Abstract
Background:
Clade C is the predominant HIV-1 strain infecting people in sub-Saharan Africa, India, and China and there is a critical need for a vaccine targeted to these areas. In this study we tested a DNA based vaccine that encodes the SIVgag, SIVpol and HIV-1 envelope clade C.
Methods:
Rhesus macaques were immunized by electroporation with the DNA plasmid encoding optimized SIVgag, SIVpol and an HIV-1 env clade C with or without the adjuvant RANTES. Animals were monitored for immune responses and challenged following the final immunization with 25 animal infectious doses (AID) of SHIV-1157ipd3N4.
Results:
We found that the vaccine induced high levels of antigen specific IFN-γ producing effector cells and the capacity for CD4+ and CD8+ to proliferate upon antigen stimulation. Importantly, we found that the vaccine induced antibody titers as high as 1/4000. These antibodies were capable of neutralizing tier 1 HIV-1 viruses. Finally, when macaques were challenged with SHIV, viral loads were controlled in vaccinated groups.
Conclusion:
We conclude that immunization with a simian/human immunodeficiency virus DNA-based vaccine delivered by electroporation can induce cellular and humoral immune responses that are able to control viral replication.
Keywords: SHIV, DNA vaccine, RANTES, Clade C
Introduction
There is evidence that the cellular immune response and antibodies will both be important in stunting HIV-1 replication. It is likely that it will be necessary to develop region specific vaccines or vaccines that incorporate multiple clade envelopes. In particular, clade C is the predominant HIV-1 strain infecting people in sub-Saharan Africa, India, and China and there is a critical need for a vaccine targeted to these areas. Vaccine studies utilizing DNA vaccines, have induced both cellular and humoral antigen specific responses (1). And recently plasmid DNA vaccines have elicited detectable immune response in humans (2). Yet, substantial research has continued to focus on increased immunogenicity through the development of adjuvants (3,4) and improving vaccine delivery with electroporation (5,6).
In this manuscript we test a DNA based vaccine that encodes SIVgag, SIVpol and HIV-1 env clade C. We incoporated several methods to maximize the DNA vaccine immunize response, electroporation and use of an adjuvant, RANTES. As an effective delivery method, electroporation has been reported to enhance the antigen-specific IFN-γ production in rhesus macaques following immunization with plasmid DNA (7, 8). Further, RANTES is a member of the CC-chemokine family and has been found to enhance cellular immune responses resulting in a more pronounced immune-modulating effect against an HIV-1-related virus in rodent and monkey models (9–12). Hadida et al have previously shown that RANTES enhances the HIV-specific cytotoxicity of CD8 T cells (13).
We found that our DNA vaccine regimens induced both an effective cellular and humoral immune response against vaccine antigens. We observed antigen-specific interferon gamma (IFN-γ) production, proliferation of lymphocytes, as well as induction of vaccine specific memory cells. Importantly, the binding antibodies were of high titer and the neutralizing antibodies demonstrated the ability to neutralize multiple clades.
Materials and Methods
Animals
Chinese rhesus macaques were housed at BIOQUAL, Inc. (Rockville, MD), in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. Animals were allowed to acclimate for at least 30 days in quarantine prior to any experimentation.
Immunization
Groups of five rhesus macaques were immunized four times with 0.5mg of SIVgag, SIVpol and HIVenv (DNA group) or 0.5mg of SIVgag, SIVpol and HIVenv co-injected with 0.5mg of RANTES (DNA+RANTES group) by electroporation. The macaques were then rested 13 weeks before performing the second set of immunizations. The second series of immunizations included an increase in dose to 1.5mg of SIVgag, SIVpol and HIVenv and 0.5mg RANTES. Five macaques were immunized with saline as a negative control.
DNA Plasmids
The SIVgag and SIVpol genes are expressed as 70-kDa and 110-kDa protein of SIVmac, respectively. These genes have been optimized for high-level expression as described (14). Consensus HIV-1 clade C Envelope was designed based on the sequences retrieved from HIV databases (http://www.hiv.lanl.gov). To enhance protein expression, viral RNA structures were disrupted by using codons typically found in human cells. The synthesized Env C gene was cloned into the expression vector pVAX (Invitrogen).
The human RANTES gene, having 384bp, was designed with a Kozak sequence, an IgE leader sequence and two stop codons. The gene was synthesized by Geneart, Inc. (Germany). The fragment was cloned into the pVAX1 vector.
Peptides
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: SIV mac239 Gag peptides (#6204), HIV Env peptides (#9499), and SIV mac239 Pol peptides (#6443).
Enzyme Linked Immunospot Assay (ELISpot) assay for IFN-γ
ELISpot assays using IFN-g reagents (MabTech, Sweden) and nitrocellulose plates (Millipore, Billerica, MA) were performed according to the manufacturer’s instructions. A positive response was defined as greater than 50 spot forming cells (SFC) per million PBMCs. Each sample was performed in triplicate with peptides.
