Abstract
The large subunit of ribosomes in Bacillus subtilis was tagged by generation of a fusion of ribosomal protein L1 to blue fluorescent protein (BFP). The fusion was fully active and localized around the nucleoids, predominantly close to the cell poles, in growing cells. However, in stationary phase cells, and in growing cells treated with rifampicin, L1-BFP was distributed throughout the cells, in contrast to cells treated with chloramphenicol, in which ribosomes still localized around nucleoids. These data show that specific localization of ribosomes is not due to nucleoid exclusion, but is a dynamic process due to active synthesis of RNA. Dual labelling of ribosomes and cold shock proteins (CSPs) tagged with green fluorescent protein (GFP) revealed colocalization of both protein classes. CSPs are implicated in coupling of transcription with translation and may bridge the spatial separation of ribosomes and nucleoid-associated RNA polymerase.
INTRODUCTION
Bacteria have traditionally been viewed as cells having a rigid cell envelope but no internal structure, except for an organization of chromosomal DNA into nucleoids that occupy the central part of the cell. This view has recently been thoroughly revised through investigation of subcellular localization of bacterial proteins. Several histidine kinases localize to foci at the cell poles, and even redistribute throughout the membrane during the cell cycle (Shapiro and Losick, 2000). Proteins involved in cell division assemble in a ring-like structure at mid cell (Lutkenhaus, 1998), and Min proteins that are important in mid cell positioning of division proteins oscillate from one pole to the other within seconds (Raskin and de Boer, 1999). These findings show that many bacterial proteins have a specific address within the cell. Moreover, bacterial chromosomes have been shown to possess a conserved layout with origin regions of replication separating rapidly towards cell poles, while termination regions localize to mid cell positions (Levin and Grossman, 1998). Chromosome sites between origins and termini localize between cell poles and mid cell, suggesting that the bacterial chromosome is roughly folded according to its physical structure (Teleman et al., 1998; Niki et al., 2000). Partition proteins localize to sites surrounding origins of replication, and show a dynamic movement towards cell poles upon chromosome segregation similar to chromosome origins or other sites on the chromosome (Levin and Grossman, 1998), suggesting that a mitotic-like apparatus segregates chromosomes in bacteria. Recently, a further and more unexpected level of organization in Bacillus subtilis and Escherichia coli was found. RNA polymerase appears to be mainly located on the nucleoid, while proteins involved in translation localize to cytosolic spaces around nucleoids, and predominantly towards cell poles (Azam et al., 2000; Lewis et al., 2000). These findings suggest that transcription and translation occur generally at different sites within bacterial cells. It has been assumed that separation of both processes is due to nucleoid exclusion of ribosomes. On the other hand, many reports have shown that transcription and translation are coupled in bacteria (Miller et al., 1970; Robinow and Kellenberger, 1994), so there appears to be a necessity to bridge the spatial separation in vivo.
A conserved family of proteins, cold shock proteins (CSPs), has been suggested to play a role in initiation of translation, or to stabilize open complex formation during transcription. CSPs exist in multigene families in many bacteria (Graumann and Marahiel, 1998; Woldringh et al., 1995), bind cooperatively to ssDNA and to RNA with little sequence specificity (Graumann and Marahiel, 1994), and destabilize secondary RNA structures in vitro (Jiang et al., 1997). In B. subtilis, CSPs perform an essential function at optimal and low growth temperature, and have a strong influence on global protein synthesis (Graumann et al., 1997). Escherichia coli CspE has been found to be associated with short transcripts in vivo (Hanna and Liu, 1998), supporting the view that CSPs may bind to nascent mRNA to prevent formation of secondary structures, which impair binding of ribosomes and thus initiation of translation (Gualerzi and Pon, 1990). Thereby, CSPs may couple mRNA synthesis with initiation of translation.
In this report, we show that a fusion of a new variant of green fluorescent protein (GFP), blue fluorescent protein (BFP), to ribosomal protein L1 localizes predominantly towards the cell poles. We found that in stationary phase and after inhibition of transcription, L1-BFP was distributed throughout cells. Thus, specific localization of ribosomes is a dynamic process in B. subtilis and is established in growing cells. We further show by means of dual labelling in vivo that DNA significantly overlaps with ribosomes, and that CSPs and ribosomes localize in an indistinguishable pattern.
