Summary
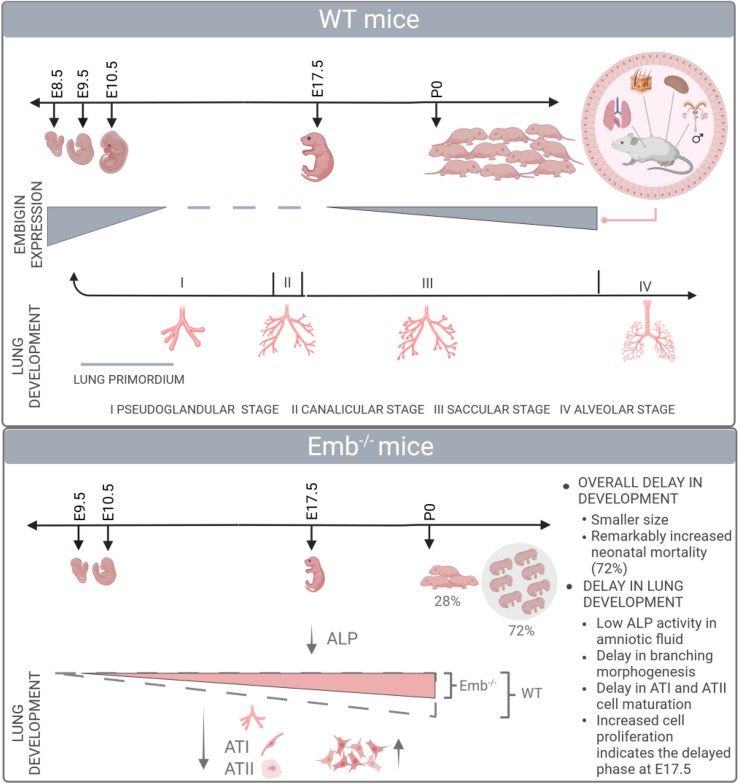
Embigin (Gp70), a receptor for fibronectin and an ancillary protein for monocarboxylate transporters, is known to regulate stem cell niches in sebaceous gland and bone marrow. Here, we show that embigin expression is at high level during early mouse embryogenesis and that embigin is essential for lung development. Markedly increased neonatal mortality of Emb−/− mice can be explained by the compromised lung maturation: in Emb−/− mice (E17.5) the number and the size of the small airways and distal airspace are significantly smaller, there are fewer ATI and ATII cells, and the alkaline phosphatase activity in amniotic fluid is lower. Emb−/− lungs show less peripheral branching already at E12.5, and embigin is highly expressed in lung primordium. Thus, embigin function is essential at early pseudoglandular stage or even earlier. Furthermore, our RNA-seq analysis and Ki67 staining results support the idea that the development of Emb−/− lungs is rather delayed than defected.
Subject areas: Developmental genetics, Developmental biology, Transcriptomics
Graphical abstract
Highlights
-
•
Embigin expression is high at early embryogenesis including the lung primordium
-
•
Maturation of the Emb−/− mice lung is delayed explaining high neonatal mortality
-
•
Embigin may execute its biological function in primordial lung cells
-
•
Embigin is detected in stem-like club cells lining bronchioles in adult mouse lungs
Developmental genetics; Developmental biology; Transcriptomics
Introduction
Embigin (Gp70) is a highly glycosylated member of the basigin subgroup in the immunoglobulin superfamily.1,2 In addition to embigin, the group includes two other type I membrane proteins, namely basigin (EMMPRIN/CD147) and neuroplastin (Np65/Gp65 and Np55/Gp55). Strong embigin mRNA expression has been reported in mouse embryos during the early phases of development.3,4 In adult mice and rats, only low levels of embigin mRNA have been found in several organs.4,5 However, at the protein level, embigin expression pattern has not been confirmed in many mouse and human tissues.
Structurally, all three members of the basigin subgroup resemble each other having an amino acid sequence identity of 37%–46%.6 They share the overall structure possessing an extracellular immunoglobulin-like (Ig-like) domain, a single hydrophobic transmembrane domain, and a short cytoplasmic tail. However, there are also significant structural differences that most probably contribute to the diverse biological roles of the proteins. One of the significant differences is the number of Ig-domain repeats in the extracellular domain: both basigin-1 and neuroplastin Np65 are composed of three Ig-like domains, while basigin-2, embigin, and neuroplastin Np55 have only two. The other two basigin isoforms found in humans, basigin-3, and basigin-4, comprise only one Ig-like domain.7 In addition to the variability in the number and the sequence of the Ig-like domains, the N-glycosylation state of these highly glycosylated proteins may contribute to the function of the protein.8,9,10,11 However, very little is known about the possible role of N-linked oligosaccharides.
Basigin has been reported to act in a wide variety of cellular processes including the development, activation, proliferation, migration, invasion, and adhesion of T lymphocytes.12 Neuroplastin, which is enriched in neurons and synapses, has basigin-like functions but in more restricted locations.8,13,14,15 All three members of the basigin group are involved in the regulation of cellular metabolism.16 They have been reported to escort monocarboxylate transporters (MCTs), the carriers of molecules such as L-lactate and pyruvate, to the plasma membrane.17 Embigin has been proposed to be the primary ancillary protein for MCT2,18 additionally, it acts as an assisting protein for MCTs 1, 3, 4, and 7.18,19,20,21 In adult mice, embigin has been identified as a bone marrow stem cell niche factor, more specifically as a hematopoietic stem/progenitor cell quiescence regulator.22 Our recent paper indicates that embigin acts also as a high affinity receptor for the extracellular matrix: the Ig1 domain of embigin binds directly to the type I repeats in the N-terminal domain of fibronectin. Thus, embigin represents a new type of adhesion receptor that combines the recognition of extracellular matrix to the MCT function and Wnt signaling.23 In the mouse sebaceous gland, Wnt-regulated embigin conducts cell differentiation by a mechanism that connects cell adhesion and lactate transport.23 Given the variety of cellular functions that the basigin family members are involved in, it is not surprising that they also have connections to pathological processes, such as cancer.24,25 To date, embigin has been reported to be a suppressor of tumorigenesis in breast cancer26 and a promoter of epithelial-mesenchymal transition in pancreatic carcinoma.27
Even though embigin has been identified as an important niche factor and the regulator of cell differentiation in adult stem cells, its role during mouse embryonic development has been unknown. Here, we report an expression pattern of the embigin protein during both embryonic development and in adult mice. An embigin knockout (Emb−/−) mouse model indicates that embigin is an essential protein for lung development and that the compromised maturation of the lungs is associated with the high neonatal mortality of Emb−/− mice. Embigin expression is at a high level from early embryogenesis until the formation of lung primordium suggesting that the main effect of embigin may occur during the early phases of both mouse and lung development. Interestingly, fibronectin, MCT-1, and MCT-4 are expressed in primordial lungs at higher levels than in lung cells at the later stages of development. Our results also indicate that in adult mouse lungs embigin expression is restricted to club cells, which are endogenous stem cells lining bronchi and bronchioles.
Results
Embigin is expressed from early mouse embryonic development into adulthood
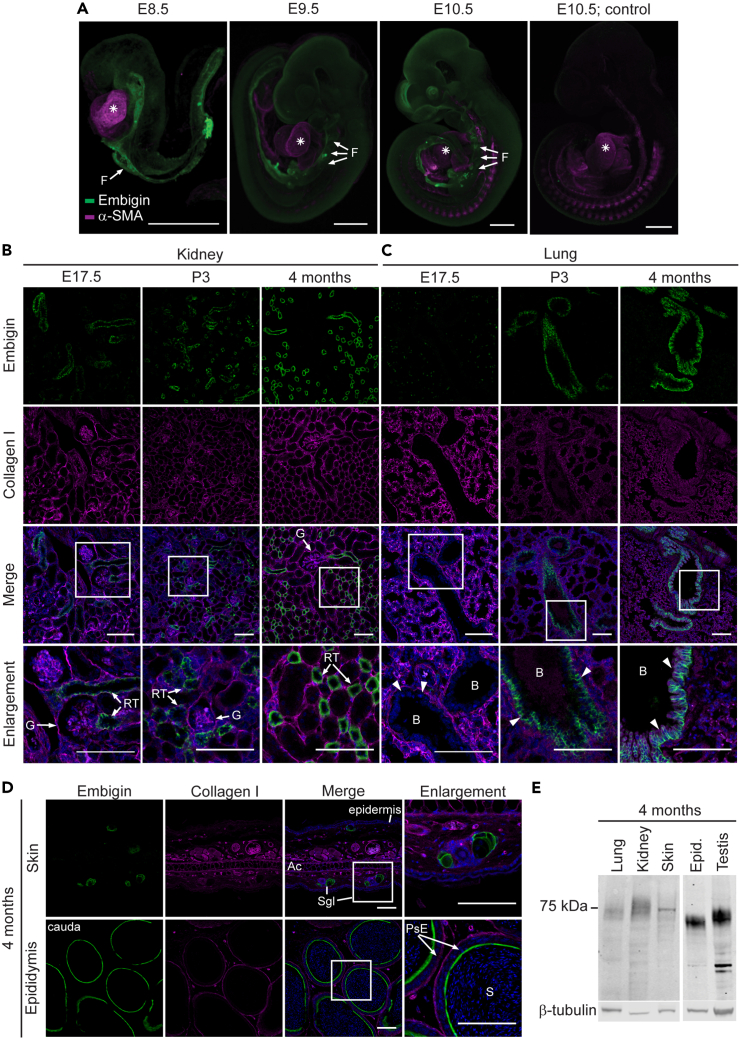
The current knowledge of embigin expression is mostly based on the determination of mRNA levels in various tissues, and thus the protein expression pattern has remained obscure. Here, we used the whole-mount immunofluorescence technique to visualize the embigin protein expression pattern in mice embryos at embryonic days E8.5–E10.5. In agreement with previous mRNA studies,3,4 the most robust embigin expression was detected at the early stage of embryogenesis, and embigin was observed to be an abundant protein specifically in the developing gut (Figure 1A). However, embigin expression did not cease after E10.5, albeit an apparent decrease in its expression was observed. At E13.5, low embigin expression was detected in restricted tissues such as kidney, lung, and small intestine (Figure S1A), and later in gestation, at E17.5, embigin expression was still observed in the kidneys while in the lung the amount of embigin hardly reached the detection level (Figure 1B). Analysis of recently published mouse organogenesis spatiotemporal transcriptomic atlas (MOSTA)28 also confirmed that embigin expression is high at E9.5 and decreases after that (Figure S1B).
Figure 1.
Embigin is widely expressed until E10.5 and after that in specific organs
(A) WT mouse embryos at embryonic days E8.5, E9.5, and E10.5 were stained with embigin and α-smooth muscle actin (α-SMA) antibodies by using the whole-mount immunostaining technique. As the negative control for embigin, only a secondary antibody was used; α-SMA was stained as a positive control, an asterisk indicates a developing heart, F: foregut. Representative images are shown. Scale bars: 500 μm.
(B–D) The paraffin sections of the kidney (B), lung (C), skin, and epididymis (D) were immunostained with embigin antibody. Collagen I was stained as a positive control and nuclei were labeled with Hoechst 33342 Fluorescent Stain. Samples were collected at E17.5. (B and C), postnatal day P3 (B and C), and four-month-old mice (B–D). Magnifications of the embigin-positive areas are shown. Scale bars: 100 μm. G: glomerulus, RT: renal tubule; B: bronchiole, arrow heads point at the epithelial cells lining bronchioles; Ac: auricular cartilage, Sgl: sebaceous gland; PsE: pseudostratified epithelia, S: sperm cells.
