Abstract
In eukaryotes, the polypeptide release factor 1 (eRF1) is involved in translation termination at all three stop codons. However, the mechanism for decoding stop codons remains unknown. A direct interaction of eRF1 with the stop codons has been postulated. Recent studies focus on eRF1 from ciliates in which some stop codons are reassigned to sense codons. Using an in vitro assay based on mammalian ribosomes, we show that eRF1 from the ciliate Euplotes aediculatus responds to UAA and UAG as stop codons and lacks the capacity to decipher the UGA codon, which encodes cysteine in this organism. This result strongly suggests that in ciliates with variant genetic codes eRF1 does not recognize the reassigned codons. Recent hypotheses describing stop codon discrimination by eRF1 are not fully consistent with the set of eRF1 sequences available so far and require direct experimental testing.
INTRODUCTION
Termination of protein synthesis is governed by the presence of a stop codon in the ribosomal A site and by polypeptide chain release factors (RFs) (reviewed by Kisselev and Buckingham, 2000). In eukaryotes, a single factor, eRF1, decodes all three stop codons, UAA, UAG and UGA, whereas in prokaryotes, RF1 responds to UAA and UAG while RF2 responds to UAA and UGA.
None of the hypotheses postulating the mechanism of decoding the termination codons has been proved directly. It is assumed that stop codons within the ribosome are recognized by class-1 termination factors RF1, RF2 and eRF1 (see Nakamura et al., 2000). The main argument is the very tight contact between class-1 RFs and stop codons within the ribosome, revealed by photocrosslinking both in prokaryotes (Brown and Tate, 1994; Poole et al., 1997) and eukaryotes (Chavatte et al., 2001). Another argument came from experiments showing that mutagenesis of class-1 RF sequences resulted in the modification of their stop codon recognition pattern (Bertram et al., 2000; Ito et al., 2000). Alternatively, it was proposed that stop codons can be recognized by specific sequences in ribosomal RNAs (see Arkov and Murgola, 1999; Ivanov et al., 2001).
A remarkable feature of some ciliate species is their use of alternative nuclear genetic codes, which have possibly arisen independently, even within a single class of ciliates (Baroin-Tourancheau et al., 1995). The known changes concern the reassignment of stop codons to sense codons. For example, Tetrahymena and Paramecium, and the hypotrichs Stylonychia and Oxytricha, translate UAA and UAG as glutamine, UGA being the only stop codon, whereas the hypotrich Euplotes translates UGA as cysteine and uses UAA and UAG as stop codons (for review see Lozupone et al., 2001). It has been postulated that in addition to changes in tRNAs, stop codon reassignment has to involve alterations of eRF1 structure. Substantial efforts were undertaken to sequence eRF1 genes from ciliate species with variant genetic codes (Karamyshev et al., 1999; Inagaki and Doolittle, 2001; Liang et al., 2001; Lozupone et al., 2001). In conjunction with the hypothesis that the N-terminal domain of eRF1 was implicated in stop codon recognition (Bertram et al., 2000), multiple sequence alignments were analyzed in an attempt to predict which amino acids of eRF1 were involved in stop codon recognition. However, depending on the number of eRF1 sequences used, different sets of amino acid residues of the N-terminal domain were chosen (Lehman, 2001; Lozupone et al., 2001; Muramatsu et al., 2001).
In this study, we verified the assumption that eRF1 from a ciliate with variant genetic code does not recognize the reassigned stop codon. Our results show that Euplotes eRF1 does not respond to UGA, which is used as a cysteine codon in this organism. We also show that introduction of new eRF1 sequences into eRF1 alignment questioned most of the recent hypotheses concerning stop codon recognition in eukaryotes.
