Abstract
Biofilm formation by the pathobiont Haemophilus influenzae is associated with human nasopharynx colonization, otitis media in children, and chronic respiratory infections in adults suffering from chronic respiratory diseases such as chronic obstructive pulmonary disease (COPD). β-lactam and quinolone antibiotics are commonly used to treat these infections. However, considering the resistance of biofilm-resident bacteria to antibiotic-mediated killing, the use of antibiotics may be insufficient and require being replaced or complemented with novel strategies. Moreover, unlike the standard minimal inhibitory concentration assay used to assess antibacterial activity against planktonic cells, standardization of methods to evaluate anti-biofilm drug activity is limited. In this work, we detail a panel of protocols for systematic analysis of drug antimicrobial effect on bacterial biofilms, customized to evaluate drug effects against H. influenzae biofilms. Testing of two cinnamaldehyde analogs, (E)-trans-2-nonenal and (E)-3-decen-2-one, demonstrated their effectiveness in both H. influenzae inhibition of biofilm formation and eradication or preformed biofilms. Assay complementarity allowed quantifying the dynamics and extent of the inhibitory effects, also observed for ampicillin resistant clinical strains forming biofilms refractory to this antibiotic. Moreover, cinnamaldehyde analog encapsulation into poly(lactic-co-glycolic acid) (PLGA) polymeric nanoparticles allowed drug vehiculization while maintaining efficacy. Overall, we demonstrate the usefulness of cinnamaldehyde analogs against H. influenzae biofilms, present a test panel that can be easily adapted to a wide range of pathogens and drugs, and highlight the benefits of drug nanoencapsulation towards safe controlled release.
Keywords: Biofilm, Haemophilus influenzae, Anti-Biofilm drugs, Cinnamaldehyde-analogs, Nanoformulation, Multimodal methods
1. Introduction
Haemophilus influenzae is a pathobiont highly adapted to the human airways. The majority of H. influenzae strains in carriage and disease are nontypeable (NTHi) isolates lacking polysaccharide capsules, and are unaffected by the protective immune response generated by the H. influenzae type b (Hib) conjugate vaccine. NTHi strains cause otitis media, conjunctivitis, sinusitis and lower respiratory infections in children; exacerbations of chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF) in adults [[1], [2], [3], [4]]. NTHi multicellular biofilm communities associate with nasopharynx carriage in healthy people, and also with persistent ear and lung infection in otitis media, chronic bronchitis, rhinosinusitis and COPD patients [[5], [6], [7]]. Moreover, H. influenzae antimicrobial resistance shows an overall increasing trend, and ampicillin resistance led to include this pathogen in the WHO list of priority pathogens [8]. Even more, biofilm architecture enhances NTHi resistance/tolerance to clinically important antibiotics including ciprofloxacin, ampicillin, amoxicillin, gentamicin or erythromycin [9,10]. Conversely, although sub-inhibitory concentrations of azithromycin decrease the formation and thickness of NTHi biofilms, clinical evidence shows that long-term, low-dose administration of this antibiotic to COPD patients triggers macrolide resistance [11,12], possibly undermining its use in chronic biofilm infections involving this pathogen. In this context, anti-biofilm drug development should contribute to expanding the global antibacterial pipeline. Moreover and closely related, standardized guidelines for anti-biofilm drug testing should be made available to parallel standard minimal inhibitory concentration (MIC) testing against planktonic cells [[13], [14], [15]].
Molecules involved in NTHi biofilm formation and/or present in the biofilm extracellular polymeric substance (EPS) are potential targets for anti-biofilm strategies. Thus, antibodies against the majority subunit of type IV pili PilA mediate dispersal of NTHi from biofilms conferring therapeutic resolution of otitis media [16]. Moreover, the nuclear-associated DNABII proteins present in the extracellular DNA (eDNA)-dependent matrix that shields sessile bacteria, bind to bent eDNA or bend such eDNA upon binding, which provides structural integrity to the biofilm. Anti-DNABII antibodies efficiently disrupt biofilms and work additively when combined with anti-PilA antibodies [[17], [18], [19], [20], [21], [22], [23], [24]]. Also, considering possible benefits of combining antimicrobial and anti-inflammatory activities, the effects of a whole range of nutraceuticals against NTHi biofilms have been reported, including those of apple polyphenols, honey, chalcones and several plant essential oils [[25], [26], [27], [28], [29]]. However, targeting quorum sensing (QS) systems is an exploited anti-biofilm approach [30] that has not been reported for H. influenzae. NTHi QS is mediated by autoinducer-2 (AI-2), synthesized from 4,5-dihydroxy-2,3-pentadione (DPD) in a reaction catalyzed by the LuxS protein, and contributes to NTHi biofilm thickness and density [[31], [32], [33], [34], [35], [36]]. Inhibition of AI-2 QS can be achieved, among others, by cinnamaldehyde analogs which comply with Lipinski's rule-of-five [37] and have an already proven therapeutic window high enough for therapeutic applications in human and animals [38]. Suitability of these drug-like molecules as NTHi anti-biofilm therapeutics is unknown. In this work, we evaluated the effects of two cinnamaldehyde analogs, (E)-trans-2-nonenal and (E)-3-decen-2-one, against H. influenzae biofilms. Detailed protocols towards standardized analysis of drug antimicrobial effect on bacterial biofilms were used, which can be easily adapted to a wide range of drugs and pathogens besides H. influenzae. We demonstrate the antimicrobial effects of these molecules against H. influenzae biofilms, further explore options for their nanoencapsulation, and provide data supporting the efficacy of cinnamaldehyde analog-loaded polymeric nanoparticles.
2. Methods and materials
Bacterial strains, media, and growth conditions. NTHi clinical strains used in this study are R2866 (blood isolate) [39], P605 and P639 (COPD sputum sample isolates) [40]. Bacteria were grown at 37 °C with 5 % CO2 on PolyViteX agar (PVX agar, Biomérieux, 43101) or on Haemophilus Test Medium agar (HTM, Oxoid, CM0898) supplemented with 10 μg/mL hemin (Merck, H9039) and 10 μg/mL nicotinamide adenine dinucleotide (NAD, Merck, N0632), referred to sHTM agar. Liquid cultures were grown at 37 °C with 5 % CO2 in supplemented brain-heart infusion (Oxoid, CM1135) referred to as sBHI. When necessary, ampicillin (Merck, A9393), (E)-trans-2-nonenal (Merck, 255653), (E)-3-decen-2-one (Merck, W353205), or (E)-trans-2-nonenal poly(lactic-co-glycolic acid) (PLGA) nanoparticles (see below) were used.
Synthesis of nanoparticles for encapsulation of cinnamaldehyde analogs.(i) Lipid nanoparticles. Nanoemulsions (NE), lipid nanoparticles constituted by oil, water, and surfactants, were used for cinnamaldehyde analog encapsulation. The synthesis of (±)-α-tocopherol NE was previously described [41]. Briefly, an ethanolic organic phase containing (±)-α-tocopherol (Merck, 10191410), octadecylamine (Merck, 124301) and Tween 80 (Croda, SD82825) was rapidly injected into ultrapure water at room temperature (RT) under continuous stirring (700 r.p.m.). The resulting NE were purified to remove solvent and non-incorporated excipients by size exclusion chromatography, using P10 desalting columns (bed size 14.5 mm × 50 mm, Cytiva, 1708501). For preparation of (E)-trans-2-nonenal-loaded NE, the active compound was added to the ethanol solution prior to injection into the aqueous phase (Table S1). (ii) Polymeric nanoparticles. Poly(D,L)-lactide-co-glycolide (PLGA, acid terminated, lactide:glycolide 75:25, Mw 4000–15,000, Merck, 719919) nanoparticles (NP) with (E)-trans-2-nonenal were synthesized by simple emulsion method. Briefly, PLGA and (E)-trans-2-nonenal were dissolved in ethyl acetate (≥99.7 %, Merck, 34858). Then, 1 % w/v PVA (Mw 9000–10,000, 80 % hydrolyzed, Merck, 360627) aqueous solution was added to the organic phase and sonicated for 1 min using a UP200S sonicator (Hielscher) with a 2 mm probe. Next, the mixture was added to a PVA 0.1 % w/v aqueous solution under magnetic stirring (500 r.p.m.). This solution was then kept under stirring until complete evaporation of ethyl acetate. NP were washed by three centrifugation cycles (14,800 r.p.m., 60 min, 4 °C) to remove excess of surfactant and non-encapsulated (E)-trans-2-nonenal, and then resuspended in Type II water. Blank NP were prepared by the same method.