(E) Embigin expression in protein samples extracted from four-month-old WT mouse lung, kidney, skin, epididymis, and testis tissues was studied by western blotting. β-tubulin was used as a control. The blot image was cut to show only embigin-positive organs.
See also Figure S1.
We also determined the precise location of embigin after the gestational period. A set of adult organs at the age of four months was analyzed using immunofluorescence microscopy. The kidney (Figure 1B), lung (Figure 1C), epididymis, and skin (Figure 1D) showed high embigin levels. In the kidneys, embigin was located in the epithelial cells lining tubular structures (Figure 1B), and in the lungs, the epithelial cells of the lung bronchioles were shown to be highly embigin-positive (Figure 1C). Furthermore, the tubular structures in the caudal pole of the epididymis were determined to have a strong embigin expression, whereas, in the skin, embigin expression was located in the sebaceous glands (Figure 1D). In both lungs and kidneys, the expression level of embigin was shown to elevate shortly after birth at P3, and the expression was observed to be the strongest in adult mice (Figures 1B, 1C, and S1C). Tabula Muris, a compendium of single-cell transcriptome data from the 3-month-old mice, supports our findings showing that embigin is present, for instance, in the epithelial cells of the lungs and the epithelial cells of the collecting duct in the kidney.29 No differences between male and female mice were observed, apart from the embigin expression in the male epididymis.
The embigin protein expression in the organs described previously was further confirmed by western blot analysis (Figure 1E). A strong signal was detected also in the mouse testis suggesting the presence of embigin in the tissue. On the contrary, the liver, spleen, small intestine, adrenal glands, ovaries, and heart were determined as embigin-negative tissues (Figure S1D). In the embigin-positive tissues, embigin was detected as a characteristic broad protein band ranging from about 60 to 90 kDa, and the complete deglycosylation of the protein resulted in a shift to 35 kDa, to the predicted size of embigin core protein (Figure S1E). Thus, the observed variation of the approximated molecular mass is due to differential glycosylation of the nine potential glycosylation sites reported in the protein.2 The highest molecular mass of embigin was observed in the kidney, while the lowest molecular mass was found in the epididymis, and the smallest variation was detected in the skin (Figure 1E). Thus, the degree of embigin glycosylation was shown to vary in a tissue-dependent manner. Yet, it is not understood, whether differences in N-linked glycosylation affect embigin function. Together, our results confirm that the embigin protein is expressed in the specific structures of the lung, kidney, skin, epididymis, and testis of four-month-old mice.
Embigin deficiency leads to an increase in neonatal mortality
To examine the role of embigin in vivo, knockout mice lacking exon 5 of the embigin gene were generated (Figure S2A). The mutated allele is predicted to either generate a truncated protein or lead to non-sense-mediated decay. The absence of embigin expression in the embigin deficient (Emb−/−) mice was confirmed both by PCR (Figure S2B) and by performing a western blot analysis of the kidney tissue (Figure S2C). In kidneys, a typical diffuse band around 75 kDa was observed only in the wild type (WT) mice (Figure S2C). The final verification of the lack of embigin in Emb−/− mice was gained through staining Emb−/− and WT E9.5 embryos with embigin and alpha smooth-muscle actin (α-SMA) antibodies (Figure S2D).
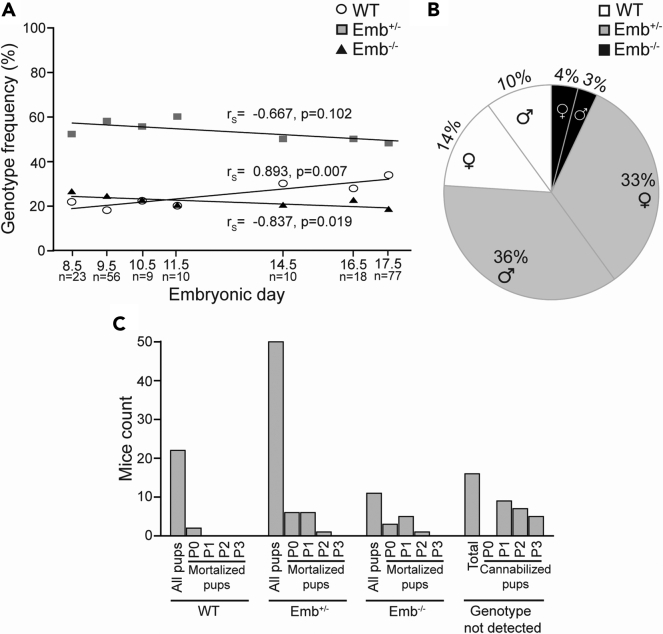
The effect of the embigin deficiency on the lifespan of affected animals was studied next using Emb−/− mice. The genotypes of 203 embryos from Emb+/− heterozygous intercrosses at the ages between E8.5 and E17.5 were analyzed. At embryonic day E8.5, 25% of all embryos were embigin deficient. Thus, the relative frequency of the genotypes at this embryonic stage was found to follow the Mendelian distribution. When embryos at E17.5 were inspected, the frequency of Emb−/− embryos was found to be only 18% instead of the expected 25% (Figure 2A). Spearman’s rank correlation analysis indicated that the small gradual decrease in the number of Emb−/− embryos as the gestation progressed, was statistically significant (rs = −0.837, p = 0.019; Figure 2A). The frequency of Emb+/− embryos was also decreasing at E17.5, but not in a statistically significant magnitude (rs = −0.667, p = 0.102; Figure 2A). Next, the genotypes of 284 pups from 40 different litters were examined between P14 and P21. Based on the study, only 7% of the pups from Emb+/− breedings were embigin deficient at P14-P21 (Figure 2B), of which 42% that survived were males and 58% females. Our results indicate that based on the Mendelian expectation, 28% of Emb−/− pups were lost already in the prenatal period, and in total, 72% of expected Emb−/− pups did not reach adulthood. The litter size had no obvious effect on the survival of the Emb−/− mice. In smaller litters (n = 4) with 3–6 pups, one Emb−/− survived and five Emb−/− mice were lost during the first three postnatal days, while in bigger litters (n = 4) with 7–10 pups, two Emb−/− mice survived and four Emb−/− were lost between days P0 and P3.
Figure 2.
Embigin deficiency increases mortality from embryonic day E8.5 to P3
(A) The relative frequency of the genotypes of the pups from heterozygous Emb+/− breedings were determined. Three litters at E8.5; 6 litters at E9.5; 1 litter at E10.5, E11.5, and E14.5; 2 litters at E16.5, and 10 litters at E17.5 were examined (n = a number of pups analyzed). The correlation between the frequency of each genotype and the embryonic stage was analyzed by calculating Spearman’s rank correlation coefficient (rs) and its statistical significance.
(B) The relative frequency of the genotypes of the pups from Emb+/− breedings was determined. 284 pups from 40 litters were analyzed at P14-P21.
(C) To determine the postnatal survival frequency of the Emb−/− mice, 100 pups from 17 litters and 6 different Emb+/− breedings were followed after birth. The genotypes were determined after the death of the pup.
To determine the time window at which the lethality of Emb−/− mice occurred, 100 pups from 17 litters from six different Emb+/− breedings were followed up after birth. The pups were genotyped after their death. The results indicate that most of the Emb−/− pups died during days 0 and 1 in postnatal life (Figure 2C). Noteworthy, the mortality of Emb+/− mice also appeared to be slightly elevated. While 81% of the born Emb−/− mice died during the first three postnatal days, 26% of Emb+/− and 9% of WT mice were lost during the period. Furthermore, 16% of the pups were fully cannibalized during postnatal days of P1-P3 before they were genotyped. While the frequency of Emb−/− embryos slightly decreased before birth, the results indicate that the major loss of Emb−/− mice occurred during the neonatal period. It cannot be excluded, however, that some pups were lost already during the parturition.
Embigin deficiency does not affect the mice after the neonatal period
To study the effect of embigin deficiency on mice that survived beyond the first three postnatal days, Emb−/− and WT mice were further analyzed at the ages of 2, 4, 6, and 12 months. The obtained results indicated that there was no difference in body weights when the adult WT and Emb−/− mice were compared (Figure S2E). Furthermore, neither the histology nor weights of specific organs were different (Figures S2F–S2I; Table S1). To assess whether embigin deficiency could affect the fertility of mice, ten pairs of Emb−/− mice were allowed to breed. Four out of ten breedings did not produce viable pups and out of the 23 litters produced, ten were fully cannibalized. On average 2.3 pups per litter survived and reached adulthood (Table 1). These observations were consistent with the high mortality rate of Emb−/− embryos and newly born mice of Emb+/− parents. Based on the data, the Emb−/− mice that survive are fertile, and they have changes neither in the typical body and organ weights nor in the histological architecture of the tissues studied.
Table 1.
Emb−/− mice are fertile
| Emb−/− breeding pairs | 10 |
| Emb−/− breeding pairs with survived litters | 6 |
| Cannibalized litters | 10 |
| Litters with survived pups | 13 |
| Survived female pups | 14 |
| Survived male pups | 16 |
| Average litter size | 2.3 |
Ten Emb−/− homozygous breedings were followed until the pups were genotyped at P14-P21.
Embigin deficiency causes delayed growth of mouse embryos
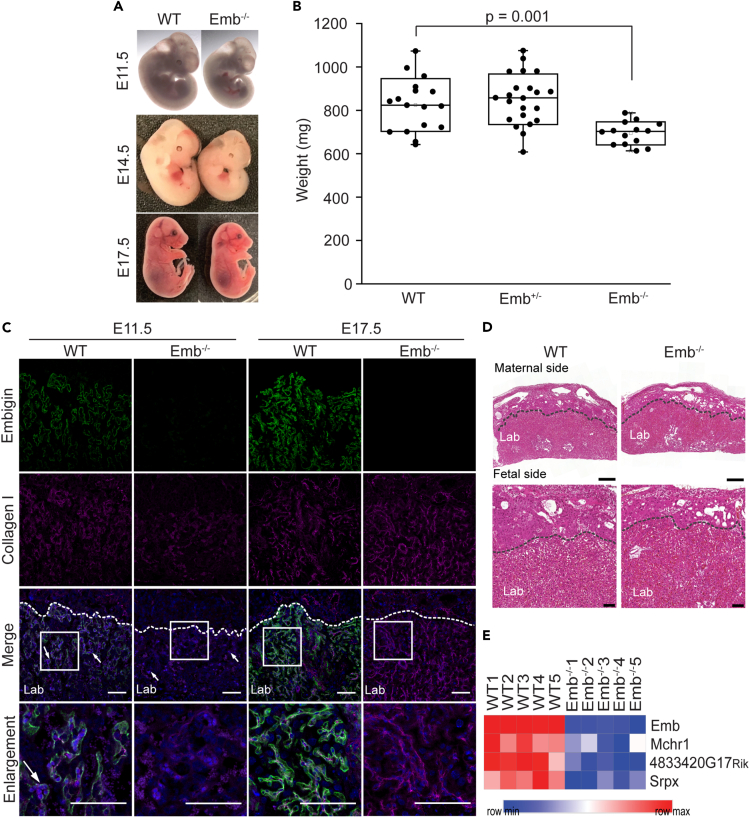
While the weights of four-month-old Emb−/− mice did not differ from WT mice, the body sizes of Emb−/− embryos tended to be smaller than their Emb+/− or WT littermates as imaged at E11.5, E14.5, and E17.5 in Figure 3A. Furthermore, the body weights of the Emb−/− embryos at E17.5 were significantly (p = 0.001) smaller than their littermates, with average body weights being 690 ± 54 mg for Emb−/− mice, 916 ± 136 mg for Emb+/−, and 834 ± 134 mg for WT embryos (Figure 3B). Keeping in mind the pivotal function of the placenta for optimal fetal growth, we decided to characterize the placentas of Emb−/− and WT embryos from Emb+/− breedings. Using an immunofluorescence staining technique, we detected an increasing embigin expression in the labyrinthine layer of the placenta in WT embryos between E11.5 and E17.5 (Figure 3C). Though intensive embigin expression was detected in the placenta, histological differences were not observed between the placentas of WT and Emb−/− embryos (Figure 3D). Furthermore, based on the RNA sequencing data, only four genes, one of them being embigin, were differentially expressed in the placentas of five Emb−/− embryos compared to the placentas of five WT embryos at E17.5 (Figure 3E). The gene was determined to be differentially expressed only if log2 of fold change value was above 0.6 or below −0.6 and Benjamini-Hochberg-corrected p value less than 0.05. Taken together, these results do not support the idea that embigin deficiency could cause placental dysfunction that would manifest as a fetal growth restriction observed in Emb−/− embryos. Therefore, other vital organs were examined next.
