Abstract
In selenoproteins, incorporation of the amino acid selenocysteine is specified by the UGA codon, usually a stop signal. The alternative decoding of UGA is conferred by an mRNA structure, the SECIS element, located in the 3′-untranslated region of the selenoprotein mRNA. Because of the non-standard use of the UGA codon, current computational gene prediction methods are unable to identify selenoproteins in the sequence of the eukaryotic genomes. Here we describe a method to predict selenoproteins in genomic sequences, which relies on the prediction of SECIS elements in coordination with the prediction of genes in which the strong codon bias characteristic of protein coding regions extends beyond a TGA codon interrupting the open reading frame. We applied the method to the Drosophila melanogaster genome, and predicted four potential selenoprotein genes. One of them belongs to a known family of selenoproteins, and we have tested experimentally two other predictions with positive results. Finally, we have characterized the expression pattern of these two novel selenoprotein genes.
INTRODUCTION
Selenoproteins are proteins that incorporate the amino acid selenocysteine, a cysteine analog in which a selenium atom is found in place of sulfur. Several components of the selenoprotein synthesis machinery are conserved between different species, suggesting an important role of selenoproteins in cell function (Low and Berry, 1996; Stadman, 1996). Incorporation of selenocysteine into selenoproteins requires an unusual translation step where UGA, normally a stop codon, specifies selenocysteine insertion. Thus, in a single mRNA, UGA can have two contrasting meanings: stop or selenocysteine. The alternative decoding of UGA is conferred by an mRNA secondary/tertiary structure (the selenocysteine insertion sequence, the SECIS element), which is located in eukaryotes in the 3′-untranslated region. SECIS structures are divided into two classes, termed form 1 and form 2, the latter having an additional small stem–loop at the top of the SECIS element. Most selenoproteins contain a single selenocysteine residue per polypeptide chain, but selenoprotein P has as many as 10–12 (Tujebajeva et al., 2000a).
Selenoproteins have been identified in Bacteria, Archaea and Eukarya. Among eukaryotes, selenoproteins appear to be more common in mammals. Thus, 19 selenoproteins have been found to date in mammals (Flohé et al., 2000), but none in the genome of Saccharomyces cerevisiae, and only one in the genome of Caenorhabditis elegans (Buettner et al., 1999; Gladyshev et al., 1999). Recently, the class 2 selenophosphate synthetase gene (sps2)—an enzyme in the pathway of selenoprotein synthesis, and a selenoprotein itself in mammals—has also been shown to be a selenoprotein in Drosophila melanogaster (Hirosawa-Takamori et al., 2000). So far, it remains the only selenoprotein identified in this organism and maps to chromosome 2L. However, pupal proteins of 68, 42 and 25 kDa have been reported to incorporate selenium (Robinson and Cooley, 1997), and a major band of 42 kDa has also been observed in protein extracts of larvae labeled with 75Se (Alsina et al., 1999). In addition, some components of the selenoprotein synthesis machinery have already been identified in the fly (Persson et al., 1997; Alsina et al., 1998; Zhou et al., 1999). Moreover, a mutation in the sps1 gene (a cysteine homolog of Sps2) leads to larval lethality, increased apoptosis and aberrant imaginal disc morphology (Alsina et al., 1998). These data strongly suggest the existence of as yet unidentified selenoproteins in the D. melanogaster genome. The recent availability of the complete DNA sequence of this genome should constitute an invaluable resource for characterizing the D. melanogaster selenoproteins.
Prediction of selenoproteins in genomic sequences, however, is particularly difficult. Without exception, computational gene prediction programs rely on the standard stop codons TAA, TAG and TGA to identify open reading frames (ORFs) and predict coding exons, through the determination of suitable splicing sites and the computation of some measure of coding likelihood, usually related to bias in codon usage (see Burge and Karlin, 1998 and Haussler, 1998 for reviews on computational gene finding). Under such an assumption, selenoprotein genes, in which TGA does not necessarily imply termination of translation, will be incorrectly predicted. Indeed, the D. melanogaster sps2 gene (dsps2) is wrongly predicted in the released annotation of the fly genome: >100 amino acids are missing from a 379 amino acids protein. Correct delineation of the exonic structure is singularly important to predict selenoprotein genes. Misprediction of only a single amino acid (the selenocysteine residue) may lead to misidentification of selenoproteins (see Results).
