. Proc. Natl Acad. Sci. USA, 85, 3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.J. and Kelly, T.J. (1984) Simian virus 40 DNA replication in vitro. Proc. Natl Acad. Sci. USA, 81, 6973–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Estrano C., Schvartzman, D.B., Krimer, D.B. and Hernandez, P. (1998) Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol., 277, 249–256. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel, B. and Gangloff, S. (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- Sander E.E., Mason, S., Munz, C. and Grummt, I. (1996) The amino-terminal domain of the transcription termination factor TTF-I causes protein oligomerization and inhibition of DNA binding. Nucleic Acids Res., 19, 3677–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl H., Dröge, P. and Knippers, R. (1986) DNA helicase activity of SV40 large tumor antigen. EMBO J., 5, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M., Schwarz, M.W. and Stahl, H. (1988) Simian virus 40 large T antigen DNA helicase. J. Biol. Chem., 263, 436–442. [PubMed] [Google Scholar]