Figure 3.
The smaller size of Emb−/− embryos is not caused by a placental failure
(A) Representative images of WT and Emb−/− embryos are shown at E11.5, E14.5, and E17.5.
(B) Six litters with 53 embryos from Emb+/− breedings were analyzed at E17.5. The significance of the weight difference between WT and Emb−/− embryos (p = 0.001) was statistically analyzed by Student’s T-test for independent samples. Data are represented as a Spear style boxplot. A square shows the mean value.
(C) WT and Emb−/− placenta paraffin sections were immunostained with embigin and collagen I antibodies at E11.5 and E17.5. Nuclei were labeled with Hoechst 33342 Fluorescent Stain. Arrowheads show fetal-derived round nuclei. Magnifications of the embigin-positive areas are shown. Lab: labyrinth zone; scale bars: 100 μm.
(D) Representative images of hematoxylin-eosin-stained WT and Emb−/− placentas at E17.5 are shown. Scale bars: 500 μm. Magnifications of the inner edges of the placental labyrinth zones are shown below. Lab: labyrinth zone; scale bars: 100 μm.
(E) Heatmap of differentially expressed genes (log2 of fold change above 0.6 or below −0.6 and Benjamini-Hochberg-corrected p value <0.05) in WT (n = 5) and Emb−/− (n = 5) placenta at E17.5 based on RNA sequencing analysis.
Embigin deficiency delays lung maturation
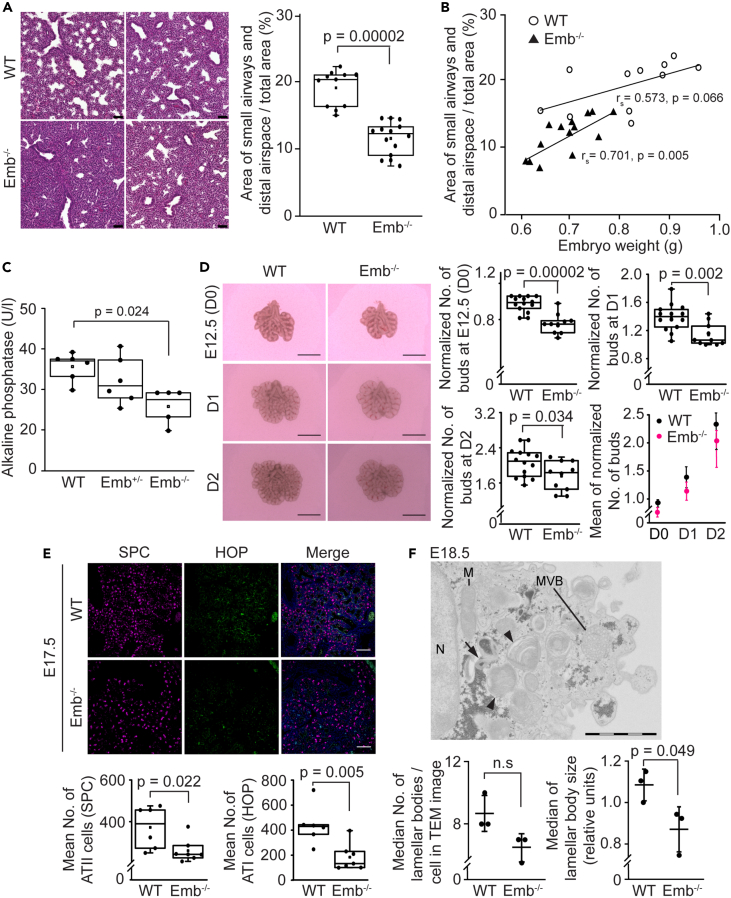
Our histological examination of the Emb−/− lungs at E17.5 unveiled the abnormal structure: the number and the size of the small airways and distal airspace were significantly smaller (p = 0.00002) when compared to the architecture of the lungs of their WT littermates (Figure 4A). In addition, the relative area of those structures in the Emb−/− embryonic lungs at E17.5 were determined to correlate strongly (rs = 0.701, p = 0.005) with the overall size of the Emb−/− embryo, (Figure 4B): the bigger the embryo, the more mature were the lungs. Noteworthy, in Emb−/− embryos, the lung development was defined to be systematically delayed when observed at E17.5: when the Emb−/− lungs were still at the canalicular stage, the WT littermates had already reached the saccular stage of the lung development.
Figure 4.
Embigin deficiency delays embryonic lung development
(A) Representative images of hematoxylin-eosin-stained lung sections from two WT and two Emb−/− E17.5 littermates are presented. Scale bars: 50 μm. The relative area of small airways and distal airspaces in the E17.5 lung sections was analyzed by ImageJ/Fiji software (n = 11 for WT and n = 14 for Emb−/−). Statistical significance (p = 0.00002) was determined by Mann-Whitney U-test. Data are represented as a Spear style boxplot. A square shows the mean value.
(B) The correlation between the body weight of the embryo and the relative area of small airways and distal airspaces in the lung sections was studied by calculating Spearman’s rank correlation coefficient (rs) and its statistical significance.
(C) Alkaline phosphatase activity was determined from the amniotic fluids of WT, Emb+/−, and Emb−/− embryos at E17.5 with VetScan Chemistry Analyzer (n = 6 for WT, and n = 5 for Emb+/− and Emb−/−). Statistical significance (p = 0.024) was determined by ANOVA followed by Dunnett’s t-test. Data are represented as a Spear style boxplot. A square shows the mean value.
(D) Lung explants from E12.5 WT and Emb−/− embryos were cultured for 2 days and imaged on days 0, 1, and 2 (D0 – D2, n = 14 for WT and n = 10 for Emb−/−). The normalized count of peripheral buds in lung explants of WT and Emb−/− at E12.5 was compared at D0, D1, and D2 (normalization: value/biggest value of the litter at D0). Statistical significances (D0: p = 0.00002, D1: p = 0.002, and D2: p = 0.034) were determined by Student’s T-test for independent samples. Data are represented as a Spear style boxplot. No statistical difference between WT and Emb−/− was observed from D0 to D2 in the progression of branching morphogenesis. Scale bars: 1000 μm.
(E) WT and Emb−/− lung paraffin sections were immunostained with SPC and HOP antibodies at E17.5 and nuclei were labeled with Hoechst 33342 Fluorescent Stain. The number of SPC (ATII) and HOP (ATI) -positive cells in WT (n = 6) and Emb−/− (n = 7) embryonic lungs at E17.5 were counted from five different areas of each lung. The amounts of SPC-positive cells in each sample were statistically compared with Mann-Whitney U-test (p = 0.022) and HOP-positive cells were analyzed by Student’s T-test for independent samples (p = 0.005). Data are represented as a Spear style boxplot. Scale bars: 100 μm.
(F) A representative transmission electron micrograph of an alveolar ATII cell from Emb−/− lungs at E18.5 is presented. Black arrows point to mature lamellar bodies and arrowheads to multilamellar bodies, N: nucleus; M: mitochondria, MVB: multivesicular body. Scale bar: 2000 μm. The amounts and the sizes of lamellar bodies of ten ATII cells in each sample were analyzed (n = 3 for WT and Emb−/−), and medians (n = 71–96/sample) were statistically analyzed by Student’s T-test for independent samples. The medians of each sample and mean ± SD are shown.
See also Figures S3 and S5.
Next, the activity of alkaline phosphatase was analyzed from the amniotic fluid at E17.5. Elevated alkaline phosphatase activity at the end of gestation has been shown to indicate increasing fetal lung maturity.30 When the alkaline phosphatase activity of amniotic fluids from WT, Emb+/−, and Emb−/− embryos were measured, the average activity was 35.8 U/l for WT, 32 U/l for Emb+/−, and 26 U/l for Emb−/− embryos. Thus, the alkaline phosphatase activity in the amniotic fluid of the Emb−/− embryos was significantly lower than the activity detected in WT embryos (p = 0.024). The average alkaline phosphatase activity in the amniotic fluid of Emb+/− embryos neither reached the same level as detected in WT embryos, but the difference was not statistically significant (p = 0.45; Figure 4C). Not only the activity of the alkaline phosphatase, but also sodium, calcium, and glucose concentrations have been reported to vary in the amniotic fluid during normal pregnancy: their concentrations increase and subsequently decrease as the gestation progresses.31 While the alkaline phosphatase activity was significantly lower in the amniotic fluid from Emb−/− embryos at E17.5, the concentrations of sodium, calcium, or glucose did not significantly differ between WT, Emb+/−, and Emb−/− embryos (Figures S3A–S3C). Only moderate changes observed in the concentrations of these factors indicate that the pregnancies of Emb−/− embryos progressed normally.
When WT and Emb−/− lungs were harvested at E12.5 and cultured as explants for 48 h, the smaller Emb−/− lungs presented significantly less (p = 0.00002) peripheral branching buds than WT lungs at E12.5 (Figure 4D). Based on the data, it is clear that lung branching was progressing also in Emb−/− lungs, albeit Emb−/− lungs stayed continuously behind the developmental schedule of WT lungs. Furthermore, the formation and maturation of distal epithelial cells were slightly behind the schedule at E17.5: alveolar type II (ATII) cells (p = 0.02) and especially alveolar type I (ATI) cells (p = 0.005) were detected at a lower number in Emb−/− than in WT lungs (Figure 4E). However, the formed ATII cells of Emb−/− lungs were able to synthesize surfactant protein C (SPC) at E17.5 (Figure 4F) and form lamellar bodies to store SPC as detected by transmission electron microscopy at E18.5 (Figure 4F). Still, the sizes of observed lamellar bodies were significantly smaller (p = 0.049) in Emb−/− lungs (Figure 4F). These results support the idea that the Emb−/− lungs can perform normal lung organogenesis but in a delayed manner. Importantly, when the lung sizes of the Emb−/−, Emb+/−, and WT embryos were compared, only Emb−/− lungs differed significantly from WTs (p = 0.031; Figure S3D). Thus, the remarkable changes in lung development are connected to the total embigin protein deficiency and are not seen when only one allele harbors the deletion of exon 5.