Although searching for potential SECIS elements has proved useful in identifying new selenoproteins in expressed sequence tag (EST) sequences (Kryukov et al., 1999; Lescure et al., 1999), this approach is impractical when applied to genomic sequences, given the high frequency of occurrence of the SECIS pattern (see Results). To reduce the number of false positive predictions, we developed a method that relies on the correlated prediction of SECIS elements and of genes in which the strong codon bias characteristic of protein coding regions extends beyond a TGA codon interrupting the ORF. Indeed, we have found that in selenoproteins the region comprised between the in-frame TGA codon and the stop codon shows codon bias comparable to that found in coding regions, while in non-selenoproteins the region comprised between the stop codon TGA and the next stop codon in-frame shows codon bias comparable to that in non-coding regions (Supplementary data). Therefore, measures of codon bias can be used to distinguish actual selenoproteins from false predictions in SECIS-positive nucleotide sequences
RESULTS
Prediction of novel selenoproteins in the D. melanogaster genome
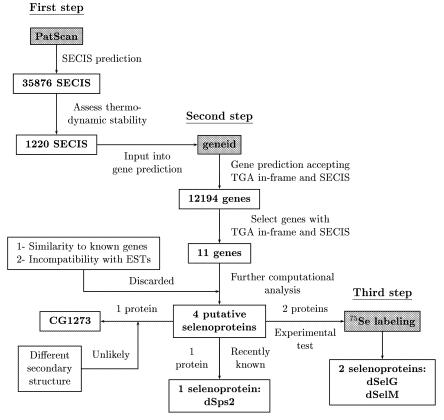
The March 24, 2000 release of the D. melanogaster genome sequence summing up 115 229 998 bp and containing 13 329 annotated genes was used (Adams et al., 2000). 37 876 potential SECIS elements were found along this sequence using the program PatScan. The minimum free energy of each putative SECIS was measured, and only those fitting an energy stability criteria were considered further. This resulted in 1220 potential SECIS. Along with the sequence, positions of these elements were given to a modified version of the program geneid, which allows for the prediction of genes interrupted by in-frame TGA codons. The restriction was enforced such that genes could not be further than 500 bp upstream from a predicted SECIS. Eleven potential selenoproteins were predicted among a total of 12 194 genes. Seven of them were discarded because the predicted exonic structure was incompatible with the exonic structure of known overlapping genes, contradicted identical EST sequences or was similar to known proteins with functions apparently unrelated to those of selenoproteins. Of the remaining four, one corresponded to the previously identified D. melanogaster selenoprotein dsps2. For another two, we identified cysteine paralogs within the set of proteins predicted in the D. melanogaster genome. No additional evidence was found for the fourth putative selenoprotein, after an exhaustive search against a number of databases of known coding sequences using the BLAST suite of programs (Altschul et al., 1997). In addition, the predicted secondary structure around the selenocysteine residue of this putative selenoprotein is not compatible with the known crystal structure of the bovine glutathione peroxidase, a eukaryotic selenoprotein. This structure appears to be common to most known selenoproteins, including the two other predictions (Supplementary data). We have thus considered this prediction to be a false positive. Incorporation of selenium was subsequently demonstrated for the two other predicted selenoproteins (which we name dselG and dselM), and their expression pattern during development was characterized.
dselG
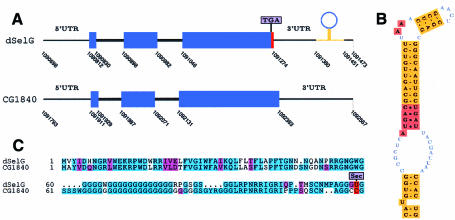
dSelG is a 110 aa protein which maps to 10F4-6 of the X chromosome. It differs in only two amino acids from the annotated protein in D. melanogaster (CG1844), the in-frame TGA lying only one codon upstream from the stop codon. dselG has a cysteine paralog, the CG1840 gene. They appear in tandem, separated by only 320 bp, and have the same exonic structure sharing 65% identity at the protein level (Figure 1).