T cell proliferation and memory T cell subset assay
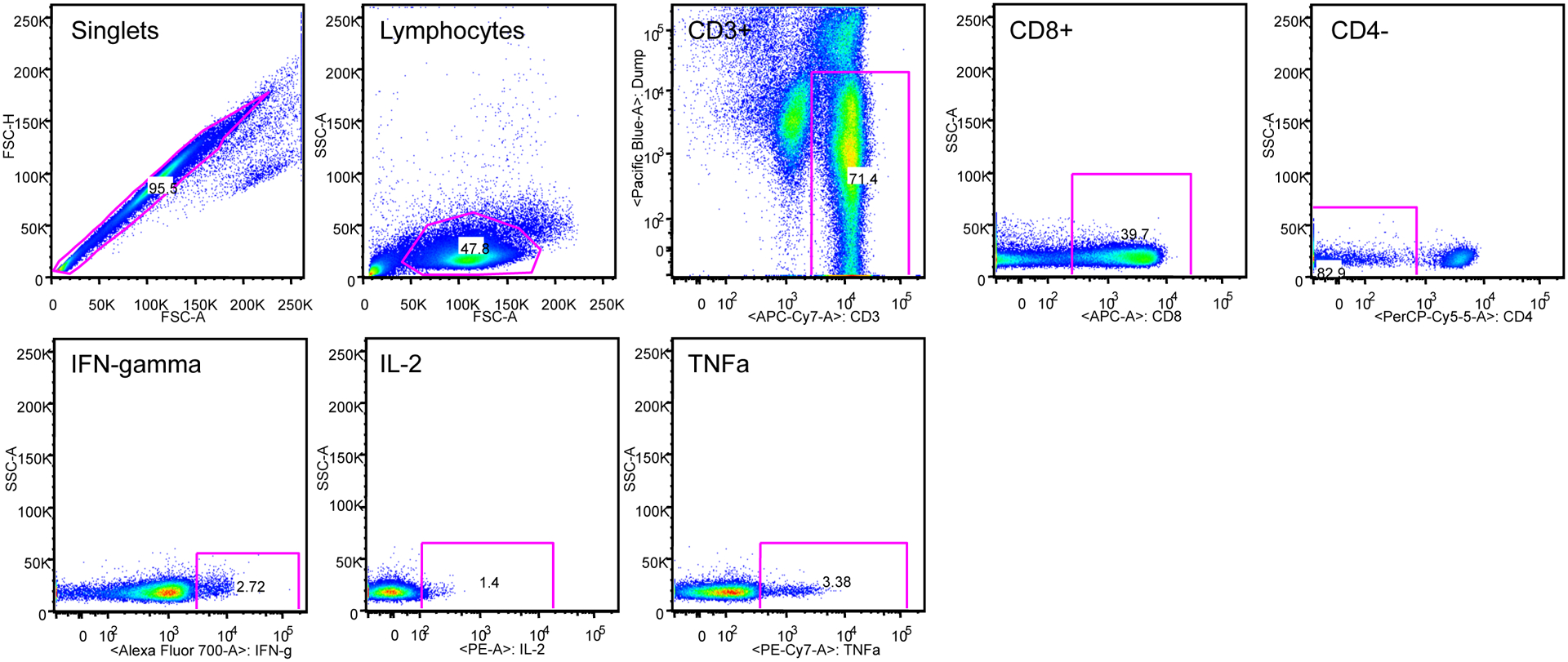
PBMCs were incubated with carboxyfluorescein succinimidyl ester (CFSE) (5 μmol/l) for 8 min at 37°C. Cells were then washed and incubated with antigens at a concentration of 5 μg/ml for 5 days at 37°C in 96-well plates. Cultures without peptides were used to determine the background proliferative responses. PBMCs were immunostained with the following mAbs: anti-CD3 APC-Cy7, anti-CD4 PerCP-Cy5.5, anti-CD8 APC and anti-CD95 PE-Cy5 (BD-Pharmingen, San Diego, CA), and anti-CD28 ECD (Beckman Coulter, Fullerton, CA). Central and effector memory T cells were defined as CD28+ CD95+ and CD28¯CD95+ respectively (14). Stained cells were washed in PBS and fixed (Cell-Fix), acquired on an LSRI instrument using CellQuest software (BD Biosciences) and analyzed with FlowJo software (Tree Star, Ashland, OR).
Intracellular Cytokine Staining
PBMCs were incubated with peptides for 5 hours at 37°C. An unstimulated and positive control (Staphylococcus enterotoxin B (SEB), 1 μg/mL; Sigma-Aldrich) was included in each assay. Following incubation, the cells were washed and stained with surface antibodies: CD4 (PerCP Cy5.5), CD3 (APC Cy7), and CD8 (APC) (BD Biosciences, San Jose, CA). Cells were then washed and fixed using the Cytofix/Cytoperm kit (BD PharMingen, San Diego, CA) according to their instructions. Following fixation, the cells were stained with antibodies against intracellular markers IL-2 (PE), IFN-γ (Alexa Fluor 700) and TNF-α (PE Cy7) (BD Biosciences, San Jose, CA). Cells were washed and fixed with PBS containing 1% paraformaldehyde. Stained and fixed cells were acquired on an LSRI instrument using CellQuest software (BD Biosciences) and analyzed with FlowJo software (Tree Star, Ashland, OR).
Binding Antibodies
A direct ELISA was used to measure serum titers of anti-HIV-1 Env antibodies. Ninety-six-well ELISA plates were coated overnight at 4°C with 100 μl of 1 μg/ml recombinant human gp120 protein, HIV-1 CN54 (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH), in PBS. The remainder of the ELISA was conducted at room temperature. Sera were serially diluted in 2% BSA/0.05% Tween-20, and added to ELISA wells. Following a 1-h incubation, the plate was washed three times and then incubated with a 1/5000 dilution of a peroxidase-conjugated affinity-purified rabbit anti-mouse secondary Ab (Jackson Laboratories) in 2% BSA/0.05% Tween-20 for 1 h. The plate was washed three times, developed with TMB (KPL, Gaithersburg, MD), stopped with 1% HCl, and analyzed at 450 nm with a Dynatech MR5000 ELISA plate reader.
TZM.bl neutralization assay
NAb responses against tier 1 and tier 2 HIV-1 Env pseudoviruses were measured by using luciferase-based virus neutralization assays with TZM.bl cells as previously described (15,16). These assays measure the reduction in luciferase reporter gene expression in TZM-bl cells following a single round of virus infection. Briefly, a fixed dose of virus containing 50,000 – 150,000 relative luminescence units (RLU) equivalents was incubated with serial 3-fold dilutions of test sample in duplicate in a total volume of 150 μl for 1 hr at 37°C in 96-well flat-bottom culture plates. Freshly trypsinized TZM-bl cells (10,000 cells in 100 μl of growth medium containing 37.5 μg/ml DEAE dextran) were added to each well. One set of control wells received cells plus virus (virus control) and another set received cells only (background control). After 48-hour incubation, 100 μl of cells was transferred to 96-well black solid plates (Costar) for measurements of luminescence using the Britelite Luminescence Reporter Gene Assay System (PerkinElmer Life Sciences). Neutralization titers are expressed as the inhibitory dilution (ID50) at which RLU were reduced by 50% compared to virus control wells after subtraction of background RLUs. Titers were derived from a 5-parameter curve-fitting model. Test viruses that exhibit a tier 1 neutralization phenotype included MW965.26 (clade C), MN (clade B), SF162.LS (clade B), BaL.26 (clade B), and W61D-TCLA.71 (clade B). Also assayed was R5 SHIV 1157ipd3N4 (Clade C) (17,18), which exhibits a tier 2 phenotype. All HIV-1 Env pseudoviruses were produced and titers were determined as previously described (15,16).
