Abstract
Biogenesis of Fe/S clusters involves a number of essential mitochondrial proteins. Here, we identify the essential Erv1p of Saccharomyces cerevisia mitochondria as a novel component that is specifically required for the maturation of Fe/S proteins in the cytosol, but not in mitochondria. Furthermore, Erv1p was found to be important for cellular iron homeostasis. The homologous mammalian protein ALR (‘augmenter of liver regeneration’), also termed hepatopoietin, can functionally replace defects in Erv1p and thus represents the mammalian orthologue of yeast Erv1p. Previously, a fragment of ALR was reported to exhibit an activity as an extracellular hepatotrophic growth factor. Both Erv1p and full-length ALR are located in the mitochondrial intermembrane space and represent the first components of this compartment with a role in the biogenesis of cytosolic Fe/S proteins. It is likely that Erv1p/ALR operates downstream of the mitochondrial ABC transporter Atm1p/ABC7/Sta1, which also executes a specific task in this essential biochemical process.
INTRODUCTION
Mitochondria perform a central task in the biogenesis of cellular iron–sulfur (Fe/S) proteins (reviewed by Craig et al., 1999; Lill et al., 1999; Lill and Kispal, 2000). They harbour a complex apparatus termed ISC (iron–sulfur cluster) assembly machinery consisting of some ten proteins. Many of these proteins resemble bacterial proteins encoded by the isc operons (Zheng et al., 1998), indicating a bacterial origin of this crucial mitochondrial process. The ISC assembly machinery is required for the biogenesis of Fe/S proteins within mitochondria including aconitase and subunits of complexes I, II and III of the respiratory chain. In addition, mitochondrial ISC assembly components are involved in the maturation of cytosolic Fe/S proteins such as isopropyl malate isomerase Leu1p (Kispal et al., 1999; Kaut et al., 2000; Lange et al., 2000; Li et al., 2001). Minute amounts of the ISC assembly component Nfs1p are present in the nucleus (Nakai et al., 2001). However, only the mitochondrial, but not the extra-mitochondrial form of Nfs1p, supports Fe/S protein assembly in the cytosol (Kispal et al., 1999). To date, only one component with a specific function in cytosolic Fe/S protein maturation has been identified, the Saccharomyces cerevisiae ABC transporter Atm1p of the mitochondrial inner membrane. The protein is present in all eukaryotes including man (ABC7; Bekri et al., 2000) and plants (Sta1; Kushnir et al., 2001), and has been proposed to export from the mitochondrial matrix a component required for assembly of Fe/S proteins in the cytosol.
Here, we report on the identification of Erv1p as a novel component of the mitochondrial intermembrane space that is specifically required for the maturation of Fe/S proteins outside mitochondria. Furthermore, the human homologous protein ALR (‘augmenter of liver regeneration’; Hagiya et al., 1994; Giorda et al., 1996), also termed hepatopoietin (Li et al., 2000), was demonstrated as the functional orthologue of yeast Erv1p.
RESULTS
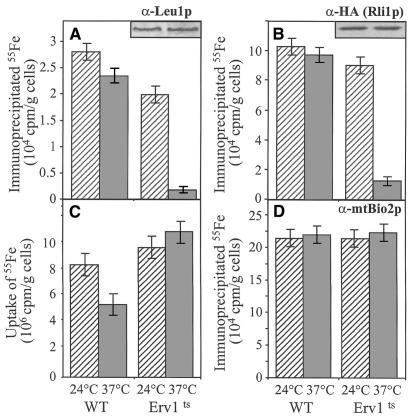
To investigate the function of Erv1p we took advantage of a temperature-sensitive erv1 mutant strain (termed Erv1ts; Lisowsky, 1992). First, we analysed in vivo the maturation of the cytosolic Fe/S proteins Leu1p (Kohlhaw, 1988) and Rli1p (containing an N-terminal ferredoxin domain; Matsubara and Saeki, 1992; Kispal et al., 1999). Erv1ts cells were grown at 24°C overnight and a fraction was shifted to the non-permissive temperature (37°C) for 5 h. Cells were labelled with radioactive 55Fe for 1 h in the presence of ascorbate, and lysates were prepared by breaking the cells with glass beads. Incorporation of 55Fe into Leu1p and HA-epitope-tagged Rli1p was followed by immunoprecipitation with specific antibodies. The amount of radiolabel associated with the immuno-beads was quantitated by scintillation counting and served as a measure for the incorporation of an Fe/S cluster. Upon inactivation of Erv1p at the non-permissive temperature, a strong impairment in the incorporation of 55Fe into both Leu1p and Rli1p was observed in comparison with wild-type cells or Erv1ts cells grown at 24°C (Figure 1A and B). The defective formation of the Fe/S clusters in Leu1p and Rli1p was not the result of a reduced synthesis of the polypeptide chains, as similar amounts of these proteins were detected by immunostaining before and after the inactivation of Erv1p (inserts). Likewise, defects in iron uptake into the cell cannot account for the Leu1p maturation defect, since the content of 55Fe in cell lysates was even higher upon inactivation of Erv1p (Figure 1C). Together, these findings demonstrate a severe defect in the maturation of cytosolic Fe/S proteins upon inactivation of Erv1p.