Fig. 1. dSelG. (A) Gene structure for dselG and in-tandem CG1840 paralog plotted using gff2ps (Abril and Guigó, 2000). Coordinates correspond to the AE002593 (X) scaffold. The extra coding region is shown in red as predicted by geneid and the annotated coding exons are in blue. (B) dselG form 2 SECIS. (C) Alignment of dSelG and CG1840 paralogs using CLUSTAL_W (Thompson et al., 1994).
dselM
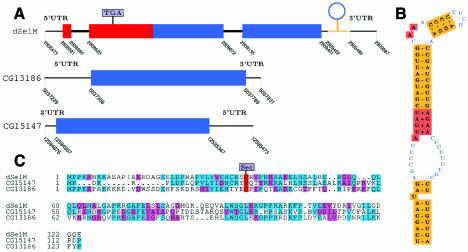
dSelM is a 249 aa protein which maps to 12A4–6 of the X chromosome. It differs substantially from the protein annotated in D. melanogaster (CG11177), the first exon, and a large fraction of the second (in which the in-frame TGA is located) having been missed. dselM has two distant paralogs, the CG13186 and CG15147 genes (Figure 2).
Fig. 2. dSelM. (A) Gene structure for dselM and single exon CG15147, CG13186 paralogs. Coordinates correspond to the AE002593 (X), AE002690 (2L) and AE002787 (2R) scaffolds, respectively. (B) dselM form 2 SECIS. (C) Alignment of dSelM and CG15147, CG13186 paralogs.
75Se labeling of the D. melanogaster selenoproteins expressed in mammalian cells
75Se labeling of HEK cells was undertaken to demonstrate that GH03581 (dselG) and SD09114 (dselM) genes encoded bona fide selenoproteins. In cells transfected with empty vector, the background pattern of endogenously expressed selenoproteins can be seen (Figure 3A, lane 1), including thioredoxin reductases (∼55 kDa), glutathione peroxidase (∼23 kDa), phospholipid hydroperoxide glutathione peroxidase (∼20 kDa), and an ∼12–14 kDa triplet. Transfection of the GH03581 expression vector resulted in an increase in labeling in the 12 kDa size range, overlapping the lower band of the endogenous triplet (Figure 3A, lane 2). Transfection of the SD09114 expression vector resulted in appearance of a prominent new band of ∼30 kDa (Figure 3A, lane 3), corresponding to the predicted size of SD09114 (∼27 kDa). SD09114 protein appears to be a D. melanogaster-specific selenoprotein, with no analog in mammalian cells of similar molecular weight.
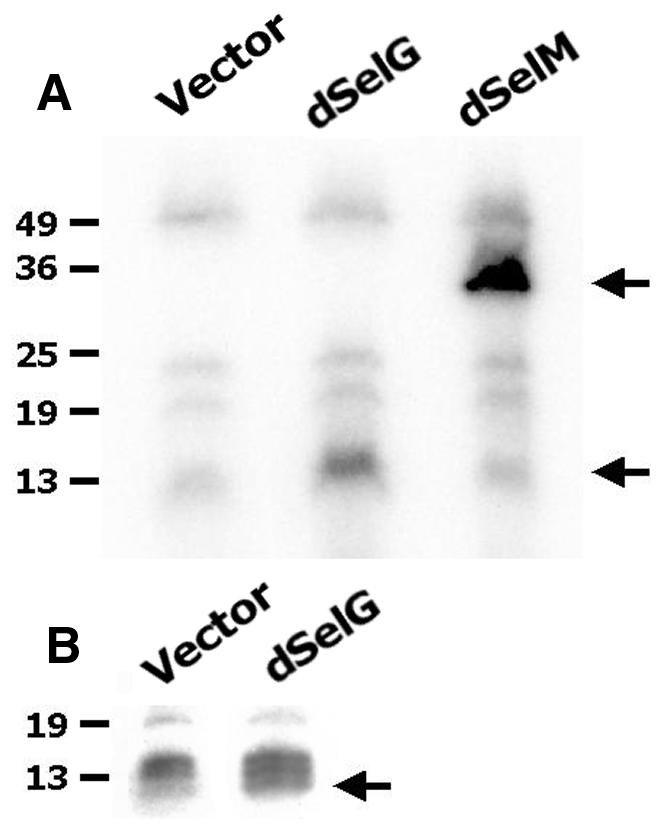
Fig. 3. 75Se-labeling of the D. melanogaster selenoproteins expressed in mammalian cells. (A) Lane 1: 75Se-labeling of cells transfected with empty vector. Lane 2: 75Se-labeling of cells transfected with dselG. Lane 3: 75Se-labeling of cells transfected with dselM. (B) High magnification of the region corresponding to the dselG labeling.