Physico-chemical characterization of (E)-trans-2-nonenal nanoformulations.(i) Particle size characterization. The hydrodynamic diameter and polydispersity index (PDI) of blank and (E)-trans-2-nonenal-loaded formulations (NE and NP) were assessed by photon correlation spectroscopy (PCS) using a Zetasizer nano ZS90 (Malvern Panalitical, UK). All measurements were performed in triplicate at RT. (ii) Determination of zeta potential. The surface charge of blank and (E)-trans-2-nonenal-loaded nanoformulations (NE and NP) was determined by Electrophoretic Light Scattering (ELS) using a Zetasizer Nano ZS90 (Malvern Instruments, UK). The nanosuspensions were diluted (1:20) in NaCl 100 mM prior to zeta potential analysis. Samples were analyzed in triplicate at RT. (iii) Stability studies. Blank and (E)-trans-2-nonenal-loaded nanoformulations (NE and NP) were subjected to stability studies in triplicate. Physical stability in ultrapure water was evaluated following storage at 4 °C and RT, by measurement of hydrodynamic size and zeta potential at pre-established time-points. Stability was also tested in sBHI at 37 °C at different dilution ranges up to 48 h. (iv) Determination of entrapment efficiency of (E)-trans-2-nonenal. The concentration of (E)-trans-2-nonenal in loaded-NE was determined before and after the purification process through HPLC-UV (Agilent 1260 system) analysis using a Zorbax Eclipse XDB-C18 column (4.6x150 3.5 μm). The chromatography was performed at a flow rate of 1 mL/min in isocratic conditions, using acetonitrile/water (60:40 v/v). NE was disrupted for analysis by dilution 1:5 in the mobile phase. Likewise, (E)-trans-2-nonenal concentration in PLGA NPs was determined through HPLC-UV (Agilent 1260 Infinity) at 226 nm using a Zorbax Eclipse Plus C18 (4.6x150 5.0 μm) at 35 °C. The analysis was performed with a flow rate of 1 mL/min, using acetonitrile/water (50:50 v/v) in isocratic conditions. NP were previously disrupted in acetonitrile. In both NE and NP, entrapment efficiency (EE) was calculated using the following formula: EE (%) = CAP/CBP X 100, where CAP is (E)-trans-2-nonenal concentration found in disrupted nanoformulation after the purification process, and CBP is (E)-trans-2-nonenal concentration found in disrupted nanoformulation before the purification process. (v) (E)-trans-2-nonenal release. Purified (E)-trans-2-nonenal-loaded PLGA NP were suspended in ultrapure water and incubated at 4, 25 or 37 °C for 80 days with continuous shaking, 150 r.p.m. At pre-determined time intervals, samples were collected, centrifuged (14,800 r.p.m., 30 min), filtered through 0.22 μm filters, and analyzed via HPLC. Drug release of the PLGA NP formulations in water, PBS and sBHI at 37 °C up to 48 h was also evaluated. Assays were conducted in triplicate, and the cumulative drug release profiles were constructed.
Drug minimal inhibitory concentration (MIC). Ampicillin, (E)-trans-2-nonenal and (E)-3-decen-2-one susceptibility was determined by microdilution broth method, following CLSI and EUCAST guidelines [42,43] with some modifications. Each drug was prepared as it follows: ampicillin was freshly prepared as a stock solution at 100 mg/mL in distilled H2O (dH2O), filtered, and diluted to 256 μg/mL in sBHI. (E)-trans-2-nonenal was prepared as a stock solution at 8.46 mg/mL in 95 % ethanol and diluted to 200 μg/mL in sBHI. (E)-3-decen-2-one was prepared as a stock solution at 8.22 mg/mL in 95 % ethanol and diluted to 400 μg/mL in sBHI. (E)-trans-2-nonenal PLGA NP were diluted to 200 μg/mL in sBHI (NP concentrations were calculated to get 200 μl/mL (E)-trans-2-nonenal in the PLGA NP starting working solution; please note that PLGA concentrations may vary from batch to batch). Next, 200 μL of each drug were added to individual wells in row A in 96-well plates (Sarstedt, 82.1581.001). One hundred-microliter aliquots of sBHI were transferred to individual wells in the rest of the plate. Next, 100 μL aliquots were serially transferred from wells in row A to wells in row B and up to row G, where 100 μL were discarded. For bacterial inoculum, a suspension of bacteria grown on PVX agar was generated with fresh sBHI, adjusted to 0.5 MacFarland (OD600 = 0.063) and diluted 1:100 in sBHI (∼1,5 × 106 colony forming units (CFU)/mL). Next, 100 μL bacterial aliquots were transferred to each well (∼1,5 × 105 CFU/well). Plates were incubated for 24 h at 37 °C, 5 % CO2, without shaking. MIC was determined as the lowest concentration of drug where not bacterial growth was observed. Bacterial growth controls (without antibiotics) were included in each case (row H). Assays were performed in triplicate on three independent occasions (n = 3).
Biofilm formation inhibition assays. Drug working solutions were prepared in sBHI as above indicated, i.e. ampicillin, 256 μg/mL; (E)-trans-2-nonenal, 200 μg/mL; (E)-3-decen-2-one, 400 μg/mL; (E)-trans-2-nonenal PLGA NP, 200 μg/mL. Suspensions of bacteria grown on PVX agar were generated with fresh sBHI, adjusted to 0.5 MacFarland (OD600 = 0.063), and diluted 1:100 in sBHI (∼1,5 × 106 CFU/mL). Drug inhibitory effects on biofilm formation were assessed with three assay types: (i) Crystal violet staining. Drugs and bacteria were disposed in 96-well plates same as specified above for MIC determination (see above section, Drug minimal inhibitory concentration (MIC)). Plates were incubated at 37 °C with 5 % CO2 for 24 h without shaking. The liquid portion in each well was then discarded, plates were washed 3 times by gently pipetting 200 μL dH2O/well and allowed to air dry. Next, 150 μL/well 0.5 % crystal violet (Merck, V5265) were added, and plates were incubated for 20 min at RT on gentle agitation, followed by plate washing as previously described. Finally, 150 μL/well 95 % ethanol were added, plates were incubated for 20 min at RT on gentle agitation, and OD570 was determined on a SynergyH1 (Biotek) microplate reader as a measure of biofilm biomass. OD570 for each strain from at least three independent assays performed in triplicate (n ≥ 3) was calculated. (ii) Confocal Laser Scanning Microscopy (CLSM). Once drug solutions and bacterial suspensions were prepared, 1 mL of each drug was added to individual wells in row A in 24-well plates (Greiner Bio-one, 662102). Five hundred-microliter aliquots of sBHI were transferred to individual wells in the rest of the plate. Next, 500 μL aliquots were serially transferred from wells in row A to wells in row B and up to row C, where 500 μL were discarded. Next, 500 μL bacterial aliquots were transferred to each well (∼7,5 × 105 CFU/well). Bacterial growth controls (without antibiotics) were included in each case (row D). Next, 250 μL of each dilution were transferred to eight-well chambered coverglass (Ibidi GmbH, 80826). Chambers were incubated at 37 °C with 5 % CO2 for 24 h without shaking. Medium was then exchanged by 200 μL saline solution (NaCl 0.85 %) with 0.3 μL SYTO9 for staining live bacteria and 0.3 μL propidium iodide (PI) for staining dead bacteria (Live-dead kit BacLight™, Invitrogen), and incubated for 20 min at RT in darkness prior CLSM analysis. CLSM image acquisition was performed using a Zeiss LSM 880 AxioObserver inverted confocal microscope equipped with a LD LCI Plan-Apochromat 40x 1.2 NA W objective. Five Z-stack images per sample were acquired at the center of each well, with a distance of 0.48 μm between planes. 488 nm and 561 nm excitation lasers were used for SYTO9 and PI, respectively. Each assay was replicated in two separate experiments, which included duplicate wells for each strain. CLSM stacks were processed with fully-automated in-house software. (iii) Real-time cell analysis. Continuous real-time biofilm monitoring was performed using an xCELLigence RTCA (Real-Time Cell Analysis) SP instrument (ACEA Biosciences, San Diego, CA, USA), following [44]. This assay is based on the ability of bacteria to impede electric current when they attach and grow on the gold electrodes placed at the bottom of 96-well E-plates (ACEA Biosciences, 300601010). Cell sensor impedance was expressed as arbitrary units (cell index, CI) which correlate with the total biofilm mass. CI at each time point is defined as (Zn−Zb)/15, where Zn is the cell electrode impedance of the well when it contains bacterial cells, and Zb is the background impedance with growth media. For assessing inhibition of biofilm growth, drug solutions and bacterial suspensions were prepared as described above. Next, 200 μL of each drug were added to individual wells in row A in 96-well plates (ampicillin, 256 μg/mL; (E)-trans-2-nonenal, 200 μg/mL; (E)-3-decen-2-one, 400 μg/mL). One hundred-microliter aliquots of sBHI were transferred to individual wells in the rest of the plate. Next, 100 μL aliquots were serially transferred from wells in row A to wells in row B and up to row G, where 100 μL were discarded. Impedance background was then measured for 5 min using the standard protocol provided by the software. Next, 100 μL bacterial aliquots were transferred to each well (∼1,5 × 105 CFU/well). Plates were positioned in the xCELLigence Real-Time Cell Analyzer MP, incubated at 37 °C and monitored on the RTCA system at 15 min time intervals for 24 h. For data analysis, CI values were normalized by the negative control impedance, and the average of four replicates was calculated for plotting. Assays were performed in quadruplicate, on at least three independent occasions (n ≥ 3).
Eradication of preformed biofilms. Drug inhibitory effects on previously formed biofilms were assessed using two assay types: (i) Viable CFU counts. Biofilms were first grown. Briefly, a suspension of bacteria grown on PVX agar was generated with fresh sBHI, adjusted to 0.5 MacFarland (OD600 = 0.063), and diluted 1:100 in sBHI (∼1,5 × 106 CFU/mL). Next, 200 μL bacterial aliquots were transferred to each well in 96-well plates (∼3 × 105 CFU/well). Plates were incubated for 24 h at 37 °C, 5 % CO2, without shaking. By using drug stock solutions prepared as detailed above, drug working solutions were prepared in phosphate buffer saline (PBS), ampicillin, 6400 μg/mL; (E)-trans-2-nonenal; 400 μg/mL; (E)-3-decen-2-one, 3200 μg/mL; (E)-trans-2-nonenal/PLGA nanoparticles, 400/1133 μg/mL. Drugs were 2-fold serially diluted in PBS in 2 mL eppendorf tubes. The liquid portion in each biofilm-containing well was then discarded, and 200 μL/well of each drug dilution were added. Bacterial growth PBS or PLGA controls were included in each case. Plates were incubated for 6 h at 37 °C, 5 % CO2. Liquid was then removed, and 200 μL PBS were added to each well. Plates were incubated at RT for 5 min and shaking (200 r.p.m.), and biofilm cells were mechanically detached using a pipet tip for 45 s. Next, 100 μL aliquots from each well were used for 10-fold serial dilution, plating on sHTM agar, and CFU counting. Assays were performed in triplicate on three independent occasions (n = 3). (ii) Real-time cell analysis. First, 90 μL sBHI aliquots were added to 96-well E-plates for background measurements. Drugs were prepared as described in (i) Viable CFU counts. Then, 90 μL bacterial aliquots (bacteria grown on PVX agar, suspension generated with fresh sBHI, adjusted to 0.5 MacFarland (OD600 = 0.063), and diluted 1:50 in sBHI, ∼3 × 106 CFU/mL; ∼2,7 × 105 CFU/well) were added to each well, and monitored on the RTCA system for 12 h. The program was then paused for drug addition (20 μL/well for each drug dilution to be tested), and biofilms were monitored for an additional 24 h. Each drug concentration was tested in duplicate; negative controls without drug were included. For data analysis, CI values for each well were normalized by the negative control impedance. Assays were repeated at least three times, with four technical replicates per assay (n ≥ 3).