SHIV Challenge
Animals were challenged three months following the last immunization with 25 animal infectious doses (AID) of SHIV-1157ipd3N4 (18) by the intrarectal route. Viral stocks were provided by Advanced BioSciences Laboratories, Inc (ABL, Inc.) in Kensington, MD and titered by Bioqual Inc. (Rockville, MD).
Viral load and CD4 number quantitation
Plasma viral load was determined by a quantitative NASBA assay provided by ABL Inc. as previously described (19). CD4 amounts were measured by Bioqual Inc. (Rockvile, MD).
Statistics
For comparisons of IFN-γ ELISpots, T cell proliferation, memory cells, intracellular cytokine staining, and viral loads two tailed Mann-Whitney tests were performed using Prism Graphpad Software. P values that were <0.05 were considered a significant difference.
Results
Induction of Cellular Immune Response
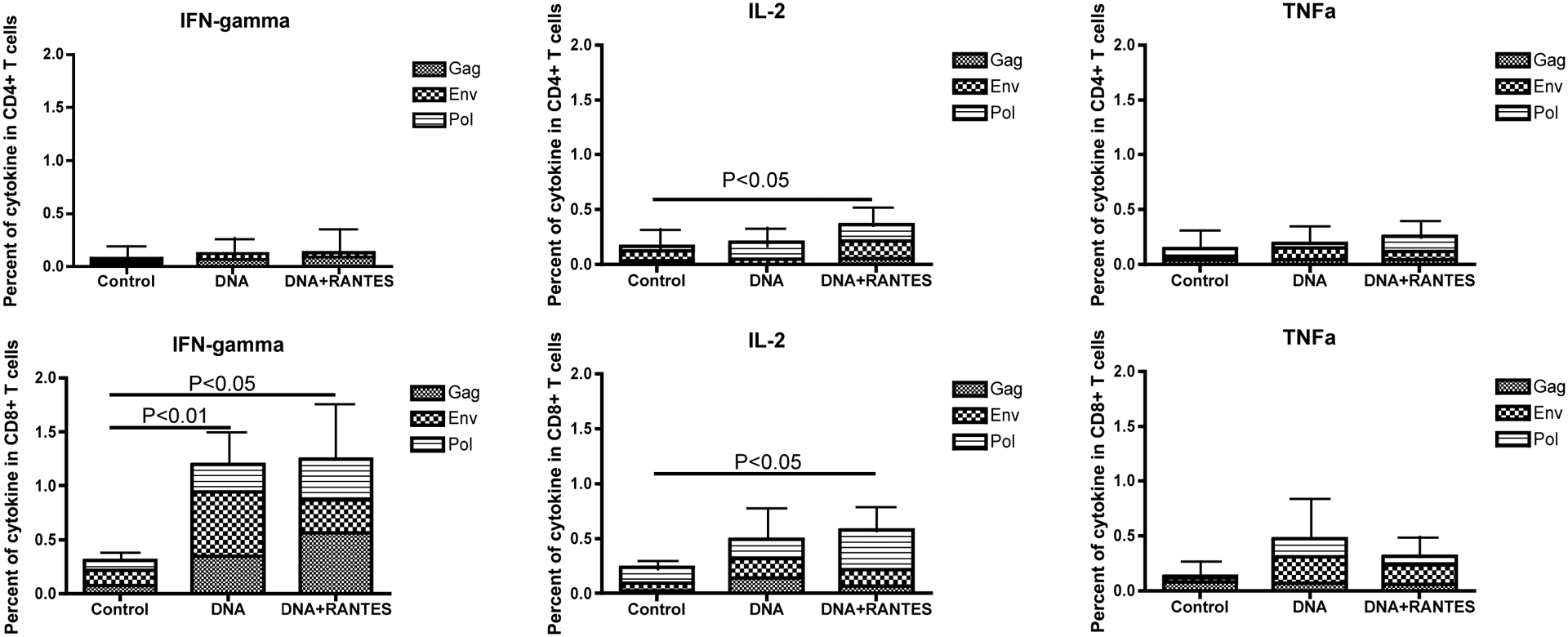
We assessed the ability of our vaccine to induce an antigen-specific cellular immune response in all macaques using an IFN-γ ELISpot assay. After one injection, macaques in DNA and DNA+RANTES groups induced an average of 488 and 518 spot-forming cells (SFC) per 106 PBMCs, respectively (Fig. 1B–D). Following the last injection, antigen-specific IFN-γ secretion reached 4885 SFC/106 PBMCs and 5636 SFC/106 PBMCs in the DNA group and DNA with RANTES group, respectively. These numbers indicate we induced a robust cellular immune response in both vaccine groups. We also determined if the antigen specific cells were CD8 or CD4. And we determined if IL-2 and TNF-alpha could be secreting in an antigen specific manner. We, therefore, obtained blood two months following the last injection stimulated as noted above with peptides and stained for the production of IFN-γ IL-2, and TNF-a followed by surface staining for CD8 and CD4. The secretion of IFN-γ, as assessed by flow cytometry, mirrored what we observed by ELISpot. Importantly, we observed that the predominant cells secreting IFN-γ were CD8 cells. A cumulative average of 1.24% and 1.2% of total CD8 cells were capable of secreting IFN-γ in the DNA + RANTES and DNA groups, respectively. We also observed CD4 and CD8 cells capable of secreting IL-2 in animals that received DNA+RANTES (Fig. 2B). In contrast, we did not observed any TNF-a production. Altogether, antigen specific IFN-γ and IL-2 expression was observed. This data showed that macaques in the vaccine groups had a high number of antigen specific cells capable of secreting IFN-γ.
Figure 1. IFN-g -producing cells after immunizations.