In situ hybridization in embryos, discs and brains
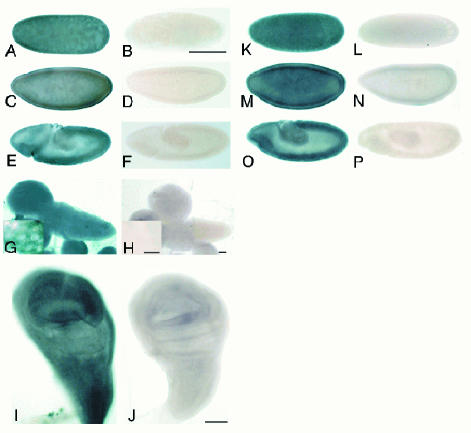
In situ hybridization experiments were performed to assess dselG and dselM mRNA expression patterns. dselM mRNA was present in all embryonic stages, especially in the blastoderm stage, suggesting that there is a strong maternal contribution (Figure 4A, C and E). Imaginal discs displayed a ubiquitous dselM expression pattern (Figure 4I), and in brain, although the staining was ubiquitous, large cells, probably neuroblasts, were highly stained (Figure 4G). dselG expression pattern was analyzed in embryos, and similarly to dselM, the mRNA was found ubiquitously in all stages (Figure 4K, M and O). Due to the high similarity at the nucleotide sequence level between dselG and the cysteine homolog (64% in the coding fraction), we have to assume that the probe used for the in situ hybridization would detect both transcripts, if present.
Fig. 4. In situ hybridization in embryos, imaginal discs and brain. (A, C and E) dselM expression pattern in sincytial blastoderm, cellular blastoderm and gastrulation embryonic stages, respectively; (B, D and F) the corresponding sense controls (scale bar is 50 mm); (G) dselM expression in brain and neuroblast staining in the inset; (H) the brain sense control (scale bar 100 mm, inset scale bar 2.5 mm); (I) dselM expression in wing disc; (J) wing disc sense control (scale bar 50 mm); (K, M and O) dselG expression pattern in sincytial blastoderm, cellular blastoderm and gastrulation embryonic stages, respectively; (L, N and P) are the corresponding sense controls (scale bar is 50 mm).
DISCUSSION
Most of the functions of selenium involve its incorporation into selenoproteins in the form of selenocysteine. Besides their putative role in regulating the redox state of the cell, selenoproteins seem to possess anticarcinogenic properties, and PHGPx plays a role in reproductive function (Ganther, 1999; Ursini et al., 1999). Drosophila provides a convenient tool for investigating selenoprotein function because of the availability of fly genetics and the already existing mutation in the sps1 gene (Alsina et al., 1998).