Fig. 1. Inactivation of Erv1p causes a specific defect in the maturation of cytosolic Fe/S protein. Wild-type (WT) and Erv1ts cells with or without a plasmid encoding an HA-epitope-tagged Rli1p were grown overnight at 24°C in iron-free minimal media, and half of them were shifted to 37°C for 5 h. Cells were radiolabelled with (55Fe) iron chloride in the presence of 1 mM ascorbate (Kispal et al., 1999) for 1 h at 24 or 37°C, cell lysates were prepared, and the uptake of 55Fe into the cells was quantitated by liquid scintillation counting (C). Immunoprecipitation was performed using antisera directed against cytosolic Leu1p (A), the HA-epitope (B) or mitochondrial Bio2p (D), and co-precipitated 55Fe was estimated by liquid scintillation counting. The background signal obtained for immunoprecipitation performed with preimmune serum (<2 × 103 c.p.m./g cells) was subtracted. Experiments were performed at least four times. The bars indicate the standard errors.
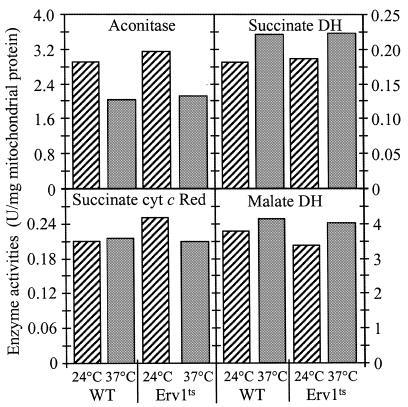
Does Erv1p, like the ISC assembly components, also play a role in the biogenesis of mitochondrial Fe/S proteins? Mitochondria were isolated from Erv1ts cells grown at 24 or 37°C, and the activities of several Fe/S cluster-containing enzymes were measured. No significant changes in the activities of aconitase, succinate dehydrogenase or cytochrome c reductase were detected upon inactivation of Erv1p (Figure 2). Furthermore, the de novo incorporation of an Fe/S cluster into the mitochondrial Fe/S protein Bio2p (biotin synthase) was measured by using the radiolabelling and immunoprecipitation assay presented above. The absence of functional Erv1p did not lead to any reduction in the maturation of Bio2p (Figure 1D). Taken together, these results indicate the importance of Erv1p for the biogenesis of cytosolic, but not of mitochondrial Fe/S proteins.
Fig. 2. Erv1p function is not required for normal activity of mitochondrial Fe/S proteins. Mitochondria were isolated from wild-type (WT) and Erv1ts cells that were grown for 5 h in lactate media containing 0.1% glucose at 24 or 37°C. Enzyme activities of the indicated Fe/S cluster-containing proteins and of malate dehydrogenase were measured. DH, dehydrogenase; Succinate cyt c Red, succinate-cytochrome c reductase.