In vivo toxicity. Larvae of Galleria mellonella were fed on an artificial diet (15 % corn flour, 15 % wheat flour, 15 % cereals, 11 % skim milk powder, 6 % dried brewer's yeast, 25 % honey, and 13 % glycerol) and reared at 34 °C. Larvae with weights ranging between 200 and 250 mg were selected and divided into groups (n = 6), ensuring that each group had similar weights (±5 mg). Larvae in the different groups were injected with different drug concentrations using a 22G syringe (Hamilton) through the top left proleg. A control group was injected with 1X PBS (Fisher Scientific, BP399), and mortality was recorded during 48 h. For data analysis, survival curves were plotted using Kaplan-Meier analysis and LD50 (median lethal dose) was calculated as the necessary drug dose to kill 50 % of the larvae within 48 h as described previously [45].
Statistical analyses. In all cases, p-value (p) < 0.05 was considered was considered statistically significant. Analyses were performed using Prism software, version 7 for Mac (GraphPad software, San Diego, CA) statistical package and are detailed in each Figure Legend.
3. Results and discussion
3.1. Cinnamaldehyde analogs inhibit H. influenzae biofilm growth independently of ampicillin resistance levels
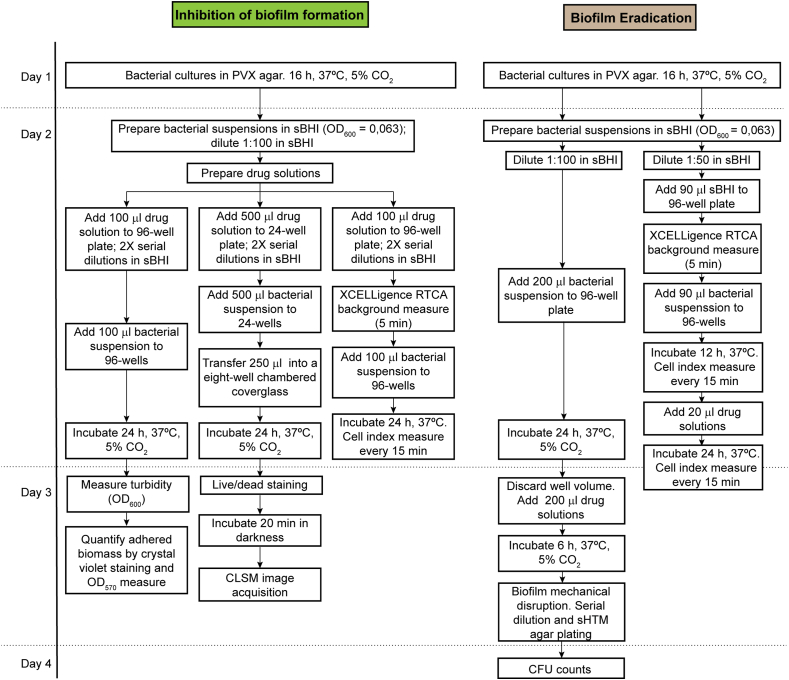
We used a panel of protocols to assess the activity of molecules against H. influenzae biofilms, by following a workflow summarized in Fig. 1. Three biofilm inhibition procedures were employed (Fig. 1, left panel). The first one relies on the use of crystal violet staining to quantify adherent biofilm biomass within the wells of microtiter plates. In a complementary manner, as crystal violet staining does not distinguish between bacterial cells and EPS, we used confocal laser scanning microscopy (CLSM) imaging to assess adherent viable bacteria on the wells of chambered coverglass-bottomed devices. Given that these two assays do not address inhibition of biofilm formation in a dynamic manner, we also used label-free electrical measurements by xCELLigence real-time cell analysis (RTCA).
Fig. 1.
Workflow of in vitro assays to test drug inhibition and eradication effects against H. influenzae bacterial biofilms. Three complementary assays were used to evaluate drug effects in terms of inhibition of biofilm formation (left panels). Two complementary assays were used to test drug efficacy on previously established biofilms (right panels).
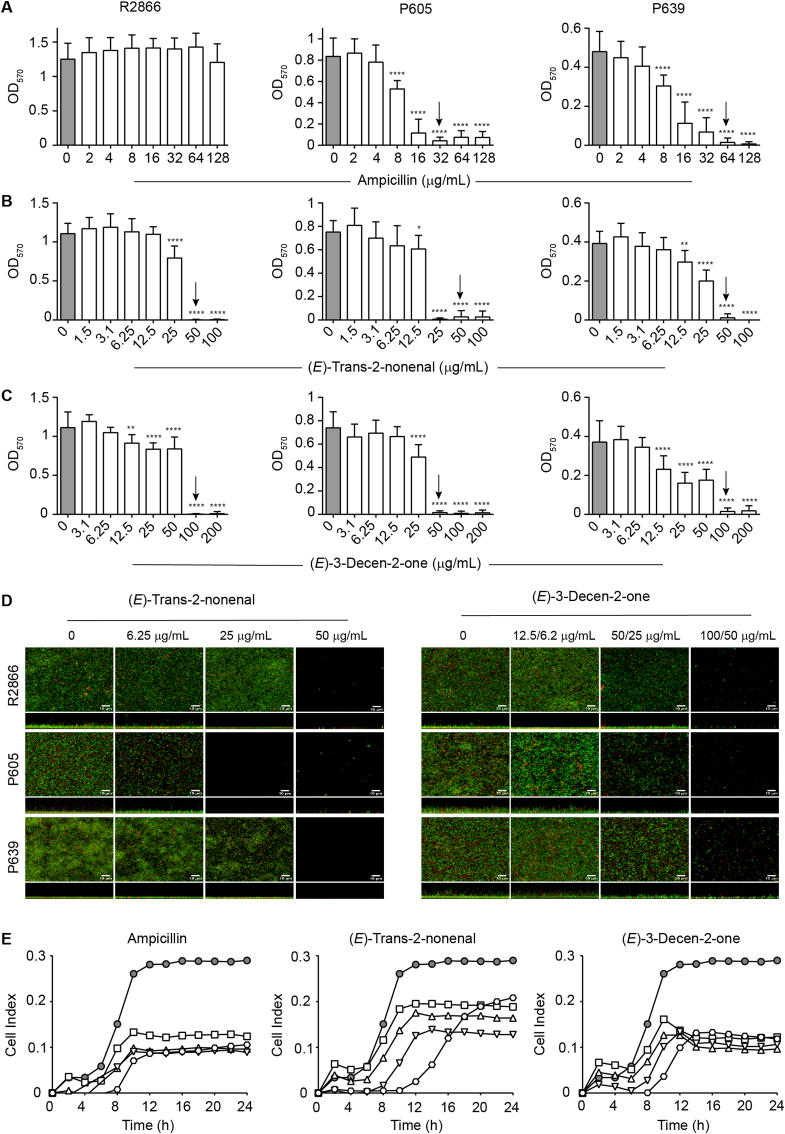
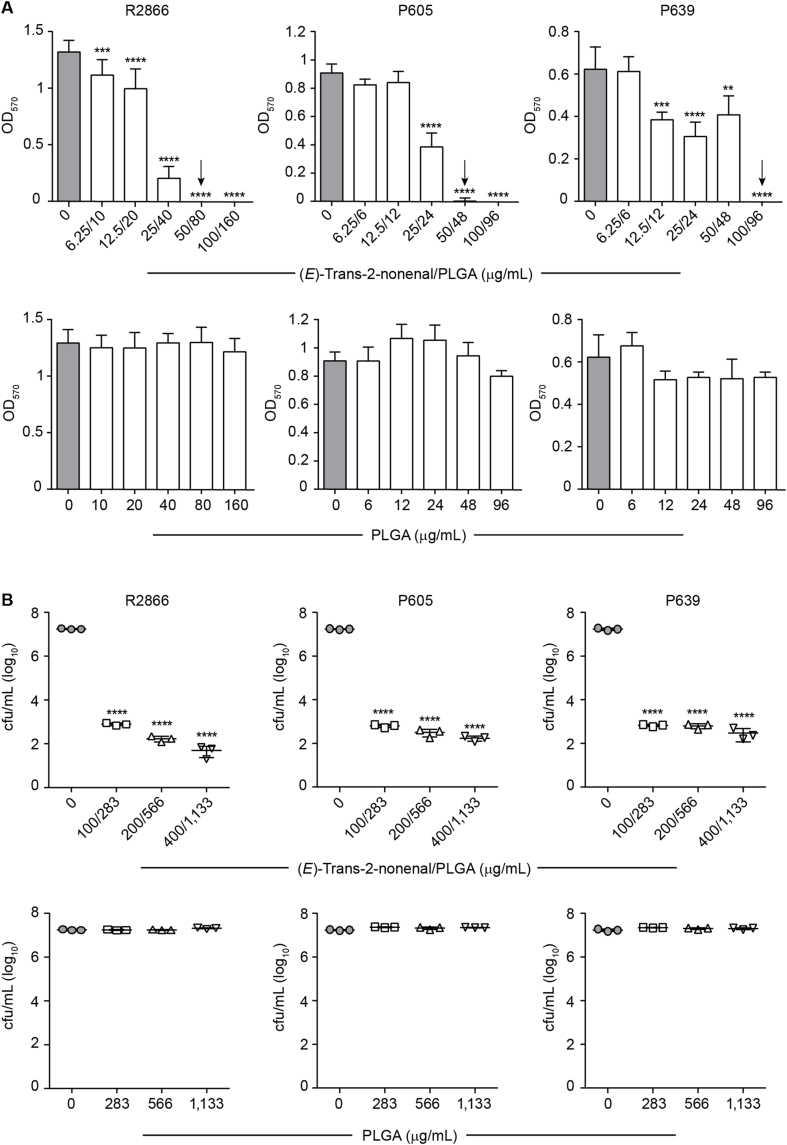
We assessed the H. influenzae biofilm inhibition by the cinnamaldehyde analogs (E)-trans-2-nonenal and (E)-3-decen-2-one, with an already proven therapeutic window [38]. Three NTHi clinical isolates were used, R2866, P605 and P639, with different biofilm-forming capacity and ampicillin susceptibility. The MICs of ampicillin for NTHi R2866, P605 and P639 were >128, 32 and 64 μg/mL, respectively. Crystal violet staining showed that ampicillin, when used up to a concentration of 128 μg/mL, did not inhibit biofilm formation by R2866; differently, an ampicillin dose-dependent inhibitory effect was shown for P605 and P639, and ampicillin 8 μg/mL was the lowest antibiotic concentration showing an inhibitory effect (Fig. 2A). The MICs of (E)-trans-2-nonenal were 50 μg/mL; the MICs of (E)-3-decen-2-one were 100, 50 and 100 μg/mL for NTHi R2866, P605 and P639, respectively. (E)-trans-2-nonenal and (E)-3-decen-2-one inhibited biofilm formation by the three strains in a dose-dependent manner. Depending on the molecule and strain, 12.5 or 25 μg/mL were cinnamaldehyde analog lowest concentrations showing inhibitory effects (Fig. 2B–C), further observed by CLSM imaging (Fig. 2D). We also evaluated the effect of these molecules on R2866 biofilm formation by real-time impedance analysis. In this assay, R2866 was the only strain forming biofilms with reproducible cell index (CI) values among replicates. The real-time dose-response experiments showed that (E)-trans-2-nonenal and (E)-3-decen-2-one were effective and prevented bacterial biofilm formation (Fig. 2E). Inhibitory effects were also observed for ampicillin, when used at 16, 32, 64 and 128 μg/mL.