Using a novel computational method we have predicted four potential selenoprotein genes in the D. melanogaster genome, with little human intervention. Three are bona fide selenoproteins: dSps2, already demonstrated as such (Hirosawa-Takamori et al., 2000), and the other two, dSelG and dSelM, shown herein by 75Se labeling. In addition, cysteine paralogs exist in D. melanogaster for these three selenoproteins. While Sps2, a selenophosphate synthetase, belongs to a known family of selenoproteins, dSelG and dSelM are novel selenoproteins, lacking sequence similarity to known proteins. dSelG has a cysteine homolog in C. elegans of unknown function, while dSelM appears to belong to a new class of selenoproteins widely distributed across the phylogenetic spectrum: we have found selenocysteine homologs to dSelM in ESTs from zebrafish, human and mouse databases, among other organisms.
It is unclear, however, how complete our characterization of the selenoprotein set in D. melanogaster is. Experimental data suggest the existence of a selenoprotein in the 60–70 kDa range (Alsina et al., 1999), for which we have not been able to find a computational prediction. Forcing the SECIS element to occur within 500 bp downstream from the selenoprotein coding region may be too restrictive. Although most mammalian selenoproteins are within this range, longer distances up to >4000 bp are possible. The fly genome is certainly more compact, and while in the human sps2 the SECIS element is 579 bp downstream from the stop codon, this distance is only 30 bp in D. melanogaster. On the other hand, exceptions to the standard eukaryotic SECIS model have recently been reported in C. elegans. In this case, a 5′-GUGA motif is present instead of AUGA (Buettner et al., 1999). Therefore, it is possible that additional selenoproteins using an alternative SECIS structure exist in the D. melanogaster genome. Relaxing the SECIS pattern to capture a more general SECIS structure results in a substantial increase in the number of predicted SECIS elements, which compounds the analysis of the search results. In this regard, the approach presented here could contribute towards systematically exploring alternative SECIS structures.
In summary, we believe that the research described here demonstrates the power of the combined in silico, in vitro and in vivo approaches towards a better understanding of living systems.
METHODS
Prediction of selenoproteins in nucleotide sequences.
The method that we have developed is described in detail in the Supplementary data, which can be found at EMBO reports Online. A general schema is shown in Figure 6. Broadly, given a query sequence, first we predict SECIS elements using the program PatScan (http://www-unix.mcs.anl.gov/compbio/PatScan/HTML/PatScan.html) (Figure 5A and B). The stability of the predicted SECIS is then assessed using the RNAfold program (Viena RNA package) using the protocol by Kryukov et al. (1999). Next, we use a modification of the program geneid (Guigó et al., 1992; Parra et al., 2000) to predict genes that may be interrupted by in-frame TGA codons. Such genes, however, can be predicted only when a putative SECIS, whose position along the genome is input into geneid during gene prediction, exists at the right distance. The modified geneid yields, in the same gene prediction, both standard genes and selenoprotein genes (Figure 5C and D).
Fig. 6. General schema for selenoprotein identification.
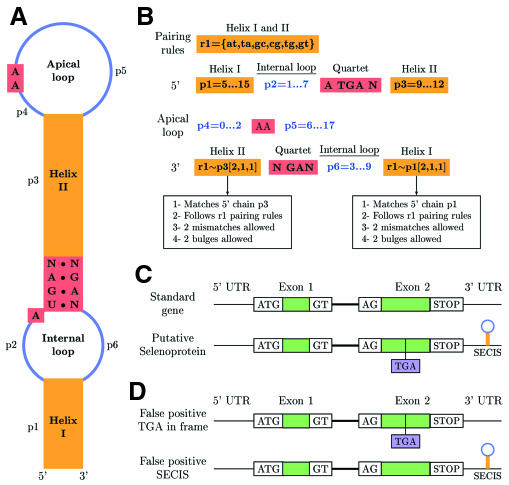
Fig. 5. SECIS and gene prediction. (A) General form 1 SECIS divided into structural units. Form 2 has an extra short stem–loop in the apical loop. (B) PatScan SECIS pattern to search for both form 1 and form 2 SECIS. The extra stem–loop in form 2 is not taken into account when searching. (C) The two possible ways of geneid prediction for an ideal two exons gene: as a normal gene or as a selenoprotein gene with a TGA in-frame and a SECIS. Exon defining signals are shown. (D) False positive selenoprotein genes with either a TGA in-frame or a SECIS. These partial predictions are not permitted in the gene prediction.