RESULTS
Isolation of the Euplotes aediculatus gene encoding eRF1
To isolate the gene encoding eRF1, we have screened a pUC18 library of E. aediculatus macronuclear DNA using Paramecium tetraurelia eRF1 gene as a probe. Four positive colonies were selected after three rounds of isolation and purification. Plasmid DNA of these clones was analyzed by Southern blotting and two of the inserts giving a strong positive signal were sequenced. The two sequences were identical and contained 5′ and 3′ telomeric 5′-dC4dA4-3′ repeats, confirming the presence of an entire transcription unit. The sequence was identified through BLAST searches (Altschul and Koonin, 1998). The results of BLASTX (e-value >10–100 with eRF1s) suggested that the selected plasmids contained a gene encoding eRF1. The sequence was analyzed using the results of BLASTX. A variant genetic code in which UGA encodes cysteine was used. The positions of ATG start and TAA stop codons were assigned based on the optimal amino acid sequence alignment and the maintenance of open reading frame (ORF); the sequence was free of introns. The coding sequence was 1314 bp long and encoded a 437-amino-acid protein with a calculated molecular mass of 49 109 Da. Four UGAs encoding cysteines presented in the ORF were mutagenized to UGC and the resulting DNA was cloned into pET21. When this work was completed, four Euplotes eRF1 genes were sequenced. The two eRF1s from E. aediculatus (Inagaki and Doolittle, 2001) shared 100 and 83.5% of amino acid identity with our sequence, and the two eRF1s from Euplotes octocarinutus (Liang et al., 2001) shared 90.6 and 78.7% of identity with our sequence. Conversely to E. octocarinatus, two eRF1 genes from E. aediculatus contain UGA encoding cysteine in their ORFs.
Release activity of Euplotes eRF1 in vitro
The release activity of the purified human and Euplotes eRF1 (Eu-eRF1) was measured with the three stop codons and the near-cognate tryptophan UGG codon in an in vitro RF assay. As known from a previous study (Frolova et al., 1994), human eRF1 in the given assay system responded to the three stop codons (Table I). However, under the same conditions, Eu-eRF1 responded only to UAA and UAG, but not to UGA, which encodes cysteine in Euplotes. No activity was observed with the sense UGG codon, with both factors (Table I) showing the maintenance of discriminating capacity of Eu-eRF1 and human eRF1 toward near-cognate codon. As in the case of vertebrate eRF1s (Frolova et al., 1994; Zhouravleva et al., 1995), Eu-eRF1 was active without eRF3 and GTP, confirming the conclusion that these components are not implicated in the hydrolysis of peptidyl-tRNA. With rabbit ribosomes, the release activity of Eu-eRF1 was slightly lower than that of human eRF1 (Table I). This insignificant difference could be associated with the use of a heterologous system in which the fitness of Eu-eRF1 for the mammalian ribosome could be not entirely perfect.
Table I. Release activity of Eu-eRF1 and human eRF1 in an in vitro RF assay.
| eRF1 | f[35S]Met released, c.p.m. | |||
|---|---|---|---|---|
| |
UGAA |
UAGA |
UAAA |
UGGA |
| Exp.1 Human | 5590 | 4640 | 5120 | 0 |
| Euplotes | 0 | 3050 | 4030 | 0 |
| Exp.2 Human | 9440 | 6420 | 7920 | 0 |
| Euplotes | 0 | 3200 | 4580 | 0 |
The amount of f[35S]Met released in the absence of tetraplet (background 500–800 c.p.m.) was subtracted from all values. Experiments 1 and 2 were performed with different f[35S]Met-tRNA preparations. In each of these experiments an average from three independent measurements is presented. The standard deviation of the measurements was 11%.
DISCUSSION
The human and frog eRF1s recognize all three stop codons and discriminate them from the near-cognate UGG codon without other factors and GTP in an in vitro translation termination system with rabbit reassociated ribosomal subunits (Frolova et al., 1994). However, there are no data available so far concerning the stop codon specificity of eRF1s from organisms with variant genetic codes. In these organisms, reassignment of a stop codon to a sense codon is governed either solely by a suppressor-like tRNA harboring the cognate anticodon, or by the concomitant presence of a cognate tRNA and a modified eRF1 that has lost its ability to recognize the reassigned stop codon, or even by the ribosome on its own. In the former case, due to the competition between suppressor tRNAs able to decipher stop codon and eRF1 (Drugeon et al., 1997; Le Goff et al., 1997), the synthesis of full-length proteins requires a high abundance of the suppressor tRNA and reduced amounts of eRF1. In the latter, one of the components of the ribosome must be involved in stop codon recognition.
To distinguish between these possibilities we have combined in vitro mammalian ribosomes with eRF1 from E. aediculatus in which UGA encodes cysteine and only UAA and UAG remain as termination signals. Thus, we addressed the question of whether two or three codons will be decoded in this heterologous system. The results of Table I show that UGA is not decoded as a stop codon by Eu-eRF1 in this system, demonstrating that Euplotes eRF1 has lost its ability to respond to the reassigned UGA and that reassignment of UGA is not mediated by tRNA competing with eRF1 within the ribosome. Moreover, these results also prove that the stop codon specificity is disclosed by the termination factor and not by the ribosome itself or ribosomal components. The ability of Eu-eRF1 to function in mammalian ribosome implies that in spite of large sequence divergence between eRF1s from organisms with canonical and variant genetic codes, the ribosome binding sites of eRF1s are well conserved and allow cross-reactivity of ribosomes and factors from evolutionarily distant species.