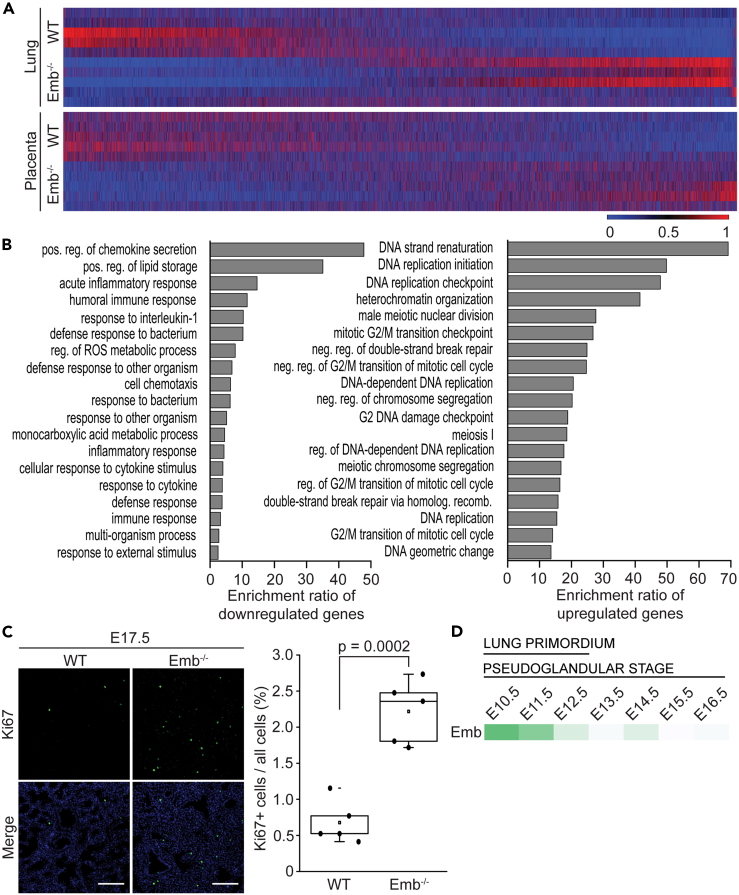
To further study the developmental delay, we analyzed transcriptomes of embryonic lungs at E17.5 by RNA sequencing. While the gene expression profiles of the placentas of Emb−/− and WT embryos resembled each other (Figures 3E and 5A), no less than 161 genes of the lungs were differentially expressed between the genotypes at E17.5 (Figures 5A and S4A). When compared to WT mice, most of the downregulated genes in Emb−/− embryos code for proteins involved in the immune response (Figure 5B), and the upregulated genes are predominated by genes that participate in cell proliferation (Figure 5B). Many of the upregulated genes are potential transcription and growth factors involved in lung development. For example, Adamts18, a pivotal effector of airway branching morphogenesis,32 and Hmga2, a protein required for Wnt signaling during lung development33 were upregulated. On the other hand, Scgb3a2, which is a cytokine-like secretory protein that functions as a growth factor in the lung promoting both early and late stages of fetal lung development,34 was downregulated. Furthermore, several cell cycle effectors, including Ccnf, Cdc6, Cdc45, Cdt1, and Gli1, were upregulated. To support the RNA sequencing data, the cell proliferation-associated nuclear antigen, Ki67, was detected at significantly (p = 0.0002) elevated levels in tissue samples from Emb−/− lungs at E17.5 (Figure 5C). The difference in cell proliferation is in a full agreement with the idea that at E17.5, the Emb−/− lungs are still at the canalicular stage, while the WT lungs have already reached the saccular stage. Importantly, RNA sequencing of E17.5 kidneys unveiled only five differentially expressed genes between WT and Emb−/− embryos (Figure S4B). Thus, the lack of embigin seems to have a remarkably larger influence on the lungs than kidneys at E17.5.
Figure 5.
Embigin deficiency leads to compromised lung function
(A) Heatmap of all expressed genes in WT (n = 5) and Emb−/− (n = 5) lung and placenta at E17.5 based on RNA sequencing analysis. The genes have been sorted from smallest to largest fold change value of Emb−/− vs. WT comparison and visualized by Morpheus.
(B) Differentially expressed genes in WT (n = 5) vs. Emb−/− embryonic lungs (n = 5) at E17.5 were analyzed from RNA sequencing data by WebGestalt. Top19 of the most enriched biological processes of those with FDR <0.05 and ≥3 overlapping genes are shown.
(C) WT and Emb−/− lung paraffin sections were immunostained with Ki67 antibody at E17.5. Nuclei were labeled with Hoechst 33342 Fluorescent Stain. Scale bars: 100 μm. Ki67-positive cells in WT (n = 5) and Emb−/− (n = 5) embryonic lungs at E17.5 were analyzed as fractions of all cells from five different areas of each lung. Statistical significance (p = 0.0002) was determined by the Student’s T-test for independent samples. Data are represented as a Spear style boxplot.
(D) Embigin mRNA expression in developing lungs from E10.5 to E16.5 based on the analysis of scRNA-seq data published in the MOSTA database.
See also Figures S4 and S6.
As the members of the basigin family, including embigin, have been shown to regulate the function of MCTs16,17,18,19,20,21 and we have recently connected embigin function to cell adhesion,23 we performed immunostainings of specific MCTs and fibronectin during the embryonic development of mouse. Neither in E10.5 embryos nor in E17.5 lungs, we could detect obvious differences in the expression levels or tissue distributions of MCT-1, MCT-4, and fibronectin in WT and Emb−/− (Figures S5A and S5B). The differential expressions of MCT-1 (WT: n = 5, mean: 10.8 ± 0.1; Emb−/−: n = 5, mean: 10.8 ± 0.11), MCT-4 (WT: n = 5, mean: 9.1 ± 0.18; Emb−/−: n = 5, mean: 9.0 ± 0.2), and fibronectin (WT: n = 5, mean: 16.2 ± 0.06; Emb−/−: n = 5, mean: 16.0 ± 0.16) were neither detected in RNA sequencing analysis of E17.5 lungs. It can also be hypothesized that basigin may compensate for the function of embigin, however, we did not find any evidence to support that since no obvious difference in the expression level of basigin was detected either in the RNA sequence analysis of lung cells at E17.5 (WT: n = 5, mean: 14.5 ± 0.08; Emb−/−: n = 5, mean: 14.3 ± 0.18) or in immunostaining in E17.5 lungs (Figure S5B). Still, we cannot exclude the possibility that during early phases of gestation the members of basigin family compensate deficiencies in each other’s functions.
All the findings suggest that embigin function is the most crucial during the early days of gestation. As described previously, the delayed lung development in Emb−/− mice was already obvious at E12.5. We could detect only very low embigin protein expression in the lungs at E13.5 (Figure S1A), and we were not able to detect a clear embigin signal in the lungs by immunofluorescence staining at E17.5 (Figure 1C). To gain more insight into embigin expression in lungs during gestation, we analyzed the MOSTA database.28 Using region-specific marker genes, Ptma, Mda, and Npm1, embigin mRNA expression was determined to be highest at the lung primordium (E10.5–E12.5; Figure 5D). In accordance with our observations during the pseudoglandular stage, embigin mRNA expression gradually decreases in the lungs, being very low at E13.5. Thus, it is probable that the embigin deficiency only affects the behavior of the primordial lung cells leading to e.g., delayed branching morphogenesis and alveolar cell (ATI and ATII) maturation seen at later developmental stages. In newborn (P0) Emb−/− pups, the number and the sizes of the small airways and the distal airspace were still smaller than in WTs (Figure S5C), though the difference was not as outstanding as in E17.5 lungs (Figure 4A). Still, the delayed lung development explains well the high perinatal mortality of the Emb−/− mice.
After the gestational period, embigin expression level rises in postnatal lungs (Figures 1C and S1C). When we analyzed previously published scRNA-seq data from the Lung Gene Expression Analysis (LGEA) database,35,36,37 it provided further confirmation to the observation. Based on LGEA, embigin expression increases in postnatal mouse lung epithelial cells when compared to embryonic lungs (Figure S6A).36,38 Here, embigin protein expression was observed to be restricted to the cells lining the bronchioles in adult lungs (Figure 1C). In bronchioles, embigin was further found to colocalize with CC10 (Scgb1a1), which is a marker for club cells. In club cells, CC10 and embigin showed polarized expression: while embigin was mainly localized on the basal side of the cell, CC10 was found on the apical side of the same cell (Figure S6B). Club cells are known as endogenous stem cells for bronchi and bronchioles being responsible for the strictly controlled cell proliferation in the lungs.39 Overall, embigin function is pivotal for the normal development of lungs, while its role in adult lungs remains open. The main results of the study have been summarized in Figure 6.
Figure 6.
Embigin plays an essential role during mouse embryogenesis, most notably affecting the development of the lungs
Embigin expression is high from early embryogenesis until E10.5, suggesting that the main effect of embigin may take place during the early phases of lung development. Based on morphological observations, RNA sequencing data, Ki67 staining, and the detected low alkaline phosphatase activity in amniotic fluid, the maturation of the Emb−/− mice lung is significantly delayed at E17.5, explaining the high neonatal mortality (72%). In adult mice, embigin is located in the epithelial cells lining tubular structures in the lung, kidney, and epididymis, in addition to skin and testis. Embigin expression in club cells lining bronchioles proposes a function of embigin in these stem-like cells in the lungs of adult mice.
Discussion
Three previous studies have connected embigin to the regulation of cellular differentiation programs. During B cell lineage commitment, the Pax5 transcription factor was first described to specifically repress embigin expression; however, the exact function of embigin in the process remained unknown.40 Lately, embigin has been shown to participate in the formation of stem cell niches in bone marrow22 and sebaceous gland.23 Our understanding of the molecular mechanism of embigin function has been also gradually progressed, and an increasing number of studies describe embigin as the facilitator and regulator of the membrane trafficking and transport activities of the proton-coupled MCTs.41 Recently, we have shown that embigin is an ECM receptor that binds to a specific domain in fibronectin23 and that the embigin function is based on its ability to link cell adhesion to the regulation of cellular metabolism through MCTs.23 Here, by using an Emb−/− mouse model, we report that embigin is required for normal lung development and that the lack of the protein leads to significantly increased mortality, especially during the perinatal period.
We focused our studies on the developing Emb−/− lungs since the structural abnormalities were the most obvious in the organ, and the compromised lung maturity explains the frequent deaths of the newborn Emb−/− pups. At a later stage of gestation (E17.5), the number and the sizes of the small airways and the distal airspace were observed to be significantly smaller in Emb−/− lungs than in WT lungs. Also, based on the RNA-seq analysis of Emb−/− and WT lungs, several cell proliferation-related genes were upregulated in Emb−/− lungs at E17.5. The increased cell proliferation, which was confirmed by Ki67 staining, suggests that at E17.5, Emb−/− lungs were still at the late canalicular stage involving high cell proliferation, while WTs had already reached the following saccular stage of lung maturation. The delayed lung maturation was further confirmed by the observation that there were fewer pulmonary ATI cells, cells required for gas exchange, in Emb−/− embryos at E17.5. Furthermore, lamellar bodies, the storage organelles for pulmonary surfactant in ATII cells, were smaller at E18.5 in the Emb−/− lungs. Finally, we showed that alkaline phosphatase activity in amniotic fluid was lower in E17.5 Emb-/- embryos, which also strongly supported the idea that the lack of embigin leads to a delay in lung development causing the death of the majority (72%) of newborn Emb−/− pups. In newborn (P0) Emb−/− pups the number and the sizes of the small airways and the distal airspace were still smaller than in WTs, but the difference was not as outstanding as in E17.5 lungs.