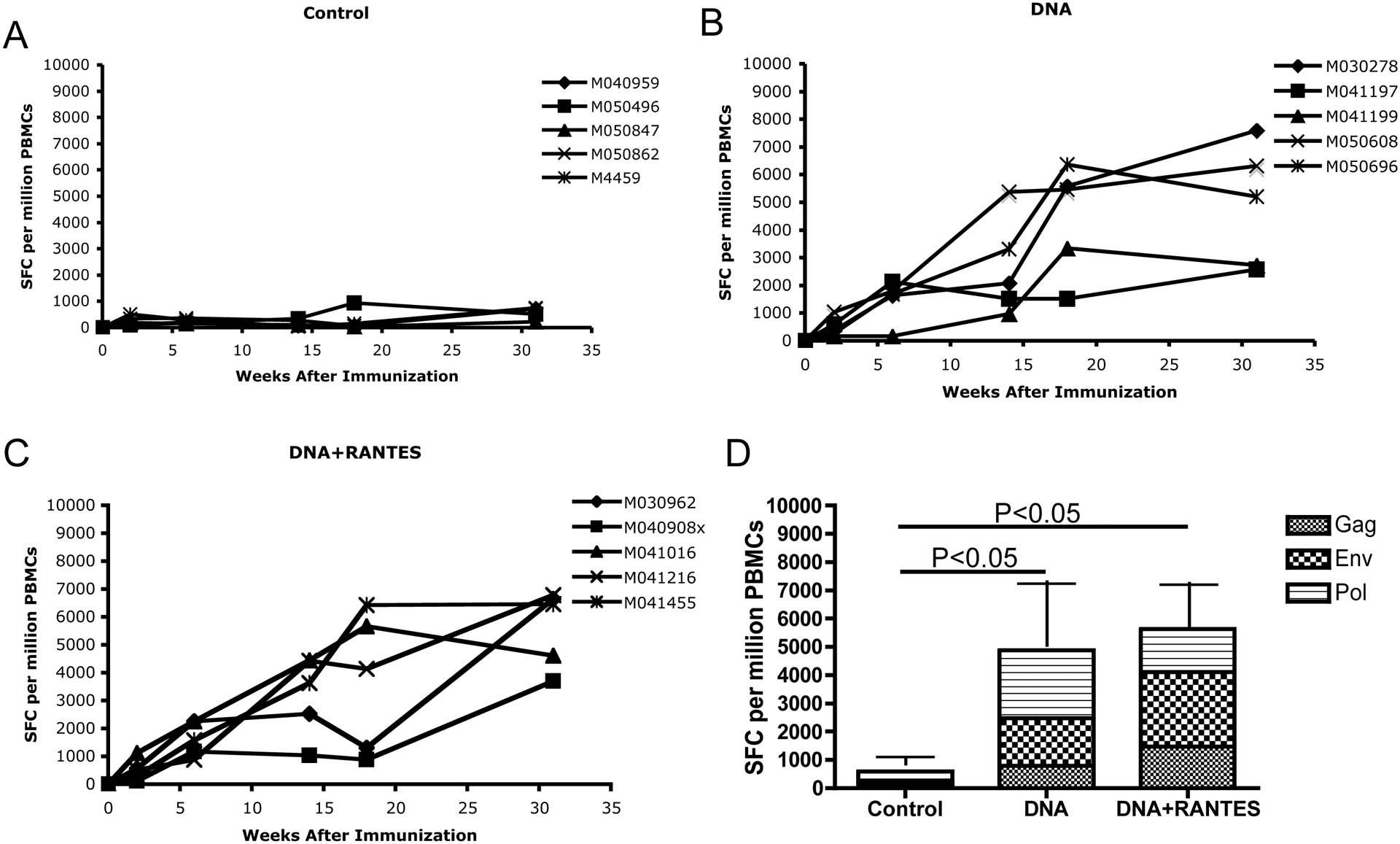
Samples were taken two weeks after each immunization. PBMCs were isolated by a standard percoll separation technique and assessed for antigen specific IFN-g immune response in the control group (A), DNA group (B), and DNA + RANTES group (C). (D) Average level of IFN-g producing cells after immunizations 1–5 with control, SHIV DNA vaccine alone, and SHIV DNA vaccine + RANTES adjuvant. The data represent the number of cells able to secrete IFN-g after each immunization.
Figure 2. Proliferative capacity of antigen specific T cells.
PBMCs from immunized macaques are stained with CFSE and stimulated with growth medium, concanavalin A (ConA), or SIV gag, SIV pol, or HIV Env peptides for 5 days. Following stimulation, cells were stained for phenotypic markers and analyzed by flow cytometry. A) Average proliferative capacity of CD4 and CD8 T cells B) Average for all primates is presented for CD4 or CD8 TCMs and TEMs. Data were acquired on an LSRI instrument analyzed with FlowJo software.
Proliferative capacity induced in the vaccine groups
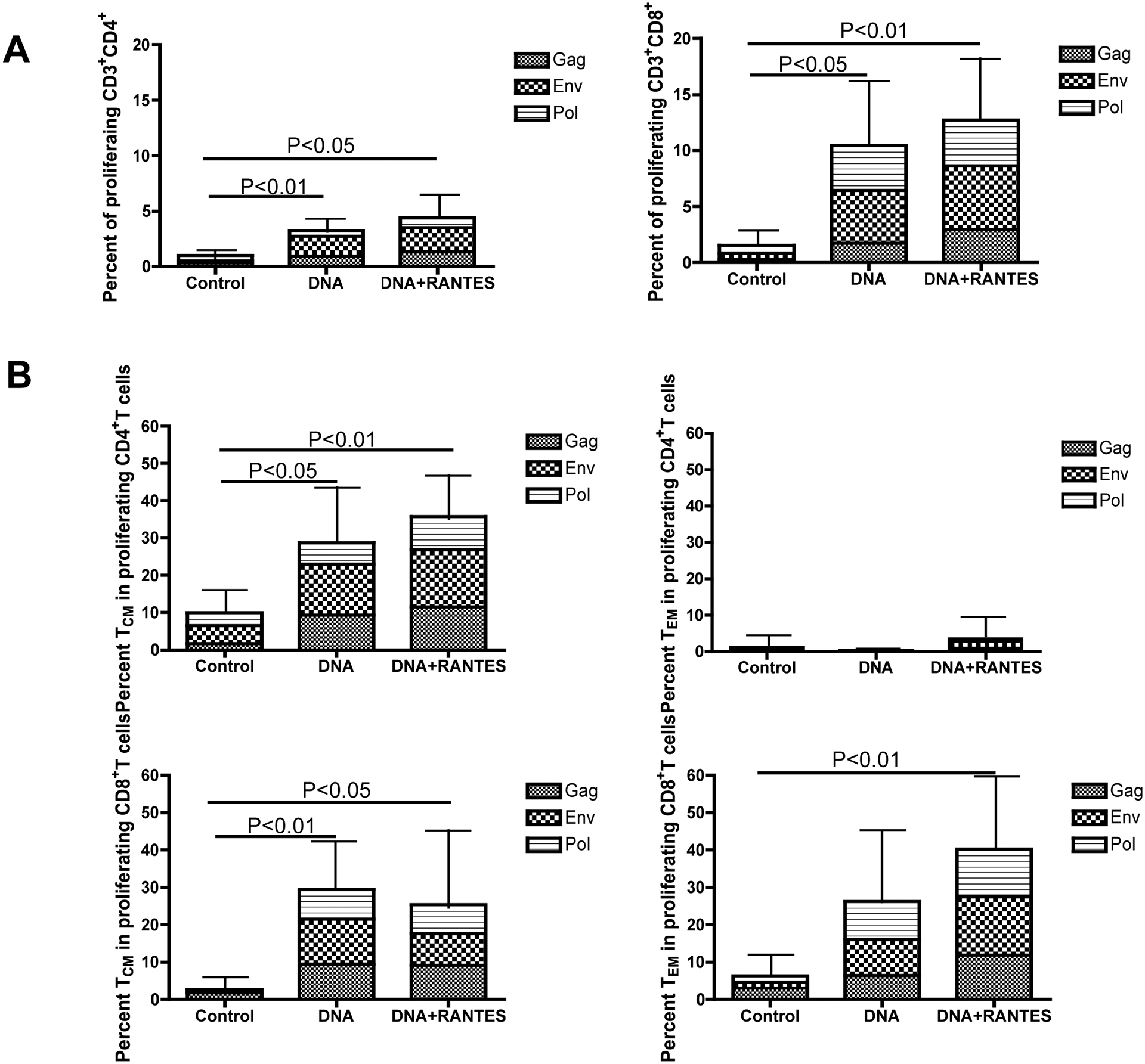
We next investigated the ability of SHIV-specific CD4+ and CD8+ effector cells to proliferate in vitro to antigen stimulation. PBMCs isolated from macaques 4 weeks after the last vaccination were incubated with CFSE, washed, and stimulated for 5 days with growth media and either gag, env or pol peptides. Following stimulation proliferation was measured by flow cytometry. We observed a proliferative response of CD4+ cells (Fig. 3A) in the DNA and DNA+RANTES group (average of 3.24% and 4.38%, respectively). However, the proliferative capacity of CD8+ T cells was stronger than that of CD4+ T cells, reaching an average of 10.4% in the DNA group and 12.7% in the DNA with RANTES group (Fig. 3A). These results demonstrate that the proliferative capacity of T cells, especially CD8+ T cells, is significant in macaques belonging to the vaccine groups.
Figure 3. Cytokine expression in CD4+ and CD8+ T cells found in blood of macaques 2 months following last immunization.
PBMCs from immunized macaques are stimulated with growth medium, ConA, SIV gag, HIV env or SIV pol peptides for 5 hours and then cells are stained for phenotypic markers and analyzed by flow cytometry. (A) Gating strategy of cytokine expression. (B) Average expression of cytokines IFN-γ, IL-2, and TNF-a in CD4+ /CD8+ T cells.
Memory T cells produced by proliferating T cells found in the vaccine groups
We then compared the frequency of proliferating memory T cell subsets among the three groups of macaques 11 weeks after the last vaccination. Proliferating memory T cells were divided into two subsets by surface marker expression: central memory T cells (TCM) were defined as CD28+CD95+ and effector memory T cells (TEM) were defined as CD28−CD95+ (18). When we evaluated the frequency of TCMs and TEMs in proliferating CD4+ cells (Fig. 2B), we detected a significantly higher percentage of TCMs in the DNA group and DNA + RANTES group compared to the control groups (Fig. 2B, upper left panel). An average of 28.7% TCMs and 35.7% TCMs were detected in proliferating CD4+ T cells isolated from animals in the DNA and DNA + RANTES group, respectively. Among the three groups, there was no significant difference in the percentages of TEMs (Fig. 3B, right panel).