Fig. 2.
Drug effects on H. influenzae biofilm formation. (A–C) Biofilm formation by three NTHi clinical strains in the absence/presence of a panel of drug concentrations, determined by OD570 measurements after crystal violet staining. Data are shown for ampicillin (A), (E)-trans-2-nonenal (B) and (E)-3-decen-2-one (C). Grey columns indicate untreated controls. Arrows indicate minimal inhibitory concentration (MIC). Each bar shows mean values ± SD; *, p < 0.05; **, p < 0.005; ****, p < 0.0001. Statistical comparisons of the means were performed with one-way ANOVA and Dunnett's multiple comparisons test. (D) Biofilm formation by three NTHi clinical strains in absence/presence of a panel of drug concentrations, determined by live-dead staining and CLSM. Data are shown for (E)-trans-2-nonenal (left) and (E)-3-decen-2-one (right). (E)-trans-2-nonenal 6.25, 25 and 50 μg/mL were used. (E)-3-decen-2-one 12.5, 50, 100 μg/mL were used for R2866 and P639 strains; E-3-decen-2-one 6.25, 25, 50 μg/mL were used for P605. For each panel, top and bottom images show XY axial and XZ lateral projections of the corresponding image stacks. (E) Real-time testing of drug effects on NTHi biofilm formation. Biofilm formation by R2866 in the absence or presence of a range of drug concentrations: ampicillin, 16 (square), 32 (triangle), 64 (inverted triangle), 128 (hexagon) μg/mL; (E)-trans-2-nonenal, 12 (square), 25 (triangle), 50 (inverted triangle), 100 (hexagon) μg/mL; (E)-3-decen-2-one, 25 (square), 50 (triangle), 100 (inverted triangle), 200 (hexagon) μg/mL. Cell index values were measured by impedance and correlated to biofilm mass. Grey circles indicate untreated controls. For ampicillin, there are significant differences in all the concentrations tested from 10 to 24 h (**, p < 0.005), compared to untreated controls. For (E)-trans-2-nonenal 50 μg/mL there are significant differences 10 to 24 h, compared to the untreated controls (*, p < 0.05), and for 100 μg/mL from 10 to 16 h (*, p < 0.05). For (E)-3-decen-2-one, there are significant differences in all the concentrations tested from 10 to 24 h (*, p < 0.05), compared to untreated controls. Statistical comparisons were performed with two-way ANOVA and Dunnett's multiple comparison test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Together, we show dose-dependent inhibitory effects for (E)-trans-2-nonenal and (E)-3-decen-2-one on H. influenzae biofilm growth, on clinical strains of different pathological origin. Regarding NTHi R2866 and ampicillin, crystal violet staining and impedance measurement did not show identical results, which could relate to the different surface materials in these two assays affecting bacterial adhesion strength. Previous work suggested that these cinnamaldehyde analogs may act as Vibrio spp. LuxR-ligands [38]. Analysis of NTHi R2866, P605 and P639 genome sequences showed that these strains do not seem to contain a LuxR encoding gene, thus suggesting different and currently unknown mechanisms of action.
3.2. Cinnamaldehyde analogs eradicate biofilms preformed by ampicillin resistant H. influenzae clinical isolates
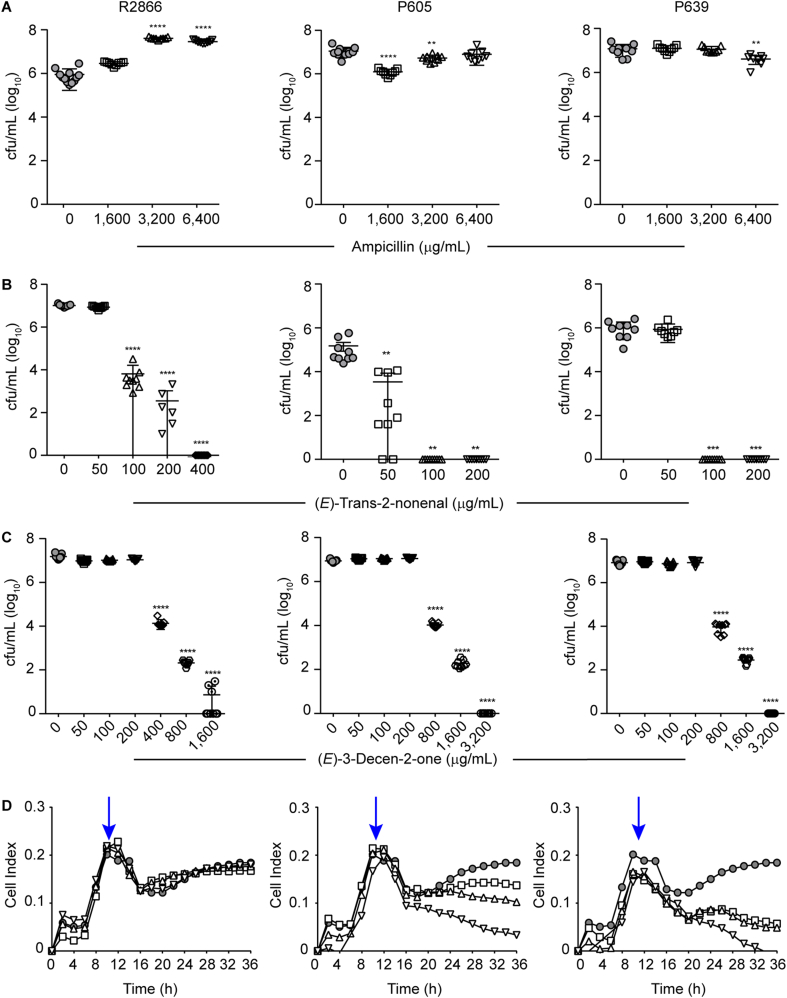
Next, we used two assays assessing the biofilm eradication potential of anti-biofilm molecules (Fig. 1, right panel). First, 96-well plates pre-established biofilms were treated with each PBS-prepared compound of interest (see Methods). Treatment duration was not extended for more than 6 h to avoid lowering bacterial viability for the strains tested, as indicated by the bacterial growth PBS controls included in each assay (data not shown). This procedure relies on mechanical removal of adhered bacterial cells after treatment and plating to determine CFU numbers, but it does not address drug effects on established biofilms over time. Thus, label-free electrical measurements by xCELLigence RTCA were also performed, by considering biofilms grown for 12 h prior drug administration for 24 additional h. We assessed the effect of (E)-trans-2-nonenal and (E)-3-decen-2-one on biofilms pre-established by strains R2866, P605 and P639, which were refractory to ampicillin treatment (Fig. 3A; Fig. 3D, left panel). Both molecules eradicated preformed biofilms in a dose-dependent manner. (E)-trans-2-nonenal concentrations were lower than those required for (E)-3-decen-2-one to be effective; in all cases, biofilm disrupting concentrations were higher than the respective MIC values (Fig. 2B–C and Fig. 3B–C). From a real-time perspective, impedance measurements complemented CFU plating viability data (Fig. 3D). Thus, the cinnamaldehyde analogs tested eradicated biofilms that are refractory to ampicillin doses up to 200 higher than the respective strain MIC. Together, we show two complementary assays testing drug efficacy to eradicate H. influenzae preformed biofilms. We acknowledge pros and cons regarding biofilm mechanical disruption and CFU counting, as drug incubation time needs to be set for each bacterial species/strain, also including appropriate bacterial growth controls in drug vehicle solution; however, although time consuming, it is highly reproducible to quantify bacterial viability.
Fig. 3.