Yeast and human cells impaired in the biogenesis of cellular Fe/S proteins were shown previously to accumulate high levels of iron within mitochondria (reviewed in Lill and Kispal, 2000). We therefore measured the amount of ‘free’ (i.e. non-heme, non-Fe/S) iron in mitochondria that were isolated from Erv1p-defective cells (5 h at the non-permissive temperature). The iron concentration within these mitochondria was 40–50 ng/mg mitochondrial protein, i.e. 20 times higher than that found in wild-type organelles (2–2.5 ng/mg mitochondrial protein). The drastic increase was comparable to the findings made for mitochondria derived from cells lacking e.g. the mitochondrial cysteine desulfurase Nfs1p (Kispal et al., 1999), the yeast homologues of adrenodoxin Yah1p (Lange et al., 2000) and adrenodoxin reductase Arh1p (Li et al., 2001), or the ABC transporter Atm1p/ABC7 (Kispal et al., 1999; Bekri et al., 2000; 50–60 ng/mg mitochondrial protein). Hence, Erv1p is a novel factor that is crucial for iron homeostasis. Interestingly, both Erv1p and Atm1p exclusively support the assembly of cytosolic Fe/S proteins (Kispal et al., 1999), raising the possibility that mitochondrial iron levels respond to the function of an Fe/S protein in the cytosol.
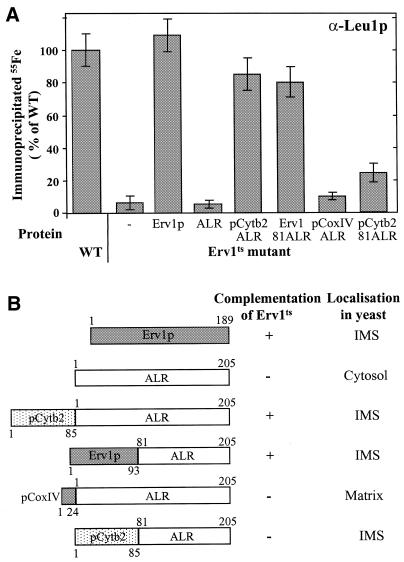
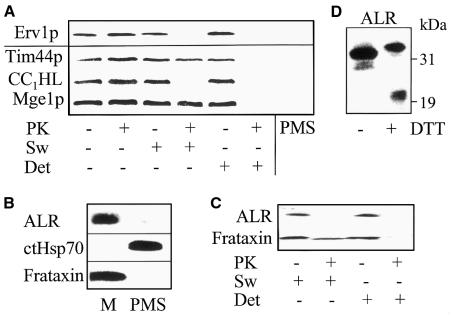
To address the question of whether human ALR and yeast Erv1p represent functional orthologues, the capacity of ALR to support the maturation of cytosolic Fe/S proteins was analysed by using the Leu1p radiolabelling assay described above. ALR and ERV1 genes were expressed in Erv1ts mutant cells. At the non-permissive temperature, ALR could not substitute for Erv1p in the maturation of Leu1p (Figure 3A). This result is in keeping with the earlier finding that ALR is unable to restore growth of yeast cells lacking Erv1p (Hofhaus et al., 1999). We reasoned that the failure in functional complementation might be due to the fact that, in yeast cells, ALR was reported to be localized in the cytosol, whereas Erv1p is associated with mitochondria (Hofhaus et al., 1999). To determine the exact subcellular localization of Erv1p and ALR, yeast and human mitochondria and post-mitochondrial supernatants were isolated, the organelles were sub-fractionated, and samples were analysed by immunostaining. Erv1p was associated with yeast mitochondria and found in the intermembrane space (Figure 4A). Mitochondrial localization was also detected for ALR using cell fractions isolated from fresh human liver (Figure 4B). Like Erv1p, ALR was located in the intermembrane space (Figure 4C). The human protein, similarly to yeast Erv1p (Lee et al., 2000), formed dimers and migrated at ∼45 kDa in SDS–polyacrylamide gels under non-reducing conditions (Figure 4D). Mitochondrial localization of ALR is in contrast to the cytosolic appearance reported for an ALR-GFP (green fluorescent protein) fusion protein (Hofhaus et al., 1999). The failure of ALR-GFP to correctly localize to the intermembrane space may result from the interference of the GFP moiety with mitochondrial targeting and/or from the overexpression of this protein in human cell culture. Taken together, both Erv1p and ALR are constituents of the mitochondrial intermembrane space.
