Abstract
Objective
This study trains a U-shaped fully convolutional neural network (U-Net) model based on peripheral contour measures to achieve rapid, accurate, automated identification and segmentation of periprostatic adipose tissue (PPAT).
Methods
Currently, no studies are using deep learning methods to discriminate and segment periprostatic adipose tissue. This paper proposes a novel and modified, U-shaped convolutional neural network contour control points on a small number of datasets of MRI T2W images of PPAT combined with its gradient images as a feature learning method to reduce feature ambiguity caused by the differences in PPAT contours of different patients. This paper adopts a supervised learning method on the labeled dataset, combining the probability and spatial distribution of control points, and proposes a weighted loss function to optimize the neural network's convergence speed and detection performance. Based on high-precision detection of control points, this paper uses a convex curve fitting to obtain the final PPAT contour. The imaging segmentation results were compared with those of a fully convolutional network (FCN), U-Net, and semantic segmentation convolutional network (SegNet) on three evaluation metrics: Dice similarity coefficient (DSC), Hausdorff distance (HD), and intersection over union ratio (IoU).
Results
Cropped images with a 270 × 270-pixel matrix had DSC, HD, and IoU values of 70.1%, 27 mm, and 56.1%, respectively; downscaled images with a 256 × 256-pixel matrix had 68.7%, 26.7 mm, and 54.1%. A U-Net network based on peripheral contour characteristics predicted the complete periprostatic adipose tissue contours on T2W images at different levels. FCN, U-Net, and SegNet could not completely predict them.
Conclusion
This U-Net convolutional neural network based on peripheral contour features can identify and