We also evaluated the frequency of TCMs and TEMs in proliferating CD8+ T cells. We found that a significantly higher percentage of TCMs in the DNA+RANTES group and DNA group (average 25.4% and 29.4%, respectively) compared to the control group (average 2.6%) (Fig. 3B, left panels). Looking at TEMs in proliferating CD8+ T cells, there was only a significant difference between the DNA+RANTES group and the control group (Fig. 2B, lower right panel). Taken together, the data shows that DNA immunization results in high proliferation of TCMs in both CD4+ and CD8+ T cells but only CD8+ T cells proliferated when looking at the TEM subset.
Binding and neutralizing antibody responses elicited by DNA Vaccine
We assessed both the binding and neutralizing antibody responses elicited by our vaccine. Binding antibody levels averaged 1:2080 for the DNA alone immunized group while the antibody titers for the DNA plus RANTES group averaged 1:4010. The antibody levels 2 weeks post-challenge were both boosted to approximately 1:60,000 with no significant difference.
It is also important to determine if the antibodies are capable of neutralizing virus. Using a panel of tier 1 viruses with a broad range of neutralization sensitivities from clades B and C we assessed the NaB responses. We defined the criteria for positivity as 50% inhibitory dose (ID50) titers that were (i) >3-fold above preimmune background levels, (ii) >2-fold above levels of a concurrent murine leukemia virus (MuLV) control, and (iii) an absolute titer of >60. Macaques immunized with our vaccine developed robust, cross-clade neutralizing activity against neutralization-sensitive tier 1 clade B, and C viruses (MN, W61D-TCLA.71, and MW965.26), with ID50 titers against MW965.26 ranging from 92 to 152 (Table 1). In contrast, no neutralization against the autologous vaccine strains was detected, presumably reflecting their inherent neutralization-resistant phenotypes.
TABLE 1.
Neutralization Titer ID50 in TNuZM-bl cells1
| Group | Bleed Wk | MW965.26 | SHIV-1157ip d3N4 | MN | SF162.LS | Bal.26 | W61D-TCLA.71 | |
|---|---|---|---|---|---|---|---|---|
| Control | Pre-immune | 20 | 21.6 | 20 | 20 | 20 | 20 | |
| 2 wks post final boost | 20 | 20 | 20 | 20 | 20 | 20 | ||
| 6 wks post challenge | 47.2 | 24.6 | 34.4 | 20 | 20 | 25.4 | ||
| DNA | Pre-immune | 20 | 25 | 20 | 20 | 20 | 20 | |
| 2 wks post final boost | 92.6 | 20.2 | 45.4 | 20 | 20 | 36.2 | ||
| 6 wks post challenge | 8252.8 | 21.2 | 11127.6 | 879.6 | 87.2 | 642.6 | ||
| DNA + RANTES | Pre-immune | 20 | 26.2 | 20 | 20 | 20 | 20 | |
| 2 wks post final boost | 152 | 21 | 90 | 23 | 20 | 36.8 | ||
| 6 wks post challenge | 5359.2 | 26.8 | 7287.6 | 1242 | 163 | 2522.6 | ||
Control of viral replication after challenge in macaques which received vaccine.