Fig. 3. Human ALR can replace the function of yeast Erv1p in the biogenesis of cytosolic Fe/S proteins. (A) Wild-type (WT) and Erv1ts mutant cells expressing Erv1p, ALR and various ALR fusion proteins were used to analyse the incorporation of 55Fe into cytosolic Leu1p by the assay described in Figure 1. Each experiment was performed at least three times. Standard errors are given by bars. (B) Schematic representation of the ALR fusion proteins. The indicated portions of various proteins were fused to the N-terminus of full-length or truncated human ALR (Hofhaus et al., 1999). Localization of the resulting fusion proteins was determined by cell fractionation (cf. Figure 4). Complementation of the growth defect of yeast Erv1ts cells by the different fusion proteins was assayed by cultivation on rich medium containing glycerol at 37°C for 3 days and was compared with growth of wild-type cells.
Fig. 4. Erv1p and ALR are located in the mitochondrial intermembrane space. (A) Mitochondria and post-mitochondrial supernatant (PMS) were isolated from yeast cells expressing an HA-epitope-tagged Erv1p. Submitochondrial localization was tested by hypotonic swelling of the organelles (Sw) leading to the rupture of the outer membrane, and by solubilization of the organelles in buffer containing 0.1% Triton X-100 (Det) followed by treatment with 50 µg/ml proteinase K (PK; Diekert et al., 2001). Samples were analysed by immunostaining using antisera raised against the HA-epitope and proteins of the mitochondrial intermembrane space (CC1HL) and the matrix (Tim44p, Mge1p). (B) Fresh human liver was used to prepare isolated mitochondria (M) and post-mitochondrial supernatant. Immunostaining was for ALR, cytosolic Hsp70 (ctHsp70) and mitochondrial frataxin. (C) Human liver mitochondria were subjected to swelling, detergent and protease treatment as in (A), and (D) they were subjected to non-reducing 4–12% gradient SDS–PAGE (Lee et al., 2000) in the presence or absence of 10 mM dithiotreitol (DTT) followed by immunostaining for ALR.
Apparently, the sequences assuring targeting of ALR to human mitochondria are not functional in yeast. We therefore fused ALR to the intermembrane space targeting signal of cytochrome b2 (to yield pCytb2-ALR) or we replaced the N-terminus of ALR by that of Erv1p (Erv1-81-ALR; Hofhaus et al., 1999; Figure 3B). The fusion proteins were located in the mitochondrial intermembrane space of yeast cells and were able to fully restore wild-type growth of Erv1ts cells at the non-permissive temperature (Figure 3B). Thus, the N-terminus of Erv1p confers targeting information for localization of ALR to the intermembrane space. Importantly, intermembrane space-localized ALR supported the assembly of the cytosolic Fe/S protein Leu1p to almost the same extent as Erv1p (Figure 3A). In contrast, no complementation of the growth defect and of the failure in the maturation of cytosolic Leu1p was observed upon targeting of ALR to the mitochondrial matrix by adding a presequence to the N-terminus of ALR (pCoxIV-ALR; Figure 3). In summary, ALR, when directed to the mitochondrial intermembrane space, may take over the essential function of yeast Erv1p in the maturation of cytosolic Fe/S proteins. We conclude that mitochondrial ALR and Erv1p are functional orthologues.
Members of the Erv1p/ALR protein family contain a conserved sulfhydryl oxidase domain at their C-terminus (Lee et al., 2000; Senkevich et al., 2000). This domain carries the point mutation rendering Erv1ts cells temperature-sensitive (Lisowsky, 1992), and hence it is crucial for Erv1p function in cytosolic Fe/S protein maturation. However, the catalytic domain is not sufficient for mediating this process, since an N-terminally truncated ALR protein (coined pCytb2-81-ALR) directed to the intermembrane space could not functionally complement the Erv1ts strain. Cells harbouring this fusion protein exhibited only weak activity in supporting the assembly of the Fe/S cluster in cytosolic Leu1p (Figure 3). In summary, both the non-conserved N-terminus and the sulfhydryl oxidase domain are crucial for Erv1p/ALR function in the maturation of cytosolic Fe/S proteins.