RESULTS AND DISCUSSION
Localization of L1-BFP to cytosolic spaces surrounding nucleoids depends on transcription but not on translation
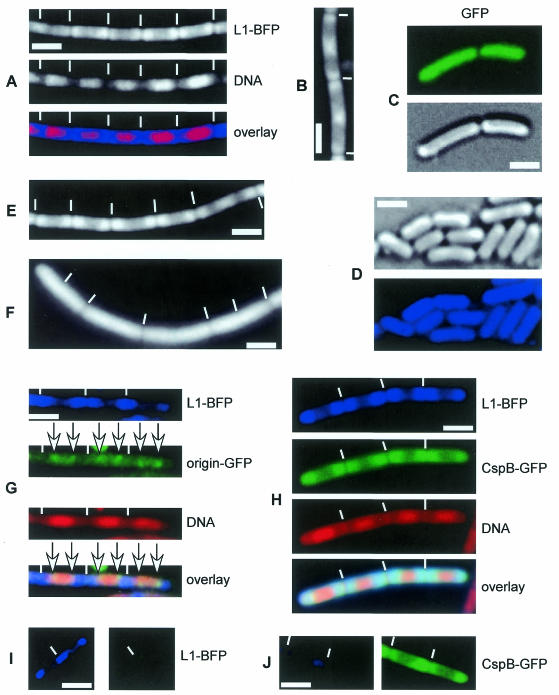
A fragment containing the 3′ half of rplA generated by PCR was fused to bfp and was integrated by single crossover into the B. subtilis chromosome. Cells of strain JM1 (rplA-bfp) grew with a generation time comparable to wild-type cells, showing that the fusion is functional. We noted that the cells grow in longer chains than the parent strain, for which we have no explanation. Blue fluorescent signals in growing JM1 cells were weak compared with green fluorescence of GFP, e.g. seen with CspB-GFP, which is also an abundant protein, indicating that emission of BFP2 is considerably lower than that of GFP. However, L1-BFP could be visualized to localize around nucleoids (most predominantly close to the poles) in growing cells (Figure 1A), in a manner similar to a fusion of S2 to GFP, reported by Lewis et al. (2000). In contrast, fluorescence of GFP alone is distributed throughout the cell (Figure 1C), likewise to non-functional fusions of GFP to several proteins in B. subtilis (Lewis et al., 2000), showing that localization of L1-BFP is not a general property of soluble proteins. Although weak staining appeared to be present within nucleoid spaces, deconvolution of Z-sections taken through live cells revealed that nucleoid-containing spaces in B. subtilis are almost completely devoid of L1 (Figure 1B). In cells with two separated nucleoids, a band of ribosomes was visible at the cell center (data not shown). These findings support the idea that generally ribosomes do not colocalize with RNA polymerase, as shown by Lewis et al. (2000), although there must be an area of overlap since transcription is coupled with translation in bacteria (e.g. transcriptional attenuation). Most likely, this overlap takes place at the periphery of the nucleoid and at DNA strands that are pulled towards membranes due to cotranslational membrane insertion of membrane and export proteins (Weber et al., 2001).
Fig. 1. Fluorescence microscopy of Bacillus subtilis cells. Blue panels show localization of L1-BFP, red panels shows SYTO59 DNA stain. Note that the gene encoding L1 is rplA. (A) Strain JM1 (rplA-bfp) growing at mid-exponential phase. (B) Deconvolution of Z-series of images taken through growing JM1 cells. (C) Growing cells of strain MW1 (amy::pxylgfp) expressing GFP only; upper panel, GFP; lower panel, Nomarski DIC. (D) JM1 cells during stationary phase; upper panel, Nomarski; lower panel, L1-BFP. (E) JM1 cells treated with chloramphenicol (inhibitor of translation) or (F) rifampicin (inhibitor of transcription) for 30 min. (G) Strain JM5 (rplA-bfp, origin GFP tag) during exponential growth. (H) Strain JM4 (rplA-bfp, cspB-gfp) expressing L1-BFP and CspB-GFP during exponential growth. (I) JM1 cells in blue or green filter. (J) MW2 (cspB-gfp) in blue or green filter sets. White lines show septa between cells, white bars indicate 2 µm.