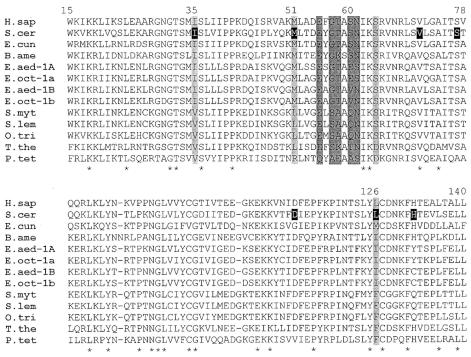
The N-terminal domain of eRF1 probably mimics the anticodon arm of tRNA (Song et al., 2000). If so, a ‘protein anticodon’ recognition model might be postulated as a way to decode stop codons (Ito et al., 2000; Nakamura et al., 2000). Bertram et al. (2000) identified mutations in yeast eRF1 that increased UAG or UGA suppression. All mutations were located in the N-terminal domain of eRF1, confirming that this domain is responsible for stop codon discrimination. Then, the challenge was to identify the amino acids of eRF1 interacting with the stop codon similarly to what was demonstrated for bacterial class-1 RFs (Ito et al., 2000). The strategy was based on the data of the eRF1 crystal structure and on the comparison of numerous eRF1 sequences from highly divergent eukaryotic species including those from ciliates (Figure 1). It was assumed that the changes in stop codon usage were mediated by the amino acids of eRF1 interacting with stop codons. The conserved NIKS motif (positions 61–64) was thought to be involved in stop codon recognition (Knight and Landweber, 2000). This assumption was abandoned when in eRF1 sequences from Stylonichia and Oxytricha, two ciliates using the same variant genetic code as Tetrahymena thermophila, the NIKS (not NIKD as in T. thermophila) motif was identified (Lozupone et al., 2001).
Fig. 1. Alignment of the N-terminal domain of eRF1 from human, S. cerevisiae, Encephalitozoon cuniculi and various ciliate species. Alignment has been performed using the CLUSTAL_W program (Thompson et al., 1994). Numbering is according to the human eRF1 sequence. Sequences were extracted from DDBJ/EMBL/GenBank [H.sap, Homo sapiens, P46055; S.cer, Saccharomyces cerevisiae, CAA51935; E.cun, Encephalitozoon cuniculi, 13560073; B.ame, Blepharisma americanum, AF317831; E.aed-1A, Euplotes aediculatus-1A, AF298831; E.oct-1a, Euplotes octocarinatus-1a, AJ272501; E.aed-1B, Euplotes aediculatus-1B, AF298832, AF334757 (this work); E.oct-1b, Euplotes octocarinatus-1b, AF245454; S.myt, Stylonychia mytilus, AF317833; S.lem, Stylonychia lemnae, AF317834; O.tri, Oxytricha trifallax, AF298830; T.the, Tetrahymena thermophila, AF298833; P.tet, Paramecium tetraurelia, AF149035 and AF149036]. The bottom line indicates the residues conserved in all sequences presented. Residues in black were identified by Bertram et al. (2000) in yeast mutants exhibiting either a UGA suppressor phenotype (mutations I35F, V71I, L126V and H131R) or a UAG suppressor phenotype (mutations M51I, S77F and D113G). Residues shaded in gray were supposed to be involved in stop codon recognition: dark gray (Liang et al., 2001; Muramatsu et al., 2001), light gray (Knight and Landweber, 2000; Lehman, 2001; Lozupone et al., 2001).
In the presumed ‘protein anticodon’ region, some residues in eRFs are conserved in all eukaryotes except ciliates, where UAR codons encode glutamine, i.e. I35V, M51L and L126F (Lozupone et al., 2001), and L126I in Euplotes, where UGA encodes cysteine (Figure 1). The fact that residues 35 and 126 are close to each other in the spatial structure (Song et al., 2000) is consistent with their potential involvement in stop codon recognition (Lehman, 2001). However, the eRF1 sequence of the microsporidia Encephalitozoon cuniculi, which uses a canonical genetic code, has methionine at position 126 (Figure 1). How can this change be accommodated with the model described above?