75Se labeling.
The pOT2 plasmids containing GH03581 (dselG) and SD09114 (dselM) cDNA clones were obtained from Research Genetics Inc. and sequenced using the Dye Terminator Cycle Sequencing method. Inserts were subcloned into pUHD10–3 vector via EcoRI and XbaI sites. Human embryonic kidney cells (HEK-293) were transiently transfected by CaPO4 DNA precipitation method (Tujebajeva et al., 2000b) with either dselG or dselM expression plasmids and co-transfected with plasmids encoding tRNA[Ser]Sec (Lee et al., 1990) and SECIS-binding protein (SBP2) (Copeland et al., 2000) to increase the efficiency of selenocysteine incorporation (Berry et al., 1994; Tujebajeva et al., 2000b). All transfection experiments were carried out with supplementation of 100 nM sodium selenite to the media. 75Se as sodium selenite (1000 mCi/mg) was added to media 1 day after transfection, and labeling proceeded for another day. Cells were harvested, sonicated in 0.25 M sucrose in PE buffer (0.1 M potassium phosphate, 1 mM EDTA pH 6.9) and analyzed by polyacrylamide gel electrophoresis, followed by autoradiography.
Whole-mount in situ hybridization.
Embryos collected from a 24-h egg-lay were dechorionated and fixed in 2% formaldehyde and 0.5 M final concentration of EGTA in PBS for 20 min. After precipitation with methanol embryos were kept at –20°C in absolute ethanol. Third-instar wild-type larvae were dissected in PBS and fixed overnight in 4% paraformaldehyde in PBS for 20 min. Further steps before hybridization and hybridization itself were performed as described by Lehner and O’Farrell (1990). Linearized pOT2 vectors containing GH03581 and SD09114 clones were used to generate a riboprobe according to the Boehringer-Mannheim protocol. Embryos and discs were then incubated with 1/2000 anti-DIG conjugated with alkaline phosphatase antibody (Boehringer-Mannheim), preabsorbed against fixed and dissected larvae. Antibody was detected using standard procedures (Boehringer-Mannheim). Embryos were postfixed and posteriorly mounted in DePeX. Discs and brains were dissected and mounted in 87% glycerol.
Data and software availability.
Sequence data and software can be found at http://www1.imim.es/databases/spdroso2001
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank E. Blanco for his assistance with geneid, and P. Higgs for helpful suggestions. R.G. thanks L.S. Shashidhara for his hospitality at the CCMB (Hyderabad), where part of this manuscript was written. This work was partially supported by grants BIO98-0443-C02-01 and PB96-1253 from ‘Plan Nacional de I+D’ (Spain). S.C. and M.M. are recipients of predoctoral fellowships from CIRIT, Generalitat de Catalunya.
REFERENCES
- Abril J.F. and Guigó, R. (2000) gff2ps: visualizing genomic annotations. Bioinformatics, 16, 743–744. [DOI] [PubMed] [Google Scholar]
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Alsina B., Serras, F., Baguña, J. and Corominas, M. (1998) Patufet, the gene encoding the Drosophila melanogaster homologue of selenophosphate synthetase, is involved in imaginal disc morphogenesis. Mol. Gen. Genet., 257, 113–123. [DOI] [PubMed] [Google Scholar]
- Alsina B., Corominas, M., Berry, M.J., Baguña, J. and Serras, F. (1999) Disruption of selenoprotein biosynthesis affects cell proliferation in the imaginal discs and brain of Drosophila melanogaster. J. Cell Sci., 112, 2875–2884. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.J., Harney, J.W., Ohama, T. and Hatfield, D.L. (1994) Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res., 22, 3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner C., Harney, J.W. and Berry, M.J. (1999) The Caenorhabditis elegans homologue of thioredoxin reductase contains a Selenocysteine Insertion Sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem., 274, 21598–21602. [DOI] [PubMed] [Google Scholar]
- Burge C.B. and Karlin, S. (1998) Finding the genes in genomic DNA. Curr. Opin. Struct. Biol., 8, 346–354. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Fletcher, J.E., Carlson, B.A., Hatfield, D.L. and Driscoll, D.M (2000). A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J., 19, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohé L., Andreesen, J.R., Brigelius-Flohé, R., Maiorino, M. and Ursini, F. (2000) Selenium, the element of the moon, in life on Earth. Life, 49, 411–420. [DOI] [PubMed] [Google Scholar]
- Ganther H.E., (1999) Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis, 20, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Gladyshev V.N., Krause, M., Xu, X.M., Korotkov, K.V., Kryukov, G.V., Sun, Q.A., Lee, B.J., Wooton, J.C. and Hatfield, D.L. (1999) Selenocysteine-containing thioredoxin reductase in C. elegans. Biochem. Biophys. Res. Commun., 259, 244–249. [DOI] [PubMed] [Google Scholar]
- Guigó R., Knudsen, S., Drake, N. and Smith, T.F. (1992) Prediction of gene structure. J. Mol. Biol., 226, 141–157. [DOI] [PubMed] [Google Scholar]
- Haussler D. (1998) Computational genefinding. Trends Biochem. Sci., Supplementary Guide to Bioinformatics, pp. 12–15.