Drug effects on preformed NTHi biofilms. (A–C) Biofilm viable counts by three clinical strains after 6 h treatment with a range of drug concentrations, determined by mechanical disruption and CFU counting. Data are shown for ampicillin (A), (E)-trans-2-nonenal (B) and (E)-3-decen-2-one (C). In each case, mean values ± SD are shown; **, p < 0.005; ***, p < 0.001; ****p < 0.0001. Statistical comparisons of the means were performed with one-way ANOVA and Dunnett's multiple comparisons test. (D) Real-time testing of drug effects on grown NTHi biofilms. Biofilms were formed by R2866 for 12 h; then, the effect of a range of drug doses was tested for 24 h. Left panel: ampicillin, 1600 (square), 3200 (triangle), 6400 (inverted triangle) μg/mL; middle panel: (E)-trans-2-nonenal, 100 (square), 200 (triangle), 400 (inverted triangle) μg/mL; right panel: (E)-3-decen-2-one, 400 (square), 800 (triangle), 1600 (inverted triangle) μg/mL). In all cases, cell index values were measured by impedance and correlated to biofilm mass. Grey circles indicate untreated controls. Blue arrows indicate the drug administration time point. After drug addition, the conductivity of the culture medium is altered due to changes in temperature and concentration of ions, resulting in a temporary increase in conductivity (drop in Cell Index) also in the control. There are no significant differences for ampicillin and (E)-trans-2-nonenal 100 μg/mL, compared with untreated controls. There are significant differences for (E)-trans-2-nonenal 200 μg/mL from 32 to 36 h (*, p < 0.05), and for 400 μg/mL from 26 to 36 h (**, p < 0.005). For (E)-3-decen-2-one 400 and 800 μg/mL there are significant differences from 28 to 36 h (*, p < 0.05), and for 1600 μg/mL from 26 to 36 h (**, p < 0.005). Statistical comparisons were performed with two-way ANOVA and Dunnett's multiple comparison test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
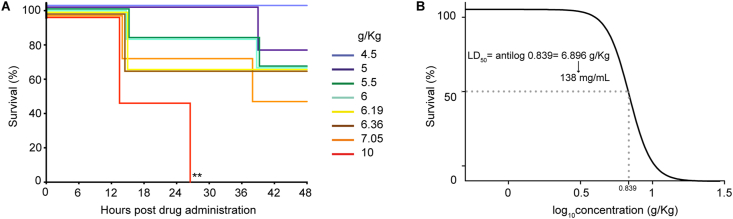
Inhibition of biofilm growth- and eradication of preformed biofilm assays needed (E)-trans-2-nonenal concentrations (25–50 and 100–400 μg/mL, respectively) lower than those required when using (E)-3-decen-2-one. (E)-trans-2-nonenal previously showed low cytotoxicity towards a MRC-5 cell line [38], and its LD50 was 138 mg/mL when tested in an in vivo toxicity Galleria mellonella larvae model, corresponding to 6.896 g (E)-trans-2-nonenal per Kg of larvae (Fig. 4). G. mellonella larvae have been widely used to evaluate the toxicity of antimicrobial compounds, exhibit a strong correlation with mammalian immune systems, and the toxicity tests conducted using them correlate effectively with mammalian models [[46], [47], [48], [49], [50], [51]]. For these overall reasons, we next focused on further exploring the (E)-trans-2-nonenal antimicrobial potential.
Fig. 4.
(E)-trans-2-nonenal toxicity in a G. mellonella larvae model. (A) Survival curve of G. mellonella larvae infected with different (E)-trans-2-nonenal concentrations. Larvae were monitored up until 48 h post-inoculation. (B) Dose-response curve of different (E)-trans-2-nonenal concentrations for lethal dose determination. Lethal dose (LD50) was determined as 6.896 g (E)-trans-2-nonenal per Kg of larvae. Assuming 0.2 g and 10 μL-volume inoculation per larvae, the (E)-trans-2-nonenal LD50 is 138 mg/mL; R2 = 0.9697. Log-rank tests were conducted between Kaplan-Meier curves to evaluate statistically significant differences. Asterisks show differences compared to the control.
From a chemical perspective, (E)-trans-2-nonenal is a small and highly volatile molecule with reduced solubility in water, which limits its efficiency. We next explored (E)-trans-2-nonenal nanoencapsulation as a procedure aiming to provide effective protection against undesirable chemical reactions and evaporation, to improve solubility, and to preserve the bioactive compound stability during storage and processing [52]. Encapsulation can also increase absorption and bioavailability improving the pharmacokinetic profile of numerous actives, and also controlling the release kinetics according to environment conditions (pH, temperature, enzymatic activity, ionic strength). Moreover, the encapsulation of antimicrobials can reduce their toxicity and overcome their inherent resistance, reducing the amount of active to be required [53]. Each encapsulation system needs to be designed ad hoc by considering its main purpose, key aspects that should be improved, and physico-chemical properties of the active molecule. The effect of nanosystems prepared by loading cinnamaldehyde into chitosan-based nanocapsules against Streptococcus mutants or Staphylococcus aureus biofilms has been previously reported [54,55]. Here, we analyzed two different (E)-trans-2-nonenal encapsulation approaches, lipid (nanoemulsions) and polymeric nanoparticles.
3.3. Vehiculization of (E)-trans-2-nonenal into nanoemulsions
Three independent batches of blank and (E)-trans-2-nonenal-loaded nanoemulsions (NE) were synthesized (see Methods section), whose composition and physico-chemical characterization are shown in Table S1. NE were <200 nm with a low PDI (PDI<0.15), which indicates homogeneity of size distribution. NE zeta potential was around +22 mV for blank NE and lower in the case of NE containing (E)-trans-2-nonenal, which may relate to the presence of (E)-trans-2-nonenal at the O/W interphase rather than in the lipidic core. To study the long-term stability, synthesized NE were stored in ultrapure water at 4 °C and RT for four months, and evaluated at pre-established time points. No significant changes in particle size were detected, indicating high stability. Zeta potential remained positive throughout the study, although a decrease in the absolute value was observed in the formulations over time, particularly those stored at RT (Figs. S1A–B). The overall stability of NE in sBHI was suitable for their use in biological assays, with a controlled increase in size beyond 24 h, also related to the dilution factor (Fig. S1C). However, despite the promising results in terms of particle size and stability behavior, the entrapment efficiency of loaded-NE was lower than expected, with only ∼20 % (E)-trans-2-nonenal incorporated, and this amount was released after few days of storage (data not shown), indicating that this type of lipid nanocarrier is not suitable for the incorporation of this cinnamaldehyde analog. This loose capacity for the entrapment of the molecule may be due to the preference of (E)-trans-2-nonenal for the interphase rather than for the lipidic core, as shown by the lower zeta potential compared to the blank-NE, favoring the exit of the compound into the surrounding media [56]. In fact, one of the main drawbacks when using NE as drug delivery systems is the premature release of the entrapped compound, which is highly influenced by core composition and chemical nature of the drug [57,58]. Based on these data, we discarded (E)-trans-2-nonenal NE for further testing.
3.4. Vehiculization of (E)-trans-2-nonenal into poly(lactic-co-glycolic acid) (PLGA) polymeric nanoparticles
Next, we explored poly(lactic-co-glycolic acid) (PLGA) as core material to encapsulate (E)-trans-2-nonenal, by combining the (E)-trans-2-nonenal hydrophobic character and PLGA co-polymer high versatility. In this regard, (E)-trans-2-nonenal shows good solubility in the organic solvents typically used in simple emulsion encapsulation methods such as ethyl acetate, dichloromethane or acetone, and good compatibility with the polymer. Variations of PLGA co-polymer composition and molecular weight allow modifying the system physico-chemical properties (size, release kinetics, surface-area ratio), and it is a highly biocompatible nanoencapsulation matrix approved by Federal Drug Administration (FDA) and European Medicines Agency (EMA), extensively studied as a drug delivery system [59].