All animals were challenged three months following the final immunization with 25 animal infectious doses (AID) of SHIV-1157ipd3N4 by the intrarectal route. The average viral load in the control group at week 2 after challenge or at the peak viral load was 5.2 logs (Fig. 5A). The peak viral loads of the DNA and DNA+RANTES groups were 4.6 logs and 4.3 logs, respectively. At week 3 post challenge, the viral load decreased in all of the groups with values of 2.8 logs for the DNA + RANTES group and 2.9 logs for the DNA group. All of which were significantly lower than that of the control group (P<0.05) (Fig. 5B). At week 6 after challenge, the group of animals that received DNA plus RANTES still exhibited a significantly lower viral load as compared to animals in the control group (P<0.05). Whereas the DNA group did not show a significant difference in the viral load level compared to that of the control group (p=0.1) (Fig. 5B). These results demonstrate that macaques co-immunized with the DNA vaccine along with RANTES induced a strong capacity to control viral replication.
Figure 5. Viral Load following Challenge with SHIV-clade C.
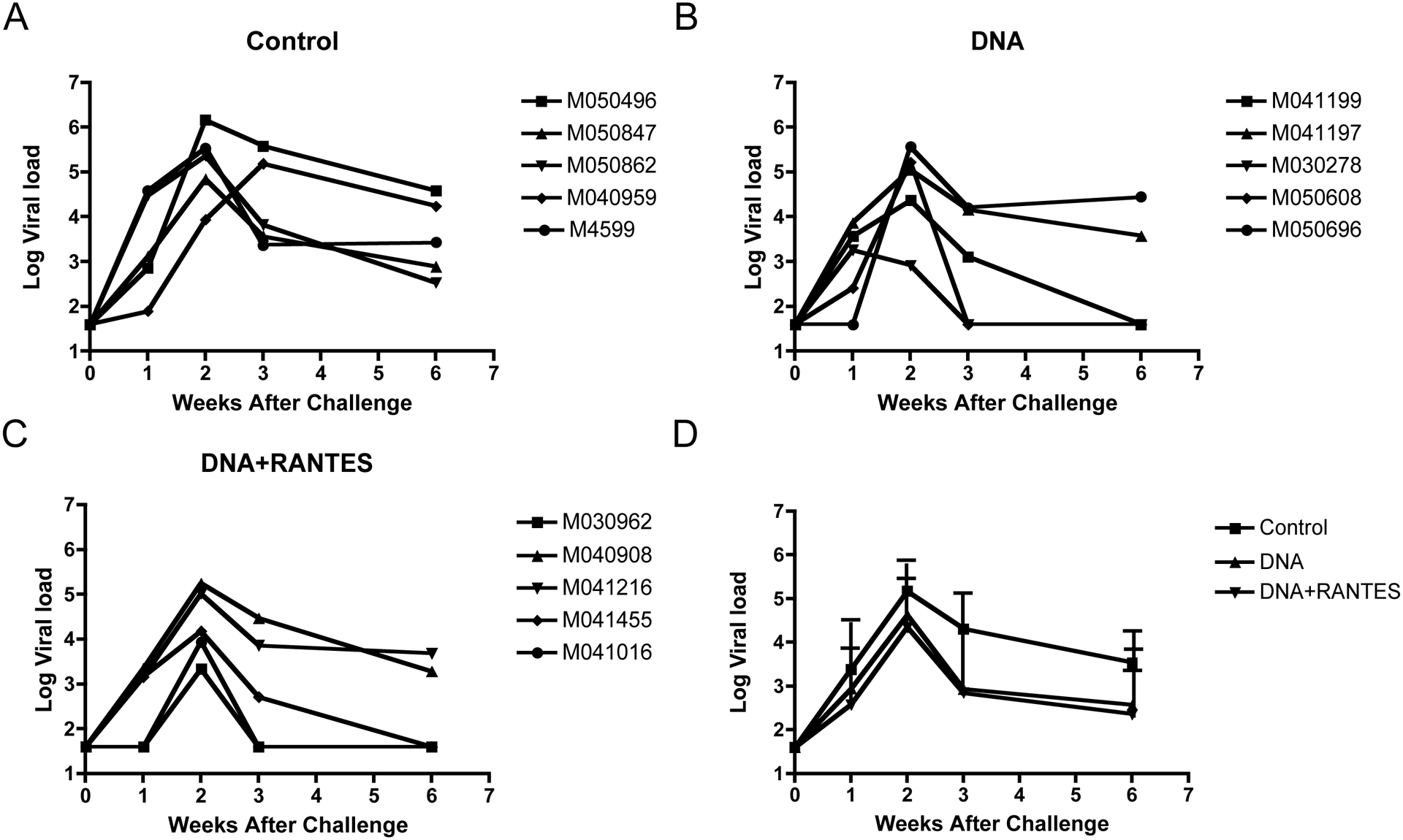
The log of the viral load for the rhesus macaques after challenge with 25 animal infectious doses (AID) of SHIV-1157ipd3N4 is presented. Viral loads are presented as individual macaques for (A) control, (B) DNA, (C) DNA+RANTES as well as (D) an average for each experimental group.
CD4+ T-cell counts after challenge
There was not a significant difference between the time points or the groups. This data showed that CD4 T cell amounts in the three groups were stable after challenge (data not shown).
Discussion
In this study, we demonstrated that immunization of rhesus macaques with a SHIV DNA-based vaccine encoding SIVgag, SIVpol and HIV-1 clade C envelope induced a cellular and antibody response. We demonstrated that macaques in the vaccine groups had high number of antigen specific IFN-γ producing cells (Figs. 1 and 2), and a high cellular proliferative responses following in vitro to SIVgag, HIVenv, and SIVpol (Fig. 3). The profile of these cells revealed more central memory in the two vaccine groups and more effector memory in DNA plus RANTES group. Furthermore, intracellular cytokine staining results showed that more IFN-γ and IL-2 were produced in animals receiving the DNA + RANTES vaccine than in the control group. Finally we observed high titer antibody responses. RANTES demonstrated a higher antibody titer pre-challenge. Fig. 4
Figure 4. Antibody Titers.