DISCUSSION
Our study identifies Erv1p/ALR as novel mitochondrial components that are specifically involved in the assembly of cytosolic Fe/S proteins (Lill and Kispal, 2000). Erv1p, like the central constituents of the yeast ISC assembly machinery, is indispensable for life (Lisowsky, 1992). This indicates that mitochondria play an essential role in the biosynthesis of extra-mitochondrial Fe/S proteins. Consistent with their specialized function in the intermembrane space, Erv1p/ALR homologues are present in virtually all eukaryotes from fungi to man (Hagiya et al., 1994; Lisowsky et al., 1995; Polimeno et al., 1999), but not in bacteria. Members of the Erv1p/ALR protein family share a conserved C-terminal domain that exhibits sulfhydryl oxidase activity, i.e. the oxygen-dependent formation of disulfide bonds in target proteins (Hoober et al., 1999; Lee et al., 2000, Lisowsky et al., 2001). To date, only the target of the viral sulfhydryl oxidase E10R has been identified, a virus-encoded glutaredoxin that in turn introduces disulfide bonds into other viral proteins as a prerequisite for virus assembly in the cytosol (Senkevich et al., 2000). While the sulfhydryl oxidase domain of Erv1p is required for function in cytosolic Fe/S protein maturation, the specific enzyme activity of Erv1p is comparatively low (Hoober et al., 1999; Lee et al., 2000). Hence, it remains to be determined whether disulfide bond formation underlies Erv1p function. We did not find evidence for stable association of Erv1p with an Fe/S cluster, even under anaerobic conditions (not shown). Future elucidation of the molecular function of Erv1p will require identification of interacting proteins and in vitro reconstitution of cytosolic Fe/S protein maturation.
The entire Erv1p/ALR protein including the poorly conserved N-terminus is needed for function in Fe/S protein assembly. In contrast, the C-terminal sulfhydryl oxidase domain of mammalian ALR suffices to support liver regeneration (Giorda et al., 1996; Li et al., 2000). The vast majority of ALR is located inside cells and ALR expression is not restricted to liver tissue (Hofhaus et al., 1999). Therefore, participation in the assembly of cytosolic Fe/S proteins appears to be the primary task of ALR. Certainly, this function is crucial for all eukaryotic cell types, whereas the proposed role of ALR as a hepatotrophic growth factor is restricted to liver cells, and may be effective only after cell damage.
According to a current working model (Lill and Kispal, 2000), the ISC components of the mitochondrial matrix pre-assemble and package Fe/S clusters, which may then be exported into the cytosol, a process possibly involving the ABC transporter Atm1p/ABC7. Since Erv1p/ALR is located in the intermembrane space, the proteins may operate subsequently to Atm1p/ABC7. Our findings provide the basis for addressing the molecular mechanism by which mitochondrial Erv1p/ALR contributes to the biogenesis of cytosolic Fe/S proteins and to cellular iron homeostasis.
METHODS
Yeast strains and cell growth. The following strains of S. cerevisiae were used: JRY 675 (MATa, ura3-52, his4-519, Δleu2) served as wild type; pet492-6A [MATa, ura3-52, Δleu2; pet492ts (Lisowsky, 1992)]. To generate a yeast strain containing a C-terminal haemagglutinin (HA)-tagged Erv1p, the ERV1 gene was amplified by PCR using AAAGAGCTCTACAATCTCAGAATTCTG and AAACCCGGGCTGATACGGCTAATTTCAAG as forward and reverse primers. The resulting DNA fragment was cloned into the YIplac211/HA integrative plasmid, which, after cleavage by EcoRI, was transformed into yeast strain W303 (MATa, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3,112). Integration of the DNA into the correct site was verified by PCR.