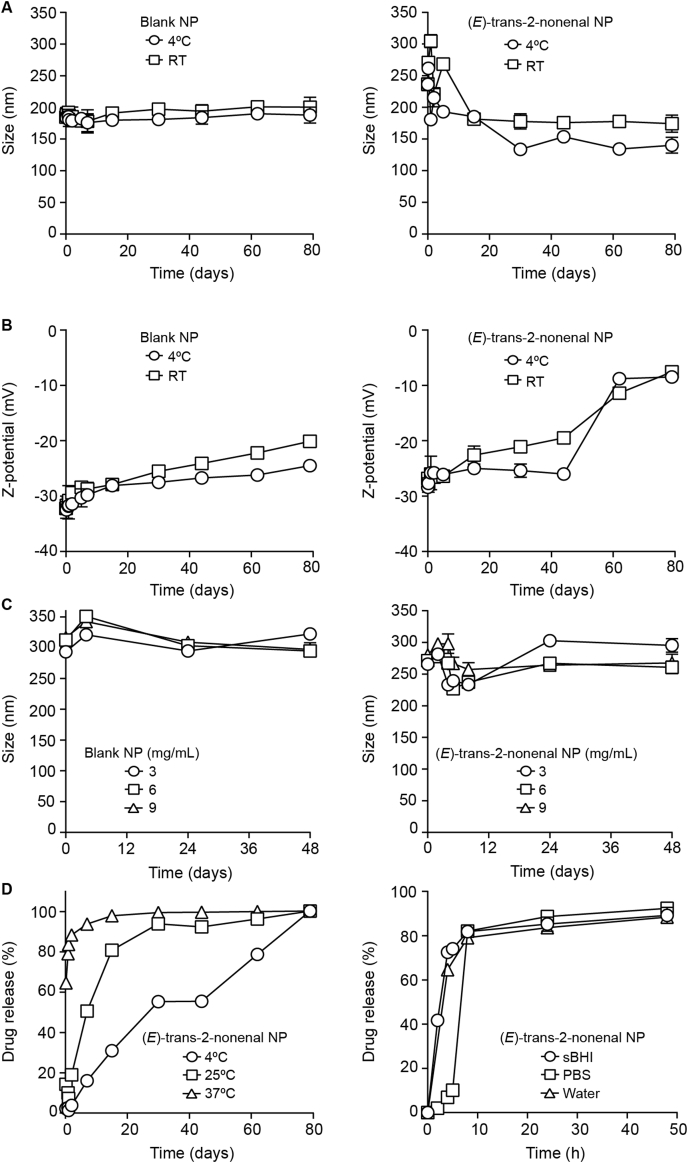
Independent batches of polymeric blank (NP) and (E)-trans-2-nonenal-loaded nanoparticles (NP) were synthesized (see Methods section), whose composition and characterization is shown in Table S2. Both blank and loaded NP were <250 nm and showed low PDI, indicating high homogeneity of their size distribution. (E)-trans-2-nonenal encapsulation reduced the zeta potential from −32 mV to −26.6 mV due to the presence of (E)-trans-2-nonenal molecules in the NP surface. (E)-trans-2-nonenal high volatility did not allow high entrapment efficiency (EE) of the molecule through the simple emulsion method, but EE values up to 58.3 % were obtained after optimization of encapsulation parameters. NP long-term stability was tested in ultrapure water at 4 °C and RT, with no significant changes in particle size (Fig. 5A), and an increase in the zeta potential value, mostly above 60 days for (E)-trans-2-nonenal-loaded NP, although it remained negative through the entire study (Fig. 5B). The stability of both blank and –loaded NP in sBHI at 37 °C was studied for 48 h, showing that they are suitable for their use in this media, with an initial controlled size increase due to swelling phenomena commonly observed in PLGA NP [60], and a later size stabilization up to 48 h (Fig. 5C). Lastly, release of (E)-trans-2-nonenal from the PLGA NP in water was evaluated for 80 days, showing to be highly dependent on temperature (Fig. 5D, left panel), as > 93 % was released within 7 days at 37 °C, while only 16 % was released at 4 °C in the same period. Likewise, media slightly affected (E)-trans-2-nonenal release kinetics, mostly at short incubation times, being between 85,2 % and 88,6 % after 24 h in sBHI and PBS, respectively (Fig. 5D, right panel). Although relatively low EE is achieved for the encapsulation of (E)-trans-2-nonenal in PLGA NP, small, homogeneous and stable particles with good physico-chemical properties were produced, suitable for further testing. When compared to previously reported encapsulation of cinnamaldehyde in chitosan, the PLGA NP developed here are of similar size and show similar fast release behavior, but (E)-trans-2-nonenal encapsulation in PLGA showed better stability and encapsulation efficiency, reaching 58.3 % compared to 39.0 % in the case of chitosan encapsulation [54,55].
Fig. 5.
Effect of storage conditions on particle size (A) and zeta potential (B) on PLGA polymeric nanoparticles (NP), blank and (E)-trans-2-nonenal-loaded, in ultrapure water (mean ± SD, n = 3). (C) Evolution of NP particle size at different concentrations in sBHI at 37 °C for 48 h, for blank (left panel), and (E)-trans-2-nonenal-loaded (right panel) NP. (D) Release profiles of (E)-trans-2-nonenal-loaded NP in water at different temperatures (left panel); effect of media on drug release at 37 °C (right panel).
As above indicated, encapsulation is a tool to overcome adverse pharmaceutical properties of some active compounds. Nevertheless, some compounds such as (E)-trans-2-nonenal are also challenging from a nano-formulation perspective due to both their lipophilic and volatile nature. Similar limitations have been observed with other volatile active compounds like terpenes, where the use of PLGA NP was shown to be an effective strategy for their therapeutic use, more promising that other kind of nanocarriers [61,62].
3.5. Anti-biofilm effects of (E)-trans-2-nonenal-loaded PLGA nanoparticles
Next, we assessed the effects of (E)-trans-2-nonenal-loaded PLGA nanoparticles (NP) on H. influenzae biofilms. NP inhibited biofilm formation by strains R2866, P605 and P639 in a concentration-dependent manner (Fig. 6A, upper panels). Effects were comparable to those observed when using the free drug, with slight differences among strains as encapsulation seemed to improve efficacy for R2866, but this was not the case for P605 and P639 strains. Efficacy on established biofilms also showed NP dose-dependent effects, although reduction of bacterial viability did not reach eradication levels (Fig. 6B, upper panels). In both assay types, (E)-trans-2-nonenal encapsulation required using PLGA concentrations that did not have inhibitory effects (Fig. 6A and B, bottom panels).
Fig. 6.
Effect of E-trans-2-nonenal-loaded PLGA NP on NTHi biofilms. (A) Biofilm formation by three NTHi strains in the absence/presence of a range of NP concentrations, determined by OD570 measurement after crystal violet staining. Grey columns indicate untreated controls. Arrows indicate MIC. Each bar shows mean values ± SD; **, p < 0.005; ***, p < 0.001; ****p < 0.0001. Statistical comparisons of the means were performed with one-way ANOVA and Dunnett's multiple comparisons test. (B) Biofilm viable counts by three strains after 6 h treatment with a panel of NP concentrations, determined by mechanical disruption and CFU counting. In each case, mean values ± SD are shown; ****, p < 0.0001. Statistical comparisons of the means were performed with one-way ANOVA and Dunnett's multiple comparisons test.
Together, we present nanoformulations consisting of (E)-trans-2-nonenal-loaded PLGA with dose-dependent inhibitory effects on H. influenzae biofilms associated to the cargo molecule. When comparing nanoformulation and free (E)-trans-2-nonenal results, some aspects deserve consideration. Regarding inhibition of biofilm formation, the effect of (E)-trans-2-nonenal matching concentrations was highly comparable (data in Figs. 2B and 6A). However, in terms of disrupting preformed biofilms, nanoformulation performance was lower than that of free drug matching concentrations, and NP did not reach a complete eradication effect (data in Fig. 3, Fig. 6B). Higher encapsulation efficiencies and prolonged stability would be desirable to determine if (E)-trans-2-nonenal encapsulation improves H. influenzae biofilms disruption compared to the free molecule. Although the demanding physico-chemical properties of this cinnamaldehyde analog limit these systems, there is some room for further improvement. For instance, adding components such as polyethylene glycol (PEG) in the composition of PLGA NP may increase their loading capacity and better control the release of (E)-trans-2-nonenal, which may be the cause of the biological results observed, as previously shown for volatile terpenes [61]. Also, more complex lipid nanoparticles, such as solid lipid nanoparticles (SLN) or nanostructured lipid carrier (NLC), could be explored to encapsulate lipophilic volatile compounds, as they are reported to overcome some limitations of NE as those encountered here [63].
Despite its challenging development, we consider that (E)-trans-2-nonenal vehiculization is an appropriate strategy to avoid its lipophilicity and volatility and, therefore, to enhance its in vivo biodistribution and bioavailability. Industrial development of this type of optimized systems would be feasible because all components of the polymeric nanosystems are considered GRAS (generally recognized as safe) [64], or have been approved by FDA for their use [65]. In this regard, in vivo biodistribution and bioavailability experiments should be performed to determine the feasibility of these nanoencapsulation systems, next to in vivo efficacy testing. Preclinical models for H. influenzae chronic infection rely on using chinchilla or Junbo mice to model otitis, or smoke-exposed ferrets modeling COPD [7,66,67]. Aiming to overcome technical challenges of such models, we tried adapting a model of chronic subcutaneous infection and abscess formation that has been previously used for Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae, Enterobacter cloacae and Escherichia coli [68]. However, it was not suitable for modeling high-density H. influenzae infection (data not shown). The use of adequate preclinical model systems will be tackled in future studies.
4. Conclusions
Biofilm-related infections are difficult to treat, making the development and testing of new effective therapeutics needed. Also, there is a pressing need for standardized methods that appreciate the biological variability of bacterial biofilms while also able of reproducibly assess the potential of anti-biofilm drugs [15]. Here, we detail simple protocols that can be performed routinely in numerous laboratories worldwide. The methods are described for H. influenzae biofilms, but can be easily adapted to other bacteria that form biofilms, and could be useful for systematization and further improvement/complementation with other assay types. Here, we showed that the cinnamaldehyde analogs (E)-trans-2-nonenal and (E)-3-decen-2-one act against H. influenzae biofilms, precluding formation or disrupting previously grown ones. (E)-trans-2-nonenal limiting chemical properties led us to evaluate two encapsulation approaches, nanoemulsions and polymeric nanoparticles, with different results, and to shortlist the later one for in vitro activity analyses. Future work will attempt improving nanoformulation capacities, explore encapsulating platforms-drug synergistic combinations, expand the range of cinnamaldehyde analog bacterial targets, and evaluate their biodistribution and efficacy in suitable in vivo model systems.
CRediT authorship contribution statement
Javier Asensio-López: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. María Lázaro-Díez: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Tania M. Hernández-Cruz: Writing – review & editing, Methodology, Formal analysis, Data curation. Núria Blanco-Cabra: Writing – review & editing, Methodology, Formal analysis, Data curation. Ioritz Sorzabal-Bellido: Methodology, Formal analysis. Eva M. Arroyo-Urea: Methodology, Investigation, Formal analysis, Data curation. Elena Buetas: Methodology. Ana González-Paredes: Writing – review & editing, Supervision, Methodology, Funding acquisition, Formal analysis, Conceptualization. Carlos Ortiz de Solórzano: Methodology, Funding acquisition. Saioa Burgui: Project administration, Methodology, Funding acquisition. Eduard Torrents: Writing – review & editing, Methodology, Formal analysis, Data curation. María Monteserín: Writing – review & editing, Project administration, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization. Junkal Garmendia: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Junkal Garmendia reports financial support was provided by Agencia Estatal de Investigación (AEI). Junkal Garmendia reports financial support was provided by Navarra Government General Directorate of Industry Energy and Strategic Projects S3. Junkal Garmendia reports financial support was provided by Sociedad Española de Neumología y Cirugía Torácica.