- Hirosawa-Takamori M., Jäckle, H. and Vorbrüggen, G. (2000) The class 2 selenophosphate synthetase gene of Drosophila contains a functional mammalian-type SECIS. EMBO Rep., 1, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov G.V., Kryukov, V.M. and Gladyshev, V.N. (1999) New mammalian selenocysteine-containing proteins identified with an algorithm that searches for Selenocysteine Insertion Sequence Elements. J. Biol. Chem., 274, 33888–33897. [DOI] [PubMed] [Google Scholar]
- Lee B.J., Rajagopalan, M., Kim, Y.S., You, K.H., Jacobson, K.B. and Hatfield, D. (1990) Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol., 10, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner C.F. and O’Farrell, P.H. (1990) Drosophila cdc2 homologues: a functional homologue is coexpressed with a cognate variant. EMBO J., 9, 3573–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure A., Gautheret, D., Carbon, P. and Krol, A. (1999) Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem., 274, 38147–38154. [DOI] [PubMed] [Google Scholar]
- Low S.C. and Berry, M.J. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryote. Trends Biochem. Sci., 21, 203–208. [PubMed] [Google Scholar]
- Parra G., Blanco, E. and Guigó, R. (2000) Geneid in Drosophila. Genome Res., 10, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B.C., Böck, A., Jäckle, H. and Vorbruggen, G. (1997) SelD homologue from Drosophila lacking selenide-dependant monoselenophosphate synthetase activity. J. Mol. Biol., 274, 174–180. [DOI] [PubMed] [Google Scholar]
- Robinson D.N. and Cooley, L. (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development, 124, 1405–1417. [DOI] [PubMed] [Google Scholar]
- Stadman T.C. (1996) Selenocysteine. Annu. Rev. Biochem., 65, 83–100. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva R.M., Ransom, D.G., Harney, J.W. and Berry, M.J. (2000a) Expression and characterization of nonmammalian selenoprotein P in the zebrafish, Danio rerio. Genes Cells, 5, 897–903. [DOI] [PubMed] [Google Scholar]
- Tujebajeva R.M, Copeland, P.R., Xu, X-M., Carlson, B.A., Harney, J.W., Driscoll, D.M., Hatfield, D.L. and Berry, M.J. (2000b) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep., 1, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F., Heim, S., Kiess, M., Maiorino, M., Roveri, A., Wissing, J. and Flohé, L. (1999) Dual function of the selenoprotein PHGPx during sperm maturation. Science, 285, 1393–1396. [DOI] [PubMed] [Google Scholar]
- Zhou X., Park, S.I., Moustafa, M.E., Carlson, B.A., Crain, F., Diamond, A.M., Hatfield, D.L. and Lee, B.J. (1999) Selenium metabolism in Drosophila. Characterization of the selenocysteine tRNA population. J. Biol. Chem., 274, 18729–18734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.