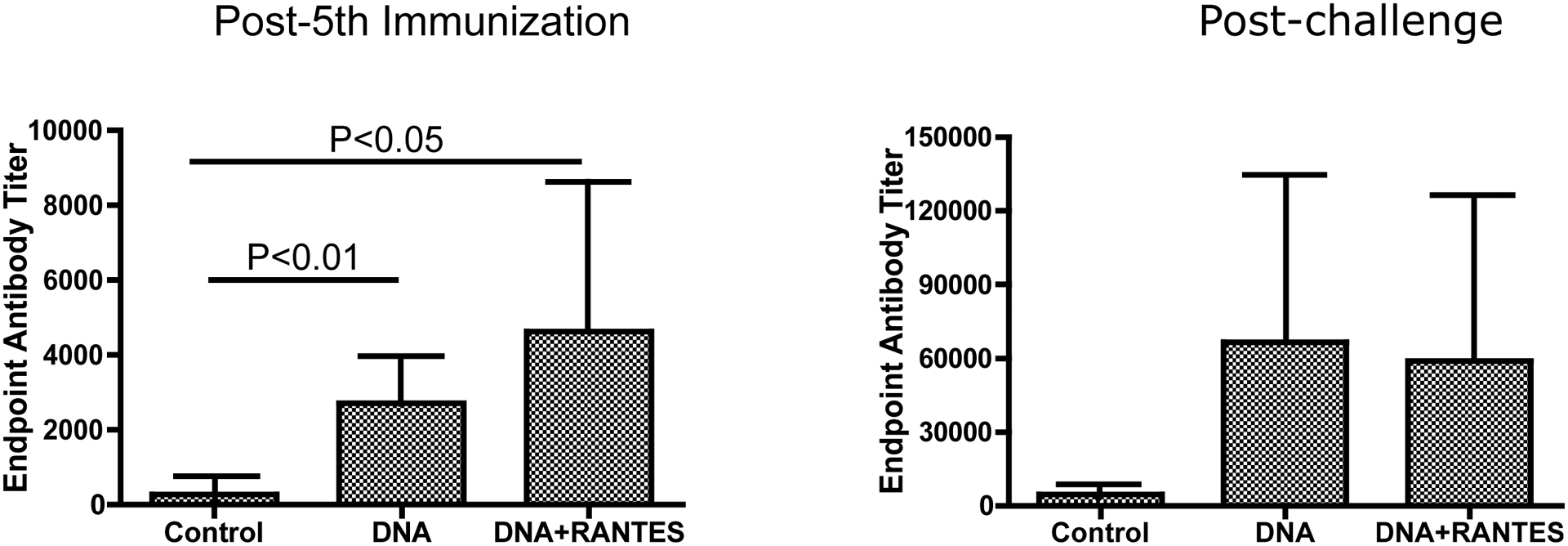
Sera were tested for antibodies capable of binding to HIV-1 envelope gp120, CN54, derived from a subject infected with clade C. Antibody titers are presented for (A) after the final immunization, prior to challenge and (B) post challenge.
RANTES, a cytokine that belongs to the chemokine family and a chemoattractant for CD4+/CD45RO T cells, can be produced by various cell types including CD8+ and CD4+ T cells as well as monocytes/macrophages. This chemokine has been shown to suppress replication of macrophage-tropic strains of HIV in CD4+ T cells (20). Our results demonstrate that co-immunized RANTES plasmid with SHIV DNA vaccine improves SHIV-specific cellular and humoral immune responses in rhesus macaques.
Sin et al demonstrated that RANTES, as an adjuvant to a DNA vaccine, can enhance antigen-specific Th1-Type CD4 T-cell-mediated protective immunity against Herpes Simplex virus in vivo (36). Shimizu et al genetically engineered a live-attenuated SHIV to express the RANTES gene (SHIV-RANTES) and found that SHIV-RANTES provided some immunity in monkeys by remarkably increasing the antigen-specific CD4+ T-cell proliferative response and by inducing an antigen-specific IFN-γ ELISpot response. Unfortunately, it failed to afford greater protection against a heterologous pathogenic SHIV (SHIV-C2/1) challenge compared to its control (21). Hadida et al have previously shown that RANTES enhances the HIV-specific cytotoxicity of CD8 T cells (13). As a result, we are the first to demonstrate that RANTES, as an adjuvant of a DNA vaccine, improved the protection against SHIV-infection in a macaque model.
In addition, the developments of new delivery methods such as: electroporation (EP) (22,23), as well as the use of agents that improve antigen uptake or presentation, and optimization of the transgene sequences are overcoming historical drawbacks. Many studies have shown that the use of EP enhances plasmid uptake and immune responses by a factor of 5:1000 in function of the specific model (24). Hence, we utilized electroporation to improve DNA vaccine delivery of a poly-optimized DNA vaccine. Our results demonstrate a 10-fold increase in the average number of IFN-γ producing cells compared to previous studies (data not shown). Importantly, in our previous studies that did not use electroporation, antibody responses were either very low or not observed pre-challenge. The RV144 study has brought a new focus to HIV-1 specific antibodies. It is believed that binding antibodies can stunt viral infection providing some impact on disease.
In conclusion, using both an adjuvant concurrently with electroporation delivery can enhance cellular and humoral immune responses. This ultimately has important implications for HIV-1 vaccine development.