Acknowledgments
We are grateful to Alex Mira (FISABIO Foundation, Valencia, Spain) for providing access to the xCELLigence RTCA instrument. Funding Agencies and grant numbers: PhD studentship from Regional Navarra Govern, Spain, reference 0011-1408-2020-000007. Margarita Salas grant from Ministerio de Universidades, Spain, funded by the EU-Next Generation EU. This work has been funded by grants from AEI MICIU RTI2018-096369-B-I00, PID2021-125801OB-I00, MICIU PDI2021-122409OB-C22; from SEPAR 875/2019; from Gobierno de Navarra PC150-151-152 and PC136-137-138; Atracción de Talento (Modalidad 1) program from Comunidad de Madrid, Spain, 2019-T1/IND-12906; Nanomedicine CSIC Hub, Spain, PIE202180E048. CIBER is an initiative from Instituto de Salud Carlos III (ISCIII), Madrid, Spain.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2024.100178.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Ahearn C.P., Gallo M.C., Murphy T.F. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis. 2017:75. doi: 10.1093/femspd/ftx042. 2017/04/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duell B.L., Su Y.C., Riesbeck K. Host–pathogen interactions of nontypeable Haemophilus influenzae: from commensal to pathogen. FEBS Lett. 2016;590:3840–3853. doi: 10.1002/1873-3468.12351. [DOI] [PubMed] [Google Scholar]
- 3.Jalalvand F., Riesbeck K. Update on non-typeable Haemophilus influenzae-mediated disease and vaccine development. Expert Rev Vaccines. 2018;17:503–512. doi: 10.1080/14760584.2018.1484286. [DOI] [PubMed] [Google Scholar]
- 4.Su Y.C., Jalalvand F., Thegerström J., Riesbeck K. The interplay between immune response and bacterial infection in COPD: focus upon non-typeable Haemophilus influenzae. Front Immunol. 2018;9:1–26. doi: 10.3389/fimmu.2018.02530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weeks J.R., Staples K.J., Spalluto C.M., Watson A., Wilkinson T.M.A. The role of non-typeable Haemophilus influenzae biofilms in chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2021;11:1–17. doi: 10.3389/fcimb.2021.720742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva M.D., Sillankorva S. Otitis media pathogens–A life entrapped in biofilm communities. Crit Rev Microbiol. 2019;45:595–612. doi: 10.1080/1040841X.2019.1660616. [DOI] [PubMed] [Google Scholar]
- 7.Hunt B.C., Stanford D., Xu X., Li J., Gaggar A., Rowe S.M., Vamsee Raju S., Edward Swords W. Haemophilus influenzae persists in biofilm communities in a smoke-exposed ferret model of COPD. ERJ Open Res. 2020;6:1–12. doi: 10.1183/23120541.00200-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., Ouellette M., Outterson K., Patel J., Cavaleri M., Cox E.M., Houchens C.R., Grayson M.L., Hansen P., Singh N., Theuretzbacher U., Magrini N., Aboderin A.O., Al-Abri S.S., Awang Jalil N., Benzonana N., Bhattacharya S., Brink A.J., Burkert F.R., Cars O., Cornaglia G., Dyar O.J., Friedrich A.W., Gales A.C., Gandra S., Giske C.G., Goff D.A., Goossens H., Gottlieb T., Guzman Blanco M., Hryniewicz W., Kattula D., Jinks T., Kanj S.S., Kerr L., Kieny M.P., Kim Y.S., Kozlov R.S., Labarca J., Laxminarayan R., Leder K., Leibovici L., Levy-Hara G., Littman J., Malhotra-Kumar S., Manchanda V., Moja L., Ndoye B., Pan A., Paterson D.L., Paul M., Qiu H., Ramon-Pardo P., Rodríguez-Baño J., Sanguinetti M., Sengupta S., Sharland M., Si-Mehand M., Silver L.L., Song W., Steinbakk M., Thomsen J., Thwaites G.E., van der Meer J.W., Van Kinh N., Vega S., Villegas M.V., Wechsler-Fördös A., Wertheim H.F.L., Wesangula E., Woodford N., Yilmaz F.O., Zorzet A. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 9.Starner T.D., Zhang N., Kim G.H., Apicella M.A., McCray P.B. Haemophilus influenzae forms biofilms on airway epithelia: Implications in cystic fibrosis. Am J Respir Crit Care Med. 2006;174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slinger R., Chan F., Ferris W., Yeung S.W., St Denis M., Gaboury I., Aaron S.D. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Carrera-salinas A., González-díaz A., Ehrlich R.L., Berbel D., Tubau F., Pomares X., et al. Genetic adaptation and acquisition of macrolide resistance in Haemophilus spp. during persistent respiratory tract colonization in chronic obstructive pulmonary disease (COPD) Microbiol Spectr. 2023;11(1) doi: 10.1128/spectrum.03860-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starner T.D., Shrout J.D., Parsek M.R., Appelbaum P.C., Kim G.H. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and diminish established biofilms. Antimicrob Agents Chemother. 2008;52:137–145. doi: 10.1128/AAC.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeters E., Nelis H.J., Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Haney E.F., Trimble M.J., Hancock R.E.W. Microtiter plate assays to assess antibiofilm activity against bacteria. Nat Protoc. 2021;16:2615–2632. doi: 10.1038/s41596-021-00515-3. [DOI] [PubMed] [Google Scholar]
- 15.Coenye T. Biofilm antimicrobial susceptibility testing: where are we and where could we be going? Clin Microbiol Rev. 2023 doi: 10.1128/cmr.00024-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novotny L.A., Jurcisek J.A., Ward M.O., Jordan Z.B., Goodman S.D., Bakaletz L.O. Antibodies against the majority subunit of type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol. 2015;96:276–292. doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurcisek J.A., Hofer L.K., Goodman S.D., Bakaletz L.O. Monoclonal antibodies that target extracellular DNABII proteins or the type IV pilus of nontypeable Haemophilus influenzae (NTHi) worked additively to disrupt 2-genera biofilms. Biofilm. 2022;4 doi: 10.1016/j.bioflm.2022.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockson M.E., Novotny L.A., Mokrzan E.M., Malhotra S., Jurcisek J.A., Akbar R., Devaraj A., Goodman S.D., Bakaletz L.O. Evaluation of the kinetics and mechanism of action of anti-integration host factor-mediated disruption of bacterial biofilms. Mol Microbiol. 2014;93:1246–1258. doi: 10.1111/mmi.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman S.D., Obergfell K.P., Jurcisek J.A., Novotny L.A., Downey J.S., Ayala E.A., Tjokro N., Li B., Justice S.S., Bakaletz L.O. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 20.Goodman S.D., Bakaletz L.O. Bacterial biofilms utilize an underlying extracellular DNA matrix structure that can be targeted for biofilm resolution. Microorganisms. 2022;10(2):466. doi: 10.3390/microorganisms10020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novotny L.A., Goodman S.D., Bakaletz L.O. Redirecting the immune response towards immunoprotective domains of a DNABII protein resolves experimental otitis media. NPJ Vaccines. 2019;4 doi: 10.1038/s41541-019-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novotny L.A., Chiang T., Goodman S.D., Elmaraghy C.A., Bakaletz L.O. Humanized anti-DNABII Fab fragments plus ofloxacin eradicated biofilms in experimental otitis media. Laryngoscope. 2021;131:E2698–E2704. doi: 10.1002/lary.29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novotny L.A., Goodman S.D., Bakaletz L.O. Targeting a bacterial DNABII protein with a chimeric peptide immunogen or humanised monoclonal antibody to prevent or treat recalcitrant biofilm-mediated infections. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novotny L.A., Jurcisek J.A., Goodman S.D., Bakaletz L.O. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine. 2016;10:33–44. doi: 10.1016/j.ebiom.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balazs V.L., Filep R., Repas F., Kerekes E., Szabo P., Kocsis B., Boszormenyi A., Krisch J., Horv G. Immortelle (Helichrysum italicum (Roth) G. Don) essential oil showed antibacterial and biofilm inhibitory activity against respiratory tract pathogens. Molecules. 2022;27:5518. doi: 10.3390/molecules27175518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birru R.L., Bein K., Bondarchuk N., Wells H., Lin Q., Di Y.P., et al. Antimicrobial and anti-inflammatory activity of apple polyphenol phloretin on respiratory pathogens associated with chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2021;11:1–9. doi: 10.3389/fcimb.2021.652944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunthalert D., Baothong S., Khetkam P., Chokchaisiri S., Suksamrarn A. A chalcone with potent inhibiting activity against biofilm formation by nontypeable Haemophilus influenzae. Microbiol Immunol. 2014;58:581–589. doi: 10.1111/1348-0421.12194. [DOI] [PubMed] [Google Scholar]