In mice, the overall embigin expression is high during the early days of embryonic development, but later it remarkably decreases. This fact has been shown at the mRNA level in the previously published embigin studies.3,4 Indeed, in the developing embryo, embigin mRNA expression drops at the same time as the primitive foregut endoderm invades the surrounding splanchnic mesoderm and forms the trachea and two lung buds. Here, we confirmed the observation at the protein level. At the developmental stage where we detected the delayed maturation of the Emb−/− lungs (E17.5), the embigin expression hardly reached the detection level when fluorescent immunohistochemistry or western blotting was used. Thus, it is unlikely that embigin would have a direct role in the regulation of cellular functions, such as cell proliferation and differentiation during the saccular and canalicular stages of lung development. We neither observed any obvious differences in the expression levels or tissue distributions of MCT-1, MCT-4, and fibronectin in WT and Emb−/− embryos at E10.5 or lungs at E17.5.
Although the overall embigin expression level significantly decreases as embryonic development progresses, embigin is expressed at a relatively high level in lung primordium until E12.5 based on the scRNA-seq data collected in MOSTA. When we harvested WT and Emb−/− lungs at E12.5 to culture them as explants for 48 h, fewer peripheral branching buds were observed at E12.5 in Emb−/− lungs. As expected, the branching progressed also in Emb−/− lungs, however, staying continuously behind the developmental schedule of WT lungs. Fibronectin, which is implicated as an essential factor in lung branching morphogenesis, is localized mostly on these branching buds.42,43,44 Based on MOSTA scRNA-seq data, not only fibronectin but also MCT-1 and MCT-4 are expressed in primordial lungs at higher levels than in lung cells at the later stages of development (Figure S6C). While embigin is known to bind fibronectin and modulate the activity and expression of several MCTs, at least MCTs 1–4 and 7,18,19,20,21 we propose that embigin executes its biological function rather in primordial lung cells than in more differentiated cell types at the later stages of lung development.
Hypothetically, basigin could partially compensate for the lack of embigin in Emb−/− mice. However, we did not detect differences in the expression level or tissue distribution of basigin in WT and Emb−/− lungs at E17.5. Effects of basigin knockout on mortality are also quite different when compared to the Emb−/− mice: the majority, about 70%, of Bsg−/− mice embryos die during the early stages of embryonic development.45 Emb−/− mice also survive better (28%) than Bsg−/− mice (14%) after birth. Furthermore, half of the survived Bsg−/− mice die due to interstitial pneumonia during the first month after birth.45 It is notable that despite significantly increased prenatal and perinatal mortality, some Emb−/− and Bsg−/− animals still reach adulthood. In Emb−/− mice, we did not find evidence that the litter size could affect the survival of the knockouts. However, we may speculate that the sufficient length of the gestation time, the time between conception and birth, is critical for the survival of Emb−/− mouse. There are also other examples of gene knockouts that lead to delayed lung development, and consequently increased perinatal mortality, but still allow a small fraction of animals to survive into adulthood.46,47
Embigin protein expression is not restricted to the gestational period, but it is present in the tubular structures of the adult mice tissues. The role of embigin in adult tissues is unknown since the morphology of all major organs is normal in Emb−/− mice. In the adult mouse lung, embigin expression is restricted to club cells that line the bronchioles. Club cells are known as endogenous stem cells that serve as the progenitors for ciliated and secretory epithelial cells. These cells have a low-level turnover during homeostasis, yet the mechanisms that regulate postnatal club cell quiescence are not fully understood.39 Based on the normal morphology of the adult Emb−/− lungs, it is possible to speculate that the potential role of embigin in the adult lung is related to the activation of stem cells e.g., after tissue injury, rather than to the direct participation in the lung function.
To conclude, the analysis of the Emb−/− mouse model demonstrated the essential role of embigin during lung development. In Emb−/− lungs, the initiation of lung organogenesis is affected, which becomes first apparent as delayed peripheral branching at E12.5, and at later stages for example as a smaller number of alveolar ATI cells. Our data do not support the idea that embigin would be critical for any specific cellular function at the later stages of lung cell differentiation. Instead, we suggest that the entire process is delayed when compared to WT lungs. During mouse embryonic development, a notable amount of embigin is present in primordial lungs, only. Thus, it is probable that the delay in the initiation of the lung development program reflects the altered behavior of the primordial cells of the Emb−/− lungs.
Limitations of the study
We have demonstrated that the lung development in Emb−/− mouse is clearly delayed and that can explain the significantly increased perinatal mortality. However, embigin expression is very low in embryos in general during later stages of gestation (after E11.5). Thus, it is unlikely that embigin would have a direct role in the regulation of cellular functions during the saccular and canalicular stages of lung development. Our data indicate that the absence of embigin affects cellular behavior at earlier stage of embryonic development, when its expression is at highest level, e.g., in primordial lungs. We have not been able to study directly the molecular mechanisms of embigin in those cell types. Indirect information suggests that embigin positive cells express MCTs, which are known to be dependent on the basigin family members. We have not managed to study MCT function in early Emb−/− embryos. Adult Emb−/− mouse lungs are morphologically normal, but the high expression level of embigin in specific structures in WT lungs suggest that embigin function could be revealed if the Emb−/− mice were challenged. However, this was not done in this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Embigin Monoclonal Antibody (clone G7.43.1), eBioscience | Thermo Fisher Scientific | Cat#14-5839-81; RRID:AB_2016582 |
| Monoclonal Anti-beta-Tubulin I antibody produced in mouse (clone SAP.4G) | Sigma-Aldrich | Cat#T7816; RRID:AB_261770 |
| Anti-GAPDH antibody | Abcam | Cat#ab9485; RRID:AB_307275 |
| Anti-alpha smooth muscle Actin antibody [1A4] (Alexa Fluor 488) (Clone 1A4) | Abcam | Cat#ab184675; RRID:AB_2832195 |
| Collagen I Antibody | Novus Biologicals | Cat#NB600-408; RRID:AB_10000511 |
| Recombinant Anti-Prosurfactant Protein C (SPC) antibody [EPR19839] | Abcam | Cat#ab211326; RRID:AB_2927746 |
| Hop Antibody (E-1) | Santa Cruz Biotechnology | Cat#sc-398703; RRID:AB_2687966 |
| Recombinant Anti-Ki67 antibody [SP6] | Abcam | Cat#ab16667; RRID:AB_302459 |
| Anti-Monocarboxylate Transporter 1 Antibody | Sigma-Aldrich | Cat#AB1286-I; RRID:N/A |
| Anti-SLC16A3/MCT 4 antibody [EPR28177-30] | Abcam | Cat#ab308528 RRID:N/A |
| Anti-Fibronectin Antibody | Sigma-Aldrich | Cat#AB2033; RRID: AB_2105702 |
| Anti-CD147/basigin antibody [EPR18008-8] | Abcam | Cat#ab188190 RRID:N/A |
| Anti-Uteroglobin/SCGB1A1/CC10 Antibody (E-11) | Santa Cruz Biotechnology | Cat#sc-365992; RRID:AB_10915481 |
| Chemicals, peptides, and recombinant proteins | ||
| AsiSI | NEB | Cat#R0630S |
| DreamTaq polymerase | Thermo Fisher Scientific | Cat#EP0702 |
| Proteinase K | Agilent | Cat#S3020 |
| Critical commercial assays | ||
| Nuclespin tissue kit | Macherey-Nagel | Cat#740952.50 |
| VetScan Comprehensive Diagnostic Profile | Abaxis | Cat#500-0038-48 |
| 8.0 μm pore size Whatman Nuclepore Track-Etched membranes | Merck | Cat#WHA10417506 |
| NucleoSpin RNA/Protein kit | Macherey-Nagel | Cat#740933.50 |
| Bead Tubes Type F | Macherey-Nagel | Cat#740816.50 |
| Pierce 660nm Protein Assay | Thermo Fisher Scientific | Cat#22660 |
| Glycoprotein Deglycosylation Kit | Sigma-Aldrich | Cat#362280 |
| NucleoSpin RNA kit | Macherey-Nagel | Cat#740955.50 |
| TruSeq Stranded mRNA Sample Preparation Kit | Illumina | Cat#20020594 |
| Deposited data | ||
| RNA-seq data | ArrayExpress database at EMBL-EBI; www.ebi.ac.uk/arrayexpress9497 | E-MTAB-10641 |
| Experimental models: Cell lines | ||
| G4 embryonic stem cells derived from 129S6/SvEvTac x C57BL/6NCrl mice | Mutant Mouse Resource and Research Center (MMRRC) | N/A |
| Neo-resistant MEF feeder cells | Applied Stem Cell | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6N mouse | Charles River Laboratories, Willmington, MA | N/A |
| Embigin knockout mouse | This paper | N/A |
| Oligonucleotides | ||
| Genotyping PCR primer pair 1 (5’-TAAGTCTCTTGTTTGCTGTG-3’; 5’-CACAACGGGTTCTTCTGTTAGTCC-3’) | Eurofins | N/A |
| Genotyping PCR primer pair 2 (5’-ACCCTTAAGTGCATGAACAAAA-3’; 5’-GGGTTCCTTGGCATTGTTACTAA-3’) | Eurofins | N/A |
| Recombinant DNA | ||
| Targeting vector for Emb gene, HTGR06008_A_1_E08 | The European Conditional Mouse Mutagenesis Program | N/A |
| Software and algorithms | ||
| ImageJ/Fiji | Schindelin et al.49 | RRID:SCR_002285; http://fiji.sc |
| ZEN blue software | Zeiss | RRID:SCR_013672; http://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html#introduction |
| Imaris | Bitplane | RRID:SCR_007370; http://www.bitplane.com/imaris/imaris |
| CaseViewer | 3D Histech | RRID:SCR_017654; https://www.3dhistech.com/caseviewer |
| Chipster | Kallio et al.50 | RRID:SCR_010939; http://chipster.csc.fi/ |
| STAR, version 2.7.3 | Dobin et al.51 | RRID:SCR_015899; https://github.com/alexdobin/STAR |
| HTSeq package, version 0.12.4 | Anders et al.52 | RRID:SCR_005514; https://htseq.readthedocs.io/en/master/ |
| edgeR R/Bioconductor package | Robinson et al.53 | RRID:SCR_012802; https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| WebGestalt: WEB-based GEne SeT AnaLysis Toolkit | Liao, Y. et al.54 | RRID:SCR_006786; http://www.webgestalt.org/ |
| DESeq2 Bioconductor package | Love et al.55 | RRID:SCR_015687; https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| SPSS Statistics software (version 25) | IMB | RRID:SCR_002865; http://www-01.ibm.com/software/uk/analytics/spss/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jyrki Heino (jyrki.heino@utu.fi).
Materials availability
A newly generated embigin knockout mouse model is available but requires an MTA for distribution.