Muramatsu et al. (2001) proposed that E55, G57/T58 and S60/N61 recognize the first, second and third base of the stop codons, respectively. We have sequenced the eRF1 gene from P. tetraurelia (S. Kervestin, unpublished), and alignment of this sequence with the other eRF1s (Figure 1) shows that the divergences observed at positions 55, 57, 58 and 60 do not fit well with this model. For instance, in Tetrahymena, Stylonichia and Oxytricha, position 57 is occupied by serine, whereas Paramecium eRF1 has alanine in this position. These substitutions are not correlated with codon reassignments in ciliate eRF1s in comparison with eRF1s from organisms with universal genetic code. Ciliate eRF1s exhibit a high evolutionary rate, which is reflected in an increased number of variable positions (Inagaki and Doolittle, 2001; David Moreira, personal communication). Thus, it can not be ruled out that either residues of eRF1 involved in stop codon recognition may occupy different variable positions in ciliate sequences, or variable positions modify the functional constraints at a few fixed positions. Additional eRF1 sequences, particularly from ciliate species, could help to select amino acids implicated in stop codon recognition.
Our data on inability of Eu-RF1 to respond to UGA strongly support the assumptions that (i) in all ciliates with variant genetic codes, eRF1 does not respond to reassigned stop codons; (ii) in these organisms, modifications of eRF1 amino acid sequences are responsible for the pattern of stop codon recognition; and (iii) presumably, ribosomes from ciliates possess the same ability to support termination of the three stop codons as ribosomes from organisms with conventional genetic code.
METHODS
Plasmids, library screening, gene manipulations, DNA sequencing and PCR amplification. The E. aediculatus eRF1 gene (Figure 2) was isolated from a macronuclear DNA pUC18 library provided by A. Baroin-Tourancheau (Université Paris-Sud). Because eRF1s of ciliates are highly divergent from other eukaryotes, the coding sequence of eRF1 from P. tetraurelia (S. Kervestin, unpublished) was used as a probe for library screening. Library screening and gene manipulations were carried out by the standard procedures (Sambrook et al., 1989). Bacterial colonies transferred on Hybond N+ filters (Amersham-Pharmacia Biotech.) were detected by hybridization at non-stringent conditions (1 h at 60°C followed by slow cooling to 30°C) and washing in 2× SSC (0.3 M NaCl, 30 mM sodium citrate) plus 0.1% SDS at 35°C. The entire nucleotide sequence of inserts was determined on both strands. PCR amplifications of DNA were carried out in 25 µl reaction mixtures containing 1 ng of plasmid DNA, 100 pmol of each primer, 200 µM each deoxynucleoside triphosphate, 1× commercial PCR buffer and 2.5 U of Pwo DNA polymerase (Roche). Amplifications were run for 20 cycles (94°C, 30 s; 50°C, 30 s; 72°C, 1 min) in a thermocycler.
Fig. 2. Euplotes aediculatus stained with an anti-tubulin antibody. Photograph courtesy of Anne Fleury, University of Paris, France.
Site-directed mutagenesis. This was performed to transform four in-frame UGA codons of the E. aediculatus eRF1 gene into the canonical cysteine UGC codon using the Transformer site-directed mutagenesis kit (Clontech). The resulting modified eRF1 DNA (from the AUG initiation codon to the last codon) was then amplified by PCR with appropriate oligonucleotides containing restriction sites (NdeI and HindIII) for direct cloning into pET21b (Novagen). The final construct, named pET-Eu-RF1-His6, contained E. aediculatus eRF1 ORF under control of T7 promotor.
Expression and purification of eRF1s. cDNA encoding the full-length human eRF1 was inserted into NdeI–XhoI sites of pET23b(+) (Novagen). Eu-eRF1 and human eRF1 containing a His-tag at the C-terminus were expressed in Escherichia coli strain BL21(DE3), and purified using Ni-NTA resin, Superflow (Qiagen), as described (Frolova et al., 1994, 2000).
Ribosomes. Rabbit reticulocyte ribosomal subunits were kindly provided by P. Simonenko (Institute of Protein Research, Pushchino). 80S ribosomes washed with 0.5 M KCl were treated with puromycin and GTP for dissociation into subunits, which were subsequently resolved by centrifugation in a 10–25% (w/v) sucrose gradient containing 0.3 M KCl, 3 mM MgCl2, 1 mM dithiothreitol and 20 mM Tris–HCl pH 7.6. Before addition to the incubation mixtures, the subunits were combined in an equimolar ratio.