Acknowledgements
This research was supported in part by National Institutes of Health (NIH) Grants N01-AI-50010, P01-A1-071739, R01-A1-071186, AK, NYS acknowledge funding in part from an NIH/NIAID/DAIDS - HVDDT award to Inovio HHSN272200800063C”- and the National Institutes of Health Intramural Research Program. J.Y. performed research, generated the data presented and wrote the paper. A.D., J.L., T.A., and M.G.L., all performed research and generated the data presented. M.A.K., J.Y., A.K., N.Y.S, and D.B.W. provided reagents and guidance. J.D.B. designed experiments and oversaw the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest; AK and NYS are full time employees of Inovio Pharmaceuticals
Reference
- 1.Barouch DH 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham BS, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Martin JE, McCluskey MM, Chakrabarti BK, Lamoreaux L, Andrews CA, Gomez PL, Mascola JR, and Nabel GJ. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J Infect Dis 194:1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, Bilska M, Craiu A, Zheng XX, Krivulka GR, Beaudry K, Lifton MA, Nickerson CE, Trigona WL, Punt K, Freed DC, Guan L, Dubey S, Casimiro D, Simon A, Davies ME, Chastain M, Strom TB, Gelman RS, Montefiori DC, Lewis MG, Emini EA, Shiver JW, and Letvin NL. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486–492. [DOI] [PubMed] [Google Scholar]
- 4.Chong SY, Egan MA, Kutzler MA, Megati S, Masood A, Roopchard V, Garcia-Hand D, Montefiori DC, Quiroz J, Rosati M, Schadeck EB, Boyer JD, Pavlakis GN, Weiner DB, Sidhu M, Eldridge JH, and Israel ZR. 2007. Comparative ability of plasmid IL-12 and IL-15 to enhance cellular and humoral immune responses elicited by a SIVgag plasmid DNA vaccine and alter disease progression following SHIV(89.6P) challenge in rhesus macaques. Vaccine 25:4967–4982. [DOI] [PubMed] [Google Scholar]
- 5.Luckay A, Sidhu MK, Kjeken R, Megati S, Chong SY, Roopchand V, Garcia-Hand D, Abdullah R, Braun R, Montefiori DC, Rosati M, Felber BK, Pavlakis GN, Mathiesen I, Israel ZR, Eldridge JH, and Egan MA. 2007. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol 81:5257–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Kjeken R, Mathiesen I, and Barouch DH. 2008. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J Virol 82:5643–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirao LA, Wu L, Khan AS, Hokey DA, Yan J, Dai A, Betts MR, Draghia-Akli R, and Weiner DB. 2008. Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine 26:3112–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prud’homme GJ, Glinka Y, Khan AS, and Draghia-Akli R. 2006. Electroporation-enhanced nonviral gene transfer for the prevention or treatment of immunological, endocrine and neoplastic diseases. Curr Gene Ther 6:243–273. [DOI] [PubMed] [Google Scholar]
- 9.Frauenschuh A, DeVico AL, Lim SP, Gallo RC, and Garzino-Demo A. 2004. Differential polarization of immune responses by co-administration of antigens with chemokines. Vaccine 23:546–554. [DOI] [PubMed] [Google Scholar]
- 10.Kim JJ, Nottingham LK, Sin JI, Tsai A, Morrison L, Oh J, Dang K, Hu Y, Kazahaya K, Bennett M, Dentchev T, Wilson DM, Chalian AA, Boyer JD, Agadjanyan MG, and Weiner DB. 1998. CD8 positive T cells influence antigen-specific immune responses through the expression of chemokines. J Clin Invest 102:1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterman PM, Kitabwalla M, Hatfield GS, Evans PS, Lu Y, Tikhonov I, Bryant JL, and Pauza CD. 2004. Effects of virus burden and chemokine expression on immunity to SHIV in nonhuman primates. Viral Immunol 17:545–557. [DOI] [PubMed] [Google Scholar]
- 12.Xin KQ, Lu Y, Hamajima K, Fukushima J, Yang J, Inamura K, and Okuda K. 1999. Immunization of RANTES expression plasmid with a DNA vaccine enhances HIV-1-specific immunity. Clin Immunol 92:90–96. [DOI] [PubMed] [Google Scholar]
- 13.Hadida F, Vieillard V, Autran B, Clark-Lewis I, Baggiolini M, and Debre P. 1998. HIV-specific T cell cytotoxicity mediated by RANTES via the chemokine receptor CCR3. J Exp Med 188:609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan J, Hokey DA, Morrow MP, Corbitt N, Harris K, Harris D, and Weiner DB. 2009. Novel SIVmac DNA vaccines encoding Env, Pol and Gag consensus proteins elicit strong cellular immune responses in cynomolgus macaques. Vaccine 27:3260–3266. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, and Montefiori DC. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol, 79:10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montefiori DC (2004) Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. Current Protocols in Immunology, (Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, and Coico R, eds.), John Wiley & Sons, 12.11.1–12.11.15. [DOI] [PubMed] [Google Scholar]
- 17.Song RJ, Chenine A-L, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li P-L, Shai-Kobiler E, Wang T, Mc Cann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. Molecularly cloned SHIV-1157pd3N4: a highly replication competent, mucosally transmissible R5 simian-human-immunodeficiency virus encoding HIV clade C env. J Virol 2006; 80:8729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddappa NB, Watkins JD, Wassermann KJ, Song R, Wang W, Kramer VG, Lakhashe S, Santosuosso M, Poznansky MC, Novembre FJ, Villinger F, Else JG, Montefiori DC, Rasmussen RA, Ruprecht RM. R5 clade C SHIV strains with tier 1 or 2 neutralization sensitivity: tools to dissect Env evolution and to develop AIDS vaccines in primate models. PLoS One, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano JW, Shurtliff RN, Dobratz E, Gibson A, Hickman K, Markham PD, and Pal R. 2000. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J Virol Methods 86:61–70. [DOI] [PubMed] [Google Scholar]
- 20.Sin J, Kim JJ, Pachuk C, Satishchandran C, and Weiner DB. 2000. DNA vaccines encoding interleukin-8 and RANTES enhance antigen-specific Th1-type CD4(+) T-cell-mediated protective immunity against herpes simplex virus type 2 in vivo. J Virol 74:11173–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu Y, Inaba K, Kaneyasu K, Ibuki K, Himeno A, Okoba M, Goto Y, Hayami M, Miura T, and Haga T. 2007. A genetically engineered live-attenuated simian-human immunodeficiency virus that co-expresses the RANTES gene improves the magnitude of cellular immunity in rhesus macaques. Virology 361:68–79. [DOI] [PubMed] [Google Scholar]
- 22.Kutzler MA, and Weiner DB. 2008. DNA vaccines: ready for prime time? Nat Rev Genet 9:776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdulhaqq SA, and Weiner DB. 2008. DNA vaccines: developing new strategies to enhance immune responses. Immunol Res 42:219–232. [DOI] [PubMed] [Google Scholar]
- 24.Bodles-Brakhop AM, and Draghia-Akli R. 2008. DNA vaccination and gene therapy: optimization and delivery for cancer therapy. Expert Rev Vaccines 7:1085–1101. [DOI] [PubMed] [Google Scholar]