- 28.Balazs V.L., Horv B., Kerekes E., Kamilla Á., Kocsis B., Böszörmenyi A., Nagy D.U., Krisch J., Szechenyi A., Horvath G. Anti-Haemophilus activity of selected essential oils detected by TLC-direct bioautography and biofilm inhibition. Molecules. 2019;24:3301. doi: 10.3390/molecules24183301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balázs V.L., Nagy-Radványi L., Filep R., Kerekes E., Kocsis B., Kocsis M., Farkas Á. In vitro antibacterial and antibiofilm activity of Hungarian honeys against respiratory tract bacteria. Foods. 2021;10:1–16. doi: 10.3390/foods10071632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santajit S., Sookrung N., Indrawattana N. Quorum sensing in ESKAPE bugs: a target for combating antimicrobial resistance and bacterial virulence. Biology. 2022;11 doi: 10.3390/biology11101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong W., Pang B., West-Barnette S., Swords W.E. Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J Bacteriol. 2007;189:8300–8307. doi: 10.1128/JB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armbruster C.E., Hong W., Pang B., Dew K.E., Juneau R.A., Byrd M.S., Love C.F., Kock N.D., Swords W.E. LuxS promotes biofilm maturation and persistence of nontypeable Haemophilus influenzae in vivo via modulation of lipooligosaccharides on the bacterial surface. Infect Immun. 2009;77:4081–4091. doi: 10.1128/IAI.00320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armbruster C.E., Pang B., Murrah K., Juneau R.A., Perez A.C., Weimer K.E.D., Swords W.E. RbsB (NTHI-0632) mediates quorum signal uptake in nontypeable Haemophilus influenzae strain 86-028NP. Mol Microbiol. 2011;82:836–850. doi: 10.1111/j.1365-2958.2011.07831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daines D.A., Bothwell M., Furrer J., Unrath W., Nelson K., Jarisch J., Melrose N., Greiner L., Apicella M., Smith A.L. Haemophilus influenzae luxS mutants form a biofilm and have increased virulence. Microb Pathog. 2005;39:87–96. doi: 10.1016/j.micpath.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Swords W.E. Quorum signaling and sensing by nontypeable Haemophilus influenzae. Front Cell Infect Microbiol. 2012;2:100. doi: 10.3389/fcimb.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swords W.E. Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front Cell Infect Microbiol. 2012;2:97. doi: 10.3389/fcimb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Brackman G., Celen S., Hillaert U., van Calenbergh S., Cos P., Maes L., et al. Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reilly T.J., Chance D.L., Smith A.L. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J Bacteriol. 1999;181:6797–6805. doi: 10.1128/jb.181.21.6797-6805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moleres J., Fernández-Calvet A., Ehrlich R.L., Martí S., Pérez-Regidor L., Euba B., Rodríguez-Arce I., Balashov S., Cuevas E., Liñares J., Ardanuy C., Martín-Santamaría S., Ehrlich G.D., Mell J.C., Garmendia J. Antagonistic pleiotropy in the bifunctional surface protein FadL (OmpP1) during adaptation of Haemophilus influenzae to chronic lung infection associated with chronic obstructive pulmonary disease. mBio. 2018;9:1–23. doi: 10.1128/mBio.01176-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arroyo-Urea E.M., Muñoz-Hernando M., Leo-Barriga M., Herranz F., González-Paredes A. A quality by design approach for the synthesis of palmitoyl-L-carnitine-loaded nanoemulsions as drug delivery systems. Drug Deliv. 2023;30 doi: 10.1080/10717544.2023.2179128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_50_Breakpoint_Table_01.pdf0-77
- 43.ninth ed. Clinical and Laboratory Standards Institute; Wayne: 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. CLSI document M7-A9. [Google Scholar]
- 44.Žiemytė M., Carda-Diéguez M., Rodríguez-Díaz J.C., Ventero M.P., Mira A., Ferrer M.D. Real-time monitoring of Pseudomonas aeruginosa biofilm growth dynamics and persister cells' eradication. Emerg Microbes Infect. 2021;10:2062–2075. doi: 10.1080/22221751.2021.1994355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Cabra N., Vega-Granados K., Moya-Andérico L., Vukomanovic M., Parra A., Álvarez De Cienfuegos L., Torrents E. Novel oleanolic and maslinic acid derivatives as a promising treatment against bacterial biofilm in nosocomial infections: an in vitro and in vivo Study. ACS Infect Dis. 2019;5:1581–1589. doi: 10.1021/acsinfecdis.9b00125. [DOI] [PubMed] [Google Scholar]
- 46.Allegra E., Titball R.W., Carter J., Champion O.L. Galleria mellonella larvae allow the discrimination of toxic and non-toxic chemicals. Chemosphere. 2018;198:469–472. doi: 10.1016/j.chemosphere.2018.01.175. [DOI] [PubMed] [Google Scholar]
- 47.Moya-Andérico L., Vukomanovic M., Cendra M. del M., Segura-Feliu M., Gil V., del Río J.A., Torrents E. Utility of Galleria mellonella larvae for evaluating nanoparticle toxicology. Chemosphere. 2021;266 doi: 10.1016/j.chemosphere.2020.129235. [DOI] [PubMed] [Google Scholar]
- 48.Pereira T.C., Barros PP de, de Oliveira Fugisaki L.R., Rossoni R.D., Ribeiro F. de C., Menezes RT de, Junqueira J.C., Scorzoni L. Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J Fungi. 2018;4 doi: 10.3390/jof4040128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trevijano-Contador N., Zaragoza O. Immune response of Galleria mellonella against human fungal pathogens. J Fungi. 2019;5:1–13. doi: 10.3390/jof5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wojda I. Immunity of the greater wax moth Galleria mellonella. Insect Sci. 2017;24:342–357. doi: 10.1111/1744-7917.12325. [DOI] [PubMed] [Google Scholar]
- 51.Ignasiak K., Maxwell A. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res Notes. 2017;10:1–8. doi: 10.1186/s13104-017-2757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pateiro M., Gómez B., Munekata P.E.S., Barba F.J., Putnik P., Kovačević D.B., Lorenzo J.M. Nanoencapsulation of promising bioactive compounds to improve their absorption, stability, functionality and the appearance of the final food products. Molecules. 2021;26:1547. doi: 10.3390/molecules26061547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco-Padilla A., Soto K.M., Hernández Iturriaga M., Mendoza S. Food antimicrobials nanocarriers. Sci World J. 2014;2014 doi: 10.1155/2014/837215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mu R., Zhang H., Zhang Z., Li X., Ji J., Wang X., Gu Y., Qin X. Trans-cinnamaldehyde loaded chitosan based nanocapsules display antibacterial and antibiofilm effects against cavity-causing Streptococcus mutans. J Oral Microbiol. 2023;15 doi: 10.1080/20002297.2023.2243067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J., Lin Q., Sheng M., Ding T., Li B., Gao Y., Tan Y. Antibiofilm effect of cinnamaldehyde-chitosan nanoparticles against the biofilm of Staphylococcus aureus. Antibiotics. 2022;11:1–13. doi: 10.3390/antibiotics11101403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hörmann K., Zimmer A. Drug delivery and drug targeting with parenteral lipid nanoemulsions - a review. J Control Release. 2016;223:85–98. doi: 10.1016/j.jconrel.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Ðoković J.B., Savić S.M., Mitrović J.R., Nikolic I., Marković B.D., Randjelović D.V., Antic-Stankovic J., Božić D., Cekić N.D., Stevanović V., Batinić B., Arandelović J., Savić M.M., Savić S.D. Curcumin loaded pegylated nanoemulsions designed for maintained antioxidant effects and improved bioavailability: a pilot study on rats. Int J Mol Sci. 2021;22:1–27. doi: 10.3390/ijms22157991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elsewedy H.S., Aldhubiab B.E., Mahdy M.A., Elnahas H.M. Brucine PEGylated nanoemulsion: in vitro and in vivo evaluation. Colloids Surfaces A Physicochem Eng Asp. 2021;608 [Google Scholar]
- 59.Alsaab H.O., Alharbi F.D., Alhibs A.S., Alanazi N.B., Alshehri B.Y., Saleh M.A., Alshehri F.S., Algarni M.A., Almugaiteeb T., Uddin M.N., Alzhrani R.M. PLGA-based nanomedicine: history of advancement and development in clinical applications of multiple diseases. Pharmaceutics. 2022;14:1–30. doi: 10.3390/pharmaceutics14122728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rapier C.E., Shea K.J., Lee A.P. Investigating PLGA microparticle swelling behavior reveals an interplay of expansive intermolecular forces. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-93785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El-Hammadi M.M., Small-Howard A.L., Jansen C., Fernández-Arévalo M., Turner H., Martín-Banderas L. Potential use for chronic pain: poly(ethylene glycol)-poly(lactic-co-glycolic acid) nanoparticles enhance the effects of cannabis-based terpenes on calcium influx in TRPV1-Expressing cells. Int J Pharm. 2022;616 doi: 10.1016/j.ijpharm.2022.121524. [DOI] [PubMed] [Google Scholar]
- 62.El-Hammadi M.M., Small-Howard A.L., Fernández-Arévalo M., Martín-Banderas L. Development of enhanced drug delivery vehicles for three cannabis-based terpenes using poly(lactic-co-glycolic acid) based nanoparticles. Ind Crops Prod. 2021;164 [Google Scholar]
- 63.Sakellari G.I., Zafeiri I., Batchelor H., Spyropoulos F. Solid lipid nanoparticles and nanostructured lipid carriers of dual functionality at emulsion interfaces. Part II: active carrying/delivery functionality. Colloids Surfaces A Physicochem Eng Asp. 2023;659 [Google Scholar]
- 64.Adams T.B., Gavin C.L., Taylor S.V., Waddell W.J., Cohen S.M., Feron V.J., Goodman J., Rietjens I.M.C.M., Marnett L.J., Portoghese P.S., Smith R.L. The FEMA GRAS assessment of α,β-unsaturated aldehydes and related substances used as flavor ingredients. Food Chem Toxicol. 2008;46:2935–2967. doi: 10.1016/j.fct.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Qin B., Xia G., Choi S.H. FDA's poly (lactic-co-glycolic acid) research program and regulatory outcomes. AAPS J. 2021;23:1–7. doi: 10.1208/s12248-021-00611-y. [DOI] [PubMed] [Google Scholar]
- 66.Harrison A., Hardison R.L., Fullen A.R., Wallace R.M., Gordon D.M., White P., Jennings R.N., Justice S.S., Mason K.M. Continuous microevolution accelerates disease progression during sequential episodes of infection. Cell Rep. 2020;30:2978–2988.e3. doi: 10.1016/j.celrep.2020.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hood D., Moxon R., Purnell T., Richter C., Williams D., Azar A., Crompton M., Wells S., Fray M., Brown S.D., Cheeseman M.T. A new model for non-typeable Haemophilus influenzae middle ear infection in the Junbo mutant mouse. Dis Model Mech. 2016;9:69–79. doi: 10.1242/dmm.021659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pletzer D., Mansour S.C., Wuerth K., Rahanjam N., Hancock R.E.W. New mouse model for chronic infections by gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. mBio. 2017;8 doi: 10.1128/mBio.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.