The following plasmids were used: YEp351-ADH-cERV1 (Lisowsky, 1992); YEp352-ADH-Erv1-81-ALR and YEp352-ADH-pCoxIV-ALR (Hofhaus et al., 1999); pRS426-GPD-ERV1 was derived from plasmid pRS426-GPD (Mumberg et al., 1994) and carries the ERV1 gene in the SmaI restriction site. Plasmids YEp352-ADH-pCytb2-ALR and YEp352-ADH-pCytb2-81-ALR encoding fusion proteins of the presequence of cytochrome b2 (residues 1–85) and full-length and N-terminally truncated ALR (cf. Figure 4) were generated by amplifying the ALR coding regions by PCR from a human cDNA clone. Forward primers were CCCAAAGATATCGATGGCGGCGCCCGGCGAGCG and CCCAAAGATATGATGCGGACGCAGCAGAAGCGG, respectively; the reverse primer was CCCGGTACCCTAGTCACAGGAGCCATCCTT. The DNA fragments were inserted into the EcoRV and KpnI sites of the vector pBSC-preCytb2. The reading frames were excised as BamHI–KpnI fragments and inserted into the respective sites of vector YEp352-ADH. The RLI1 gene and an additional 300 upstream nucleotides were amplified by PCR using forward (AAAGAGCTCGACTATACGCTGCTTCAGTCA) and reverse (AAAGTCGACTACCGGTGTTATCCAAGAAA) primers. The PCR product was cloned into the SacI–SalI site of plasmid Yep351/3HA introducing the coding region of a C-terminal HA tag. This chimera was again amplified with forward (AAAGAGCTCGACTATACGCTGCTTCAGTCA) and reverse (AAACTCGAGTCAGCGGCCGCACTGAGCAG) primers and cloned into the SacI–XhoI sites of pRS426 plasmid (Mumberg et al., 1994). The desired sequence was verified by DNA sequencing. Growth of yeast cells was detailed elsewhere (Kispal et al., 1999).
Miscellaneous methods. The following published methods were used (unless stated otherwise, see Kispal et al., 1999): manipulation of DNA and PCR; transformation of yeast cells; isolation of mitochondria from yeast and human liver (Robinson et al., 1987); whole cell lysates by breaking cells with glassbeads; determination of ‘free’ (i.e. non-heme, non-Fe/S) iron by the bathophenanthroline method; measurement of radioactive 55Fe incorporation into cytosolic and mitochondrial Fe/S proteins; enzyme activities of malate dehydrogenase, aconitase, succinate dehydrogenase and succinate-cytochrome c reductase. The standard errors for determination of enzyme activities varied between 5 and 15%. For raising antibodies in rabbits against human ALR, a purified His6-tagged fragment of ALR (residues 81–205) was used.
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Richter for expert technical assistance. Our work was generously supported by grants of the Sonderforschungsbereiche 189, 286 and 575 of the Deutsche Forschungsgemeinschaft, Deutsches Humangenomprojekt, the Volkswagen-Stiftung, the Fonds der Chemischen Industrie and the Hungarian Funds OKTA.
REFERENCES
- Bekri S., Kispal, G., Lange, H., Fitzsimons, E., Tolmie, J., Lill, R. and Bishop, D.F. (2000) Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia (XLSA/A) with disruption of cytosolic iron–sulfur protein maturation. Blood, 96, 3256–3264. [PubMed] [Google Scholar]
- Craig E.A., Voisine, C. and Schilke, B. (1999) Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol. Chem., 380, 1167–1173. [DOI] [PubMed] [Google Scholar]
- Diekert K., deKroon, A.I.P.M., Kispal, G. and Lill, R. (2001) Isolation and sub-fractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol., 65, 37–51. [DOI] [PubMed] [Google Scholar]
- Giorda R., Hagiya, M., Seki, T., Shimonishi, M., Sakai, H., Michaelson, J., Francavilla, A., Starzl, T.E. and Trucco, M. (1996) Analysis of the structure and expression of the augmenter of liver regeneration (ALR) gene. Mol. Med., 2, 97–108. [PMC free article] [PubMed] [Google Scholar]
- Hagiya M., Francavilla, A., Polimeno, L., Ihara, I., Sakai, H., Seki, T., Shimonishi, M., Porter, K.A. and Starzl, T.E. (1994) Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc. Natl Acad. Sci. USA, 91, 8142–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhaus G., Stein, G., Polimeno, L., Francavilla, A. and Lisowsky, T. (1999) Highly divergent amino termini of the homologous human ALR and yeast scERV1 gene products define species specific differences in cellular localization. Eur. J. Cell Biol., 78, 349–356. [DOI] [PubMed] [Google Scholar]