Data and code availability
-
•
The RNA-seq data have been deposited at the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) and are publicly available as of the date of publication. Accession number is listed in the key resource table. All the data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported will be shared by the lead contact upon request.
Experimental model and study participant details
Animal models
C57BL/6N mice (Mus musculus, Charles River Laboratories, Willmington, MA) and the generated embigin knockout mice were maintained in Central Animal Laboratory at the University of Turku, Finland (study approval numbers: KEK/2016-0111-Heino and KEK/2016-2411-TCDM-gm kannat). All animal experiments were formally reviewed and approved by the Ethical Committee for Animal Experimentation in Finland, complying with international guidelines on the care and use of laboratory animals. Emb-/- mice were generated in collaboration with Turku Center for Disease Modeling. The mouse embryos were examined between embryonic days E8.5-E18.5 and pups at postnatal days P0-P3. Both male and female adult mice were studied at the age of 2, 4, or 6 months.
Cell lines
G4 embryonic stem cells derived from 129S6/SvEvTac x C57BL/6NCrl mice (Mutant Mouse Resource & Research Center (MMRRC)) were cultured on neomycin-resistant primary embryonic fibroblast (Neo-resistant MEF feeder cells, Applied StemCell) feeder layer in KnockOut DMEM medium (Gibco, Thermo Fisher Scientific) supplemented with 10% ES screened fetal bovine serum, heat-inactivated (Cytiva). The cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2.
Method details
Antibodies
Following antibodies were used in our studies: Embigin Monoclonal Antibody, clone G7.43.1, 14-5839-81, Lot#4343173, eBioscience, Thermo Fisher Scientific (Western blotting 1:1000, whole-mount and immunofluorescence 1:200); Monoclonal Anti-β-Tubulin I antibody produced in mouse, clone SAP.4G, T7816, Lot#068M4850V, Sigma-Aldrich (Western blotting 1:20 000); Anti-GAPDH antibody, ab9485, Lot#GR276202-1, Abcam (Western blotting 1:2500); Anti-α smooth muscle Actin (α-SMA) antibody [1A4] (Alexa Fluor 488), ab184675, Lot#GR316286-7, Abcam (whole-mount: 1:250); Collagen I Antibody, NB600-408, Lot#41476, Novus Biologicals (immunofluorescence 1:300); Recombinant Anti-Prosurfactant Protein C (SPC) antibody [EPR19839] (ab211326), Lot#GR3355590-5, Abcam (immunofluorescence 1:500); Hop Antibody (E-1), sc-398703, Lot#J1422, Santa Cruz Biotechnology (immunofluorescence 1:100); Recombinant Anti-Ki67 antibody [SP6], ab16667, Lot#GR3341233-1, Abcam (immunofluorescence 1:200); Anti-Monocarboxylate Transporter 1 Antibody, AB1286-I, Lot#3117317, Sigma-Aldrich (whole-mount and immunofluorescence 1:200); Anti-SLC16A3/MCT 4 antibody [EPR28177-30], ab308528, Lot#1047657-4, Abcam (whole-mount and immunofluorescence 1:100); Anti-Fibronectin Antibody, AB2033, Lot#2907625, Sigma-Aldrich (whole-mount and immunofluorescence 1:80); Anti-CD147/basigin antibody [EPR18008-8], ab188190, Lot#GR3372422-6, Abcam (immunofluorescence 1:250); Anti-Uteroglobin/SCGB1A1/CC10 Antibody (E-11), sc-365992, Lot#H3021, Santa Cruz Biotechnology (immunofluorescence 1:200); IRDye secondary antibodies, LI-COR Biosciences (Western blotting 1:15 000); and Alexa Fluor secondary antibodies, Thermo Fisher Scientific (whole-mount and immunofluorescence 1:400).
Generation of embigin deficient (Emb-/-) mice
First, a targeting vector for Emb gene, HTGR06008_A_1_E08 from The European Conditional Mouse Mutagenesis Program, was linearized with AsiSI restriction enzyme (R0630S, NEB). The construct was then transfected by electroporation into G4 embryonic stem cells derived from 129S6/SvEvTac x C57BL/6NCrl mice and cultured on a neomycin-resistant primary embryonic fibroblast feeder layer for 7-9 days. To ensure the occurrence of the correct homologous recombination, positive ES cell clones were screened by PCR and sequencing. ES cells were injected into C57BL/6N mouse blastocysts to generate chimeric mice. Germline transmission was achieved by cross-breeding male chimeras with C57BL/6N females.
Genotype determination
Genomic DNA was extracted with a gDNA Nucleospin tissue kit (Mache rey-Nagel) and the genotypes of the mice were determined from genomic DNA by using a PCR primer pair 1 (5’-TAAGTCTCTTGTTTGCTGTG-3’; 5’-CACAACGGGTTCTTCTGTTAGTCC-3’) to detect embigin knockout allele and a PCR primer pair 2 (5’-ACCCTTAAGTGCATGAACAAAA-3’; 5’-GGGTTCCTTGGCATTGTTACTAA-3’) to detect embigin WT allele. DreamTaq polymerase (Thermo Fisher Scientific) was used according to the manufacturer’s instructions using the following reaction settings: 95°C, 2 min; 35 x [95°C, 30 s; 50°C, 30 s; 72°C, 1 min]; 72°C, 10 min.
Timed mating
In timed mating, the day of the vaginal plug appearance was considered embryonic day 0.5 (E0.5). To analyze the survival of the Emb-/- embryos, the genotypes from 203 pups from 25 litters and 25 Emb+/- breedings were determined between embryonic days E8.5-E17.5. Furthermore, the genotypes of 100 pups from 17 litters from six different Emb+/- breedings were analyzed between postnatal days P0-P3; and the genotypes from 284 pups from 40 different litters from Emb+/- breedings were analyzed at P14-P21. The body weights of 53 embryos from six litters of Emb+/- breedings were analyzed at E17.5. Additionally, the lungs of 29 embryos from three E17.5 Emb+/- breedings were weighted and imaged with AxioZoom.V16 stereo microscope (Zeiss) using AxioCam 105 Colour camera and 1.0x PlanApo Z objective. The fertility of Emb-/- mice was studied with ten Emb-/- breedings. The crossings were followed until the pups were genotyped at the age of P14 - P21. Viable pups and average litter size were examined. Adult mice were studied at the ages of 2, 4, 6, and 12 months.
Alkaline phosphatase in amniotic fluid
Amniotic fluids were collected from six WT, five Emb-/-, and five Emb+/- embryos at E17.5 and analyzed with VetScan Comprehensive Diagnostic Profile reagent rotor (Abaxis) used with the VetScan VS2 Chemistry Analyzer (Abaxis). Alkaline phosphatase activity (U/l) and the molar concentrations (mmol/l) of sodium, calcium, and glucose were determined.
Lung explant culture
The lung developmental branching in WT and Emb-/- embryos was compared in lung explant cultures at E12.5. The lungs were harvested in PBS and the explants were cultured on 8.0 μm pore size Whatman Nuclepore Track-Etched membranes (WHA10417506, Merck) in DMEM/F12 medium (Gibco, Thermo Fisher Scientific) supplemented with 0.5% ES screened fetal bovine serum, heat-inactivated (Cytiva) and Penicillin-Streptomycin 200U/ml (Lonza). The lung explants were cultured at 37°C in a humidified atmosphere with 5% CO2 for 2 days. The samples were imaged on days 0, 1, and 2 with AxioZoom.V16 stereo microscope (Zeiss) using AxioCam 105 Colour camera and 1.0x PlanApo Z objective. Image linear brightness and contrast adjustments were performed with ImageJ/Fiji software. The number of peripheral buds in lung explants was calculated and normalization was done to remove the size variation between different litters (normalization: value / biggest value of the litter at day 0).
Whole-mount fluorescent immunohistochemistry
The embryonic embigin expression was analyzed using a whole-mount immunostaining technique as described previously in detail.48 However, PBS-MT solution was replaced with PBS-BSA-T (1% (w/v) bovine serum albumin (BSA) and 0.4% (v/v) Triton X-100 in PBS). 1% (v/v) normal mouse serum (10410, Invitrogen) and 0.5% (v/v) fetal calf serum solution (PromoCell) in PBS-BSA-T was used as the blocking solution. WT embryos at E8.5, E9.5, and E10.5 and Emb-/- embryos at E9.5 were stained with embigin and α-SMA antibodies. α-SMA was used as a positive control. Additionally, the expression levels of embigin, MCT-1, MCT-4, and fibronectin were compared in WT and Emb-/- embryos at E10.5. In all experiments, a secondary antibody without a primary antibody was applied as the negative control. The embryos were imaged with LSM 880 confocal microscope (Zeiss) using Plan-Apochromat 20x/0.8 M27 objective for E8.5 embryos and Plan-Apochromat 10x/0.3 M27 for E9.5 and E10.5 embryos. Image stacking, background subtractions, linear brightness, and contrast adjustments were performed with Zeiss ZEN blue software and Imaris (Bitplane).
Immunofluorescence staining of paraffin sections
Embigin expression was analyzed in the paraffin sections of the whole embryo at E13.5 in addition to the paraffin sections of the lung and kidney at E17.5 and postnatal day P3. Placental paraffin sections were examined at E11.5 and E17.5. The expression was also examined in the paraffin sections of the lung, kidney, skin, heart, liver, spleen, small intestine, adrenal gland, epididymis, testis, and ovary from three four-month-old males and females. Additionally, the localization of embigin in mice lungs was confirmed by immunostaining lung paraffin sections with CC10 club cell marker at four months of age. The number of ATII and ATI cells was analyzed using SPC and HOP antibodies in WT and Emb-/- lung paraffin sections at E17.5. Furthermore, cell proliferation was studied with the Ki67 antibody in WT and Emb-/- lung paraffin sections at E17.5. Moreover, the expression levels of MCT-1, MCT-4, fibronectin, and basigin were compared in WT and Emb-/- lung paraffin sections at E17.5. Formalin-fixed samples were fixed in paraffin and 4 μm sections were cut using an RM2255 microtome (Leica) and immobilized to adhesion slides (SuperFrost Plus, Thermo Fisher Scientific) overnight at 37°C. The sections were deparaffinized and rehydrated. The antigen retrieval was achieved with 3 min proteinase K treatment (S3020, Agilent). For the SPC, HOP, Ki67, MCT-1, MCT-4, fibronectin, and basigin antibodies, heat-mediated antigen retrieval was done with sodium citrate buffer (pH 6.0): microwave-heating for 20 min and cooling under running tap water for 10 min. After antigen retrieval, the sections were washed in PBS. The samples were blocked with 1% (v/v) BSA in PBS for 1 h at RT, and stained with primary antibodies in a blocking buffer overnight at 4°C. In all experiments, a secondary antibody without a primary antibody was applied as the negative control. The samples were washed with PBS and incubated with Alexa Fluor secondary antibodies in a blocking buffer for 1 h at RT. The sections were washed with PBS and nuclei were labeled with Hoechst 33342 (1:5000 in PBS, Thermo Fisher Scientific) for 10 min at RT. The sections were rinsed in PBS and finally in dH2O and mounted in Mowiol (Calbiochem) containing 25 mg/ml DABCO anti-fading reagent (Sigma). The samples were imaged with either LSM 880 confocal microscope (Zeiss) using a Plan-Apochromat 20x/0.8 M27 objective or Eclipse Ti2-E widefield microscope (Nikon) using a CFI Plan Apo Lambda 20x/0.75 objective. Image stacking, background subtractions, linear brightness, and contrast adjustments were performed with ImageJ/Fiji software. Ki67-positive cells in WT and Emb-/- embryonic lungs at E17.5 were analyzed as a fraction of all cells from five different areas of each lung.