In vitro RF assay. The eRF1 activity was measured as described (Caskey et al., 1974; Frolova et al., 1994) at saturation levels (50 µM) of one out of the three stop-codon-containing tetraplets or UGGA tetraplet containing codon for tryptophan. The incubation mixture (25 µl) contained 20 mM Tris–HCl pH 7.5, 15 mM MgCl2, 8 mM NH4Cl, 1.5 pmol f[35S]-Met-tRNAfMet–AUG–ribosome complex and eRF1 (0.2–0.3 µg). AUG and ribotetraplets were synthesized by A. Veniaminova and M. Ryabkova (Institute of Biorganic Chemistry, Novosibirsk).
Acknowledgments
ACKNOWLEDGEMENTS
The initial step of Eu-eRF1 purification and characterization was performed in Rennes 1 University and we are grateful to M. Philippe for support and encouragement. We thank A. Baroin-Tourancheau for the E. aediculatus library and A. Seit Nebi for the pET23b-human eRF1 plasmid. O.J.-J. was supported by grant No. 5511 from the Association pour la Recherche sur le Cancer. L.K. and L.F. were supported by Human Frontier Science Program (grant 96-032), by Russian Foundation for Basic Research and by Program of Support for leading Russian scientific schools.
REFERENCES
- Altschul S.F. and Koonin, E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- Arkov A.L. and Murgola, E.J. (1999) Ribosomal RNAs in translation termination: facts and hypotheses. Biochemistry (Mosc.), 64, 1354–1359. [PubMed] [Google Scholar]
- Baroin-Tourancheau A., Tsao, N., Klobutcher, L.A., Pearlman, R.E. and Adoutte, A. (1995) Genetic code deviations in the ciliates: evidence for multiple and independent events. EMBO J., 14, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Bell, H.A., Ritchie, D.W., Fullerton, G. and Stansfield, I. (2000) Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M. and Tate, W.P. (1994) Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem., 269, 33164–33170. [PubMed] [Google Scholar]
- Caskey C.T., Beadet, A.L. and Tate, W.P. (1974) Mammalian release factor: in vitro assay and purification. Methods Enzymol., 30, 293–303. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Frolova, L., Kisselev, L. and Favre, A. (2001) The polypeptide chain release factor eRF1 specifically contacts the sUGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem., 268, 2896–2904. [DOI] [PubMed] [Google Scholar]
- Drugeon G., Jean-Jean, O., Frolova, L., Le Goff, X., Philippe, M., Kisselev, L. and Haenni, A.L. (1997) Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res., 25, 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L., Merkulova, T.I. and Kisselev, L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y. and Doolittle, W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno, M. and Nakamura, Y. (2000) A tripeptide anticodon deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Ivanov V.I., Beniaminov, A.D., Miheev, A. and Minyat, E.E. (2001) Why terminator tRNAs have not been found? Because they are hidden in large ribosomal RNA. Mol. Biol. (Mosc.), 35, 987–998. [PubMed] [Google Scholar]
- Karamyshev A.L., Ito, K. and Nakamura, Y. (1999) Polypeptide release factor eRF1 from Tetrahymena thermophila: cDNA cloning, purification and complex formation with yeast eRF3. FEBS Lett., 457, 483–488. [DOI] [PubMed] [Google Scholar]
- Kisselev L. and Buckingham, R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- Knight R.D. and Landweber, L.F. (2000) The early evolution of the genetic code. Cell, 101, 569–572. [DOI] [PubMed] [Google Scholar]
- Le Goff X., Philippe, M. and Jean-Jean, O. (1997) Overexpression of human release factor 1 (eRF1) alone has an antisuppressor effect in human cells. Mol. Cell. Biol., 17, 3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman N. (2001) Please release me, genetic code. Curr. Biol., 11, R63–R66. [DOI] [PubMed] [Google Scholar]
- Liang A., Brunen-Nieweler, C., Muramatsu, T., Kuchino, Y., Beier, H. and Heckmann, K. (2001) The ciliate Euplotes octocarinatus expresses two polypeptide release factors of the type eRF1. Gene, 262, 161–168. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Knight, R.D. and Landweber, L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Heckmann, K., Kitanaka, C. and Kuchino, Y. (2001) Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett., 488, 105–109. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito, K. and Ehrenberg, M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brimacombe, R. and Tate, W.P. (1997) Decoding the translational termination signal: the polypeptide chain release factor in Escherichia coli crosslinks to the base following the stop codon. RNA, 3, 974–982. [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Song H., Mugnier, P., Webb, H.M., Evans, D.R., Tuite, M.F., Hemmings, B.A. and Barford, D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova, L., Le Goff, X., Le Guellec, R., Inge-Vechtomov, S., Kisselev, L. and Philippe, M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]