- Hoober K.L., Glynn, N.M., Burnside, J., Coppock, D.L. and Thorpe, C. (1999) Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J. Biol. Chem., 274, 31759–31762. [DOI] [PubMed] [Google Scholar]
- Kaut A., Lange, H., Diekert, K., Kispal, G. and Lill, R. (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron–sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem., 275, 15955–15961. [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere, P., Prohl, C. and Lill, R. (1999) The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J., 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw G.B. (1988) Isopropylmalate dehydratase from yeast. Methods Enzymol., 166, 423–429. [DOI] [PubMed] [Google Scholar]
- Kushnir S. et al. (2001) A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell, 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H., Kispal, G., Kaut, A. and Lill, R. (2000) A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc. Natl Acad. Sci. USA, 97, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Hofhaus, G. and Lisowsky, T. (2000) Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett., 477, 62–66. [DOI] [PubMed] [Google Scholar]
- Li J., Saxena, S., Pain, D. and Dancis, A. (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem., 276, 1503–1509. [DOI] [PubMed] [Google Scholar]
- Li Y. et al. (2000) Stimulation of the mitogen-activated protein kinase cascade and tyrosine phosphorylation of the epidermal growth factor receptor by hepatopoietin. J. Biol. Chem., 275, 37443–37447. [DOI] [PubMed] [Google Scholar]
- Lill R. and Kispal, G. (2000) Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem. Sci., 25, 352–356. [DOI] [PubMed] [Google Scholar]
- Lill R., Diekert, K., Kaut, A., Lange, H., Pelzer, W., Prohl, C. and Kispal, G. (1999) The essential role of mitochondria in the biogenesis of cellular iron–sulfur proteins. Biol. Chem., 380, 1157–1166. [DOI] [PubMed] [Google Scholar]
- Lisowsky T. (1992) Dual function of a new nuclear gene for oxidative phosphorylation and vegetative growth in yeast. Mol. Gen. Genet., 232, 58–64. [DOI] [PubMed] [Google Scholar]
- Lisowsky T., Weinstat-Saslow, D.L., Barton, N., Reeders, S.T. and Schneider, M.C. (1995) A new human gene located in the PKD1 region of chromosome 16 is a functional homologue to ERV1 of yeast. Genomics, 29, 690–697. [DOI] [PubMed] [Google Scholar]
- Lisowsky T., Lee, J.E., Polimeno, L., Francavilla, A. and Hofhaus, G. (2001) Mammalian augmenter of liver regeneration is a sulfhydryl oxidase. Dig. Liver Dis., 33, 173–180. [DOI] [PubMed] [Google Scholar]
- Matsubara H. and Saeki, K. (1992) In Cammack, R. (ed.), Structural and Functional Diversity of Ferredoxins and Related Proteins. Academic Press, San Diego, CA, Vol. 38, pp. 223–280.
- Mumberg D., Müller, R. and Funk, M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y., Nakai, M., Hayashi, H. and Kagamiyama, H. (2001) Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem., 276, 8314–8320. [DOI] [PubMed] [Google Scholar]
- Polimeno L., Lisowsky, T. and Francavilla, A. (1999) From yeast to man—from mitochondria to liver regeneration: a new essential gene family. Ital. J. Gastroenterol. Hepatol., 31, 494–500. [PubMed] [Google Scholar]
- Robinson J.B., Inman, L., Sumegi, B. and Srere, P.A. (1987) Further characterization of the Krebs tricarboxylic acid cycle metabolon. J. Biol. Chem., 262, 1786–1790. [PubMed] [Google Scholar]
- Senkevich T.G., White, C.L., Koonin, E.V. and Moss, B. (2000) A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl Acad. Sci. USA, 97, 12068–12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Cash, V.L., Flint, D.H. and Dean, D.R. (1998) Assembly of iron–sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem., 273, 13264–13272. [DOI] [PubMed] [Google Scholar]