Histological staining
Male and female WT and Emb-/- mice were examined at the age of 2, 4, and 6 months. Three to four mice were included in each independent study group. Additionally, one male and one female Emb-/- mouse were analyzed at the age of 1 year. The mice and the specific organs: heart, lung, liver, spleen, kidney, epididymis, testis, and ovary, were weighted and prepared for histological analysis. Also, the samples of skin, small intestine, and adrenal glands were prepared for histological analysis. Lungs were additionally analyzed from 11 WT and 14 Emb-/- embryos from six E17.5 Emb+/- breedings and 10 WT and 7 Emb-/- pups from four P0 Emb+/- breedings. Placentas were analyzed at E17.5. Sections (4 μm) were cut and immobilized to adhesion slides as mentioned earlier. The sections were deparaffinized, rehydrated, and stained with conventional hematoxylin and eosin (H&E), imaged with Pannoramic 250 Flash III slide scanner (3D Histech), and analyzed with the CaseViewer program (3D Histech). In the case of H&E-stained histological lung sections from E17.5 embryos and P0 pups, three images per organ section at 20x magnification were selected with the CaseViewer program. The relative area of airways in the lung section images was analyzed with ImageJ/Fiji.49
Transmission electron microscopy
Lungs from E18.5 WT and Emb−/− embryos (n = 3) were collected and fixed in 5% glutaraldehyde in 0.16 M s-collidine buffer, pH 7.4. The samples were post-fixed for 2 h with 2% OsO4 containing 3% potassium ferrocyanide, dehydrated with a series of increasing ethanol concentrations (70%, 96%, and twice at 100%), and embedded in 45359 Fluka Epoxy Embedding Medium kit. 70-nm sections were cut with an ultramicrotome, and stained with 1% uranyl acetate and 0.3% lead citrate. The sections were examined with a JEOL JEM-1400 Plus transmission electron microscope. Lamellar bodies in ten cells of each section were counted (n = 71-96) and areas were measured with ImageJ/Fiji software.
Protein extraction
Protein samples were extracted from organs with the NucleoSpin RNA/Protein kit (Macherey-Nagel). Macherey-Nagel Bead Tubes Type F was used in tissue homogenization. Protein concentrations were measured with Pierce 660nm Protein Assay (Thermo Scientific). When embigin expression in lung and kidney tissues from WT and Emb-/- were analyzed by Western blotting at different time points (E17.5, P3 and 4 months), organs from 3-5 individuals were pooled at E17.5 and at P3 time points before protein extraction.
Deglycosylation
50 μg of protein extracts from either lung, kidney, skin, epididymis, testis, or liver of WT mice at 4 months of age were deglycosylated for 4 hours at 37°C by using Glycoprotein Deglycosylation Kit (Sigma-Aldrich). The results were analyzed by Western blotting.
Western blotting
10–20 μg of protein were loaded on 8% or 10% SDS-PAGE gel or 4–20% FastGene SDS-PAGE gradient gels (Nippon Genetics). Embigin and β-tubulin or GAPDH (diluted in 5% milk and 0.1% Tween-20 in TBS) were stained in the membrane for 2 hours at RT. IRDye secondary antibodies and Odyssey CLx imager (LI-COR Biosciences) were used for signal detection.
RNA sequencing
Five E17.5 WT and Emb-/- lungs, kidneys, and placentas were dissected, and total RNA was isolated by using the NucleoSpin RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. The libraries were prepared from 300 ng of RNA / sample using TruSeq Stranded mRNA HT Kit and TruSeq Stranded mRNA Sample Preparation protocol 15031047 (Illumina). Sequencing was performed with NovaSeq 6000 SP Sequencing System (Illumina) using paired-end sequencing chemistry and 2 x 50 bp read length. The reads obtained from the instrument were base called using bcl2fastq2 conversion software. Raw data were obtained as fastq-files, which were uploaded to Chipster.50 The reads were aligned against the reference genome (Mus musculus GRCm38.95, available in Chipster) using STAR, version 2.7.3.51 The reads associated with each gene were counted using the HTSeq package, version 0.12.4.52
The edgeR R/Bioconductor package53 was used to normalize gene-wise read counts by the TMM normalization method and to perform statistical tests between groups. The results were filtered to have a minimum of 50 reads per gene in at least one sample. The gene was determined as differentially expressed if the following conditions were met: log2 of fold change value was above 0.6 or below -0.6 and Benjamini-Hochberg-corrected p-value less than 0.05. WebGestalt, http://www.webgestalt.org/,54 was used to perform an over-representation analysis of differentially expressed genes against biological process gene ontology. Morpheus (https://software.broadinstitute.org/morpheus) was used to generate heatmaps of the differentially expressed genes. For generating heatmaps by Morpheus (https://software.broadinstitute.org/morpheus), the raw counts were first transformed by using a deseq-transform function of the DEseq2 package.55
Quantification and statistical analysis
Statistical analysis
IBM SPSS Statistics software (version 25, IBM) was used for all statistical analyses. The correlation between the frequency of each genotype and embryonic stage, as well as the correlation between the body weight of the embryo and the relative area of airways in the lung sections, were analyzed with Spearman's rank correlation test. Statistical significance of the differences in the relative area of the airways between WT and Emb-/- lungs was determined with the Mann-Whitney U-test for the E17.5 lungs and the Student’s T-test for independent samples for the P0 lungs. The significance of the weight difference between WT and Emb-/- embryos was analyzed statistically by Student’s T-test for independent samples. When amniotic fluids were analyzed, statistical significance in alkaline phosphatase activity was determined by ANOVA followed by Dunnett’s t-test, ANOVA only was used for Ca2+ data, and the Kruskal-Wallis H-test with exact p-value was used for glucose and sodium data. The significance of the weight difference between WT and Emb-/- embryos and lungs at E17.5 was statistically analyzed by Student’s T-test for independent samples. The normalized number of peripheral buds in lung explants of WT and Emb-/- at E12.5 were analyzed statistically at days 0, 1, and 2 by Student’s T-test for independent samples. The number of ATII (SPC)- and ATI (HOP) -positive cells between WT and Emb-/- embryonic lungs at E17.5 was statistically compared: The number of ATII cells was statistically compared with Mann-Whitney U-test. The number of ATI cells was analyzed statistically by Student’s T-test for independent samples. The medians of the lamellar body count and area from WT and Emb-/- embryo lungs at E18.5 were analyzed statistically by Student’s T-test for independent samples. The fractions of Ki67-positive cells of all cells in WT and Emb-/- embryonic lungs were statistically compared by Student’s T-test for independent samples. A p-value < 0.05 was considered statistically significant. Statistical details of the experiments can be found in the figures and the figure legends.
Acknowledgments
The authors would like to thank the personnel of the Turku Center for Disease Modeling for their assistance. We are also grateful for the personnel in the Central Animal Laboratory of the University of Turku and in the Laboratory of Electron Microscopy at the University of Turku. Maria Tuominen is acknowledged for technical assistance and Anna-Brita Puranen for proofreading and text editing. This study has been financially supported by grants from the Academy of Finland (259769, J.H.), the Sigrid Jusélius Foundation (J.H.), the Cancer Foundation Finland (J.H.), the Finnish Foundation for Cardiovascular Research (J.H.), Research Council of Finland’s Flagship InFLAMES (337530 and 357910, J.H.), K. Albin Johanssons stiftelse (S.T.), and Orion Research Foundation sr (S.T.). The graphical abstract and summary figure (Figure 6) were created with BioRender.com.
Author contributions
S.T.: conceptualization; data curation; formal analysis; funding acquisition; investigation; validation; visualization; writing – original draft; writing – review and editing. J.J.: conceptualization; data curation; formal analysis; funding acquisition; investigation; validation; visualization; writing – original draft; writing – review and editing. K.S.: conceptualization; writing – review and editing. P.R.: formal analysis; data curation; writing – original draft; writing – review and editing. F-P.Z.: methodology; resources; writing – review and editing. M.P.: methodology; resources; writing – original draft; writing – review and editing. P.R.: conceptualization; funding acquisition; resources; writing – original draft; writing – review and editing. J.H.: conceptualization; funding acquisition; project administration; supervision; resources; writing – original draft; writing – review and editing.
Declaration of interests
The authors declare no competing interests.
Published: January 15, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.108914.
Supplemental information
References
- 1.Huang R.P., Ozawa M., Kadomatsu K., Muramatsu T. Embigin, a member of the immunoglobulin superfamily expressed in embryonic cells, enhances cell-substratum adhesion. Dev. Biol. 1993;155:307–314. doi: 10.1006/dbio.1993.1030. [DOI] [PubMed] [Google Scholar]
- 2.Ozawa M., Huang R.P., Furukawa T., Muramatsu T. A teratocarcinoma glycoprotein carrying a developmentally regulated carbohydrate marker is a member of the immunoglobulin gene superfamily. J. Biol. Chem. 1988;263:3059–3062. [PubMed] [Google Scholar]
- 3.Fan Q.W., Kadomatsu K., Uchimura K., Muramatsu T. Embigin/basigin subgroup of the immunoglobulin superfamily: different modes of expression during mouse embryogenesis and correlated expression with carbohydrate antigenic markers. Dev. Growth Differ. 1998;40:277–286. doi: 10.1046/j.1440-169x.1998.t01-1-00003.x. [DOI] [PubMed] [Google Scholar]
- 4.Huang R.P., Ozawa M., Kadomatsu K., Muramatsu T. Developmentally regulated expression of embigin, a member of the immunoglobulin superfamily found in embryonal carcinoma cells. Differentiation. 1990;45:76–83. doi: 10.1111/j.1432-0436.1990.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 5.Guenette R.S., Sridhar S., Herley M., Mooibroek M., Wong P., Tenniswood M. Embigin, a developmentally expressed member of the immunoglobulin super family, is also expressed during regression of prostate and mammary gland. Dev. Genet. 1997;21:268–278. doi: 10.1002/(SICI)1520-6408(1997)21:4<268::AID-DVG4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Hanna S.M., Kirk P., Holt O.J., Puklavec M.J., Brown M.H., Barclay A.N. A novel form of the membrane protein CD147 that contains an extra Ig-like domain and interacts homophilically. BMC Biochem. 2003;4:17. doi: 10.1186/1471-2091-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao C.-G., Kong L.-M., Song F., Xing J.-L., Wang L.-X., Sun Z.-J., Tang H., Yao H., Zhang Y., Wang L., et al. Characterization of basigin isoforms and the inhibitory function of basigin-3 in human hepatocellular carcinoma proliferation and invasion. Mol. Cell Biol. 2011;31:2591–2604. doi: 10.1128/MCB.05160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langnaese K., Beesley P.W., Gundelfinger E.D. Synaptic membrane glycoproteins gp65 and gp55 are new members of the immunoglobulin superfamily. J. Biol. Chem. 1997;272:821–827. doi: 10.1074/jbc.272.2.821. [DOI] [PubMed] [Google Scholar]
- 9.Ochrietor J.D., Moroz T.P., van Ekeris L., Clamp M.F., Jefferson S.C., deCarvalho A.C., Fadool J.M., Wistow G., Muramatsu T., Linser P.J. Retina-specific expression of 5A11/Basigin-2, a member of the immunoglobulin gene superfamily. Invest. Ophthalmol. Vis. Sci. 2003;44:4086–4096. doi: 10.1167/iovs.02-0995. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida S., Shibata M., Yamamoto S., Hagihara M., Asai N., Takahashi M., Mizutani S., Muramatsu T., Kadomatsu K. Homo-oligomer formation by basigin, an immunoglobulin superfamily member, via its N-terminal immunoglobulin domain. Eur. J. Biochem. 2000;267:4372–4380. doi: 10.1046/j.1432-1327.2000.01482.x. [DOI] [PubMed] [Google Scholar]
- 11.Yu X.-L., Hu T., Du J.-M., Ding J.-P., Yang X.-M., Zhang J., Yang B., Shen X., Zhang Z., Zhong W.-D., et al. Crystal structure of HAb18G/CD147: implications for immunoglobulin superfamily homophilic adhesion. J. Biol. Chem. 2008;283:18056–18065. doi: 10.1074/jbc.M802694200. [DOI] [PubMed] [Google Scholar]
- 12.Hahn J.N., Kaushik D.K., Yong V.W. The role of EMMPRIN in T cell biology and immunological diseases. J. Leukoc. Biol. 2015;98:33–48. doi: 10.1189/jlb.3RU0215-045R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beesley P.W., Herrera-Molina R., Smalla K.-H., Seidenbecher C. The Neuroplastin adhesion molecules: key regulators of neuronal plasticity and synaptic function. J. Neurochem. 2014;131:268–283. doi: 10.1111/jnc.12816. [DOI] [PubMed] [Google Scholar]
- 14.Hill I.E., Selkirk C.P., Hawkes R.B., Beesley P.W. Characterization of novel glycoprotein components of synaptic membranes and postsynaptic densities, gp65 and gp55, with a monoclonal antibody. Brain Res. 1988;461:27–43. doi: 10.1016/0006-8993(88)90722-6. [DOI] [PubMed] [Google Scholar]
- 15.Smalla K.H., Matthies H., Langnäse K., Shabir S., Böckers T.M., Wyneken U., Staak S., Krug M., Beesley P.W., Gundelfinger E.D. The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. Proc. Natl. Acad. Sci. USA. 2000;97:4327–4332. doi: 10.1073/pnas.080389297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirk P., Wilson M.C., Heddle C., Brown M.H., Barclay A.N., Halestrap A.P. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000;19:3896–3904. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisel P., Schaeffeler E., Schwab M. Clinical and Functional Relevance of the Monocarboxylate Transporter Family in Disease Pathophysiology and Drug Therapy. Clin. Transl. Sci. 2018;11:352–364. doi: 10.1111/cts.12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson M.C., Meredith D., Fox J.E.M., Manoharan C., Davies A.J., Halestrap A.P. Basigin (CD147) is the target for organomercurial inhibition of monocarboxylate transporter isoforms 1 and 4: the ancillary protein for the insensitive MCT2 is EMBIGIN (gp70) J. Biol. Chem. 2005;280:27213–27221. doi: 10.1074/jbc.M411950200. [DOI] [PubMed] [Google Scholar]
- 19.Skiba N.P., Cady M.A., Molday L., Han J.Y.S., Lewis T.R., Spencer W.J., Thompson W.J., Hiles S., Philp N.J., Molday R.S., Arshavsky V.Y. TMEM67, TMEM237, and embigin in complex with monocarboxylate transporter MCT1 are unique components of the photoreceptor outer segment plasma membrane. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halestrap A.P. The SLC16 gene family - structure, role and regulation in health and disease. Mol. Aspect. Med. 2013;34:337–349. doi: 10.1016/j.mam.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi K., Sugiyama K., Tomabechi R., Kishimoto H., Inoue K. Mammalian monocarboxylate transporter 7 (MCT7/Slc16a6) is a novel facilitative taurine transporter. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silberstein L., Goncalves K.A., Kharchenko P.V., Turcotte R., Kfoury Y., Mercier F., Baryawno N., Severe N., Bachand J., Spencer J.A., et al. Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell. 2016;19:530–543. doi: 10.1016/j.stem.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sipilä K., Rognoni E., Jokinen J., Tewary M., Vietri Rudan M., Talvi S., Jokinen V., Dahlström K.M., Liakath-Ali K., Mobasseri A., et al. Embigin is a fibronectin receptor that affects sebaceous gland differentiation and metabolism. Dev. Cell. 2022;57:1453–1465.e7. doi: 10.1016/j.devcel.2022.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nabeshima K., Iwasaki H., Koga K., Hojo H., Suzumiya J., Kikuchi M. Emmprin (basigin/CD147): matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol. Int. 2006;56:359–367. doi: 10.1111/j.1440-1827.2006.01972.x. [DOI] [PubMed] [Google Scholar]
- 25.Riethdorf S., Reimers N., Assmann V., Kornfeld J.-W., Terracciano L., Sauter G., Pantel K. High incidence of EMMPRIN expression in human tumors. Int. J. Cancer. 2006;119:1800–1810. doi: 10.1002/ijc.22062. [DOI] [PubMed] [Google Scholar]
- 26.Chao F., Zhang J., Zhang Y., Liu H., Yang C., Wang J., Guo Y., Wen X., Zhang K., Huang B., et al. Embigin, regulated by HOXC8, plays a suppressive role in breast tumorigenesis. Oncotarget. 2015;6:23496–23509. doi: 10.18632/oncotarget.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung D.E., Kim J.M., Kim C., Song S.Y. Embigin is overexpressed in pancreatic ductal adenocarcinoma and regulates cell motility through epithelial to mesenchymal transition via the TGF-β pathway. Mol. Carcinog. 2016;55:633–645. doi: 10.1002/mc.22309. [DOI] [PubMed] [Google Scholar]
- 28.Chen A., Liao S., Cheng M., Ma K., Wu L., Lai Y., Qiu X., Yang J., Xu J., Hao S., et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022;185:1777–1792.e21. doi: 10.1016/j.cell.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Tabula Muris Consortium. Overall coordination. Logistical coordination. Organ collection and processing. Library preparation and sequencing. Computational data analysis. Cell type annotation. Writing group. Supplemental text writing group. Principal investigators Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brocklehurst D., Wilde C.E. Amniotic fluid alkaline phosphatase, gamma-glutamyltransferase, and 5’-nucleotidase activity from 13 to 40 weeks’ gestation, and alkaline phosphatase as an index of fetal lung maturity. Clin. Chem. 1980;26:588–591. [PubMed] [Google Scholar]
- 31.Cheung C.Y., Brace R.A. Amniotic fluid volume and composition in mouse pregnancy. J. Soc. Gynecol. Invest. 2005;12:558–562. doi: 10.1016/j.jsgi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Lu T., Lin X., Pan Y.-H., Yang N., Ye S., Zhang Q., Wang C., Zhu R., Zhang T., Wisniewski T.M., et al. ADAMTS18 Deficiency Leads to Pulmonary Hypoplasia and Bronchial Microfibril Accumulation. iScience. 2020;23 doi: 10.1016/j.isci.2020.101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh I., Mehta A., Contreras A., Boettger T., Carraro G., Wheeler M., Cabrera-Fuentes H.A., Bellusci S., Seeger W., Braun T., Barreto G. Hmga2 is required for canonical WNT signaling during lung development. BMC Biol. 2014;12:21. doi: 10.1186/1741-7007-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurotani R., Tomita T., Yang Q., Carlson B.A., Chen C., Kimura S. Role of Secretoglobin 3A2 in Lung Development. Am. J. Respir. Crit. Care Med. 2008;178:389–398. doi: 10.1164/rccm.200707-1104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Y., Guo M., Whitsett J.A., Xu Y. “LungGENS”: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70:1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du Y., Kitzmiller J.A., Sridharan A., Perl A.K., Bridges J.P., Misra R.S., Pryhuber G.S., Mariani T.J., Bhattacharya S., Guo M., et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax. 2017;72:481–484. doi: 10.1136/thoraxjnl-2016-209598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Y., Ouyang W., Kitzmiller J.A., Guo M., Zhao S., Whitsett J.A., Xu Y. Lung Gene Expression Analysis Web Portal Version 3: Lung-at-a-Glance. Am. J. Respir. Cell Mol. Biol. 2021;64:146–149. doi: 10.1165/rcmb.2020-0308LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawlins E.L., Okubo T., Xue Y., Brass D.M., Auten R.L., Hasegawa H., Wang F., Hogan B.L.M. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pridans C., Holmes M.L., Polli M., Wettenhall J.M., Dakic A., Corcoran L.M., Smyth G.K., Nutt S.L. Identification of Pax5 target genes in early B cell differentiation. J. Immunol. 2008;180:1719–1728. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- 41.Xu B., Zhang M., Zhang B., Chi W., Ma X., Zhang W., Dong M., Sheng L., Zhang Y., Jiao W., et al. Embigin facilitates monocarboxylate transporter 1 localization to the plasma membrane and transition to a decoupling state. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111343. [DOI] [PubMed] [Google Scholar]
- 42.Roman J., McDonald J.A. Expression of fibronectin, the integrin alpha 5, and alpha-smooth muscle actin in heart and lung development. Am. J. Respir. Cell Mol. Biol. 1992;6:472–480. doi: 10.1165/ajrcmb/6.5.472. [DOI] [PubMed] [Google Scholar]
- 43.De Langhe S.P., Sala F.G., Del Moral P.M., Fairbanks T.J., Yamada K.M., Warburton D., Burns R.C., Bellusci S. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev. Biol. 2005;277:316–331. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Varner V.D., Nelson C.M. Cellular and physical mechanisms of branching morphogenesis. Development. 2014;141:2750–2759. doi: 10.1242/dev.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Igakura T., Kadomatsu K., Kaname T., Muramatsu H., Fan Q.W., Miyauchi T., Toyama Y., Kuno N., Yuasa S., Takahashi M., et al. A null mutation in basigin, an immunoglobulin superfamily member, indicates its important roles in peri-implantation development and spermatogenesis. Dev. Biol. 1998;194:152–165. doi: 10.1006/dbio.1997.8819. [DOI] [PubMed] [Google Scholar]
- 46.Manwani N., Gagnon S., Post M., Joza S., Muglia L., Cornejo S., Kaplan F., Sweezey N.B. Reduced Viability of Mice with Lung Epithelial-Specific Knockout of Glucocorticoid Receptor. Am. J. Respir. Cell Mol. Biol. 2010;43:599–606. doi: 10.1165/rcmb.2009-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin N., Cho S.-N., Raso M.G., Wistuba I., Smith Y., Yang Y., Kurie J.M., Yen R., Evans C.M., Ludwig T., et al. Mig-6 is required for appropriate lung development and to ensure normal adult lung homeostasis. Development. 2009;136:3347–3356. doi: 10.1242/dev.032979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokomizo T., Yamada-Inagawa T., Yzaguirre A.D., Chen M.J., Speck N.A., Dzierzak E. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 2012;7:421–431. doi: 10.1038/nprot.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kallio M.A., Tuimala J.T., Hupponen T., Klemelä P., Gentile M., Scheinin I., Koski M., Käki J., Korpelainen E.I. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-seq data have been deposited at the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) and are publicly available as of the date of publication. Accession number is listed in the key resource table. All the data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported will be shared by the lead contact upon request.