Lewis et al. (2000) observed that in cells entering stationary phase, polar localization of ribosomes is still apparent but becomes more uniform. In contrast, we found that in stationary phase cells, in which nucleoids are more condensed than in growing cells (data not shown), L1-BFP fluorescence was distributed throughout the cytosol (Figure 1D). It is possible that an S2-GFP fusion investigated by Lewis et al. (2000) delays adaptation to stationary phase. Therefore, specific localization of ribosomes is not due to nucleoid exclusion. To investigate whether transcription or translation or both processes are needed for specific localization of L1-BFP in growing cells, we treated exponentially growing cells with rifampicin, an inhibitor of transcription, or with chloramphenicol or puromycin, two translational inhibitors. While addition of chloramphenicol or of puromycin retained localization of L1-BFP around nucleoids (Figure 1E) that were more condensed than in non-treated cells (data not shown), inhibition of transcription led to localization of the protein fusion throughout the cell (Figure 1F). In the latter cells, nucleoids decondensed slightly compared with control cells (data not shown). Addition of ethanol or methanol in a concentration used for chloramphenicol or rifampicin, respectively, did not detectably affect localization of L1-BFP (data not shown). Interestingly, a functional fusion of CspB to GFP localizes in a similar manner to L1-BFP, likewise dependent on active transcription (Weber et al., 2001). This observation argues against an artefact of the localization of the L1 fusion used here, and suggests that specific localization of L1 may apply to all proteins involved in translation.
Dual labelling of proteins in B. subtilis using GFP and BFP reveals partial overlap of origin regions of chromosomes with ribosomes
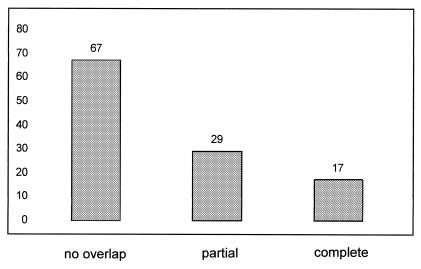
We wished to employ the BFP tag as a tool for dual labelling of proteins in live cells. To test BFP, rplA-bfp was moved into strain AT63 (Webb et al., 1998), which carries a tandem array of lactose operators inserted close to the origin of replication decorated with a GFP fusion to lactose repressor (gfp-lacI at veg locus). The resulting strain JM5 (rplA-bfp, gfp-lacI, lacO cassette at 359°) showed typical bipolar position of origin regions to the edges of nucleoids in the green filter set (Figure 1G, green panel and overlay), which was undetectable in the blue filter set, in which a normal pattern of L1-BFP localization around nucleoids was apparent (Figure 1G, blue panel). Conversely, L1-BFP was undetectable in the green filter (Figure 1I). Thus, BFP allows separate detection of two different proteins in B. subtilis. Analysis of overlays of origin signals and ribosomes (Figure 1G) showed that while origin signals generally localized within areas devoid of ribosomes (Figure 2, no overlap), in a significant number of cells (14% of 113 origin signals analysed) origins were found to be present within the polar area occupied by ribosomes (Figure 2, overlap). Frequently, origins were found to partially overlap with ribosomes in vivo (Figure 2, partial overlap). This finding shows that while the bulk of DNA lies within nucleoids, individual regions can lie outside of the regions stained by DNA selective probes such as DAPI or SYTO59. Thus, a significant overlap of DNA and ribosomes exists within live cells, indicating that a fraction of RNA polymerase may be present in spaces where ribosomes are abundant. Approximately 10–20% of transcription is devoted to mRNA, while remaining transcription leads to synthesis of stable RNA. Transcription of mRNA could occur at the boundary between nucleoids and surrounding spaces, while synthesis of stable mRNA might occur within nucleoids. This is supported by the data of Lewis et al. (2000), who observed accumulation of RNA polymerase within nucleoids at high growth rate, when the level of synthesis of stable RNA is especially high.
Fig. 2. Chart showing number of chromosome origins found not coincident, partially overlapping or completely overlapping with spaces occupied by ribosomes.
Colocalization of ribosomes and CSPs
We combined a fusion of the major cold shock protein, CspB, to GFP with L1-BFP, generating strain JM4 (rplA-bfp, cspB-gfp), to visualize localization of CSPs and ribosomes in the same cell. CSPs were also shown to localize to sites surrounding nucleoids in B. subtilis (Weber et al., 2001). Figure 1H shows that both blue and green fluorescence show a similar pattern in JM4 cells. A caveat in this experiment is the fact that fluorescence of CspB-GFP is very strong, and can be faintly observed in the blue channel in cells with a high signal of CspB-GFP. However, in the bulk of cells with average green fluorescence, the signal in the blue channel is much lower than that of L1-BFP (Figure 1J). Figure 1H therefore shows cells with a medium level of green fluorescence due to CspB-GFP, in which the signal in the blue channel is almost exclusively due to L1-BFP. We therefore conclude that CSPs localize to cytosolic spaces in which ribosomes are present. This finding indicates a main function of CSPs in translation initiation, rather than in transcription. If bulk transcription takes place on the periphery of the nucleoid, CSPs could bind to nascent mRNA until ribosomes bind to the transcript and displace CSPs, whose affinity to mRNA is only moderate (Graumann et al., 1997; Jiang et al., 1997). In support of this model, we noticed that overlays of L1-BFP, CspB-GFP and DNA stained in red frequently show a faint yellow zone at the border between nucleoids and surrounding spaces (Figure 1H). This suggests the existence of a space in which CSPs and DNA overlap (red and green), but which is devoid of ribosomes.
In this report, we provide three principal findings. We show that specific localization of ribosomes to sites surrounding nucleoids is not effected by nucleoid exclusion, but it is an active process established in growing cells due to ongoing transcription. Possibly, newly synthesized mRNA rapidly loops out of the nucleoid, such that ribosomes are present at the cellular sites of their binding targets. However, ribosomes and DNA are not completely separated, but overlap significantly, which we observed through dual visualization of origin sites of chromosomes and ribosomes. Coupling of transcription and translation may therefore occur at the interphase between nucleoids and surrounding spaces. Our third observation of colocalization of ribosomes and CSPs supports a main function of CSPs in translation initiation, possibly through prevention of secondary structure formation in mRNA.
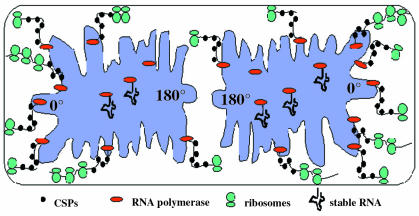
Taken together, all available data suggest a model in which in growing cells, transcription of mRNA takes place at the nucleoid periphery, while synthesis of stable RNA occupies central spaces of nucleoids (Figure 3). CSPs may bind to nascent mRNA at the nucleoid border, and ensure proper translation initiation of mRNA that is expelled from the nucleoid. Ribosomes bind to linear mRNA located between nucleoids and membranes, and predominantly close to cell poles, due to diffusion of mRNA to these sites. Therefore, chromosome structure may strongly influence transcription, if an altered organization of nucleoids reduces the flexibility of DNA and interferes with a proposed positioning of transcribed genes to the nucleoid periphery.
Fig. 3. Model for coupling of spatially separated sites of transcription and translation in bacteria.
METHODS
Bacterial strains and growth conditions. Plasmid cloning was performed using E. coli strain XL1-Blue (Stratagene, CA) grown at 37°C. Bacillus subtilis strain PY79 was used as parent strain. Bacillus cells were grown at 25°C in LB rich medium and in S750 minimal medium (Jaacks et al., 1989) for microscopy. For inhibition studies, chloramphenicol was added at 200 µg/ml, puromycin at 100 µg/ml and rifampicin at 200 µg/ml.
Construction of plasmids and strains. The pBFP2 vector was obtained from Clontech (CA). A 3′ fragment of the rplA gene was amplified from chromosomal DNA of PY79 using primers 5′-CGGATCCGTTCTCGTTTCGCAAAAGG-3′ (forward) and 5′GGGGTACCCCACCTTTTACGTTAAAAGTTGAAGAAGAGTC-3′ (reverse). The amplified product was digested with BamHI and KpnI, purified and ligated into pBFP2 vector. A spectinomycin cassette from pDG1726 was cloned in the resulting vector pBr1 using HindIII and BamHI restriction sites. Resulting plasmid pBrsp was transformed into PY79 using LB agar containing spectinomycin (100 µg/ml) to obtain strain JM1 (rplA-bfp). Single crossover integration of pBrsp into rplA was verified by PCR using primers 5′-GCTTACGACGTCTCTGAAG-3′ and 5′-CAGGATCCGTTTGTATAGTTCATCCATGCC-3′, which amplify complete rplA fused to bfp. Strain JM4 (rplA-bfp, cspB-gfp) was generated by transformation of JM1 with chromosomal DNA from MW2 (cspB-gfp) selecting for spectinomycin and Mls (5 µg/ml erythromycin, 25 µg/ml lincomycin) resistance. Strain AT63 (lacO cassette at 359°, pveg-gfp-lacI) was transformed with chromosomal DNA from JM1, selecting for spectinomycin, Mls and chloramphenicol (5 mg/ml) resistance to generate strain JM5 (rplA-bfp, lacO cassette at 359°, pveg-gfp-lacI).
Microscopy. For image generation, cells were mounted on agarose pads containing S750 medium as described in Webb et al. (1998). Fluorescence microscopy was performed on an Olympus AX70 system using a MicroMax CCD camera. Olympus filters MNU (blue) and MNIV (violet) were used for BFP (MNU was necessary for distinction between blue and green labelling because of GFP leaking into the violet spectrum), WIB for GFP and SGA for red labelling. SYTO59 (Molecular Probes, The Netherlands) was used for red cell permeable DNA stain at 100 nM. 3D image reconstruction and deconvolution were carried out using Metamorph 4.0.
Acknowledgments
ACKNOWLEDGEMENTS
This work has been supported by the Deutsche Forschungsgemeinschaft (Emmy Noether Programm and Sonderforschungsbereich 395).
REFERENCES
- Azam T.A., Hiraga, S. and Ishihama, A. (2000) Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells, 5, 613–626. [DOI] [PubMed] [Google Scholar]
- Graumann P. and Marahiel, M.A. (1994) The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG and CCAAT sequences in single stranded oligonucleotides. FEBS Lett., 338, 157–160. [DOI] [PubMed] [Google Scholar]
- Graumann P.L. and Marahiel, M.A. (1998) A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci., 23, 286–290. [DOI] [PubMed] [Google Scholar]
- Graumann P., Wendrich, T.M., Weber, M.H.W., Schröder, K. and Marahiel, M.A. (1997) A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol., 25, 741–756. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon, C.L. (1990) Initiation of mRNA translation in procaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Hanna M.M. and Liu, K. (1998) Nascent RNA in transcription complexes interacts with CspE, a small protein in E. coli implicated in chromatin condensation. J. Mol. Biol., 282, 227–239. [DOI] [PubMed] [Google Scholar]
- Jaacks K.J., Healy, J., Losick, R. and Grossman, A.D. (1989) Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol., 171, 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Hou, Y. and Inouye, M. (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem., 272, 196–202. [DOI] [PubMed] [Google Scholar]
- Levin P.A. and Grossman, A.D. (1998) Cell cycle: the bacterial approach to coordination. Curr. Biol., 8, R28–R31. [DOI] [PubMed] [Google Scholar]
- Lewis P.J., Thaker, S.D. and Errington, J. (2000) Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J., 19, 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. (1998) The regulation of bacterial cell division: a time and place for it. Curr. Opin. Microbiol., 1, 210–215. [DOI] [PubMed] [Google Scholar]
- Miller O.L. Jr,, Hamkalo, B.A. and Thomas, C.A., Jr (1970) Visualization of bacterial genes in action. Science, 169, 392–395. [DOI] [PubMed] [Google Scholar]
- Niki H., Yamaichi, Y. and Hiraga, S. (2000) Dynamic organization of chromosomal DNA in Escherichia coli.Genes Dev., 14, 212–223. [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer, P.A. (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C. and Kellenberger, E. (1994) The bacterial nucleoid revisited. Microbiol. Rev., 58, 211–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. and Losick, R. (2000) Dynamic spatial regulation in the bacterial cell. Cell, 100, 89–98. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Graumann, P.L., Lin, D.C.H., Grossman, A.D. and Losick, R. (1998) Chromosome arrangement within a bacterium. Curr. Biol., 8, 1102–1109. [DOI] [PubMed] [Google Scholar]
- Webb C.D., Graumann, P.L., Kahana, J., Teleman, A.A., Silver, P. and Losick, R. (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol., 28, 883–892. [DOI] [PubMed] [Google Scholar]
- Weber M.H.W., Volkov, A., Fricke, I., Marahiel, M.A. and Graumann, P.L. (2001) Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C.L., Jensen, P.R. and Westerhoff, H.V. (1995) Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett., 131, 235–242. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Fang, L. and Inouye, M. (1998) The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol., 27, 247–255. [DOI] [PubMed] [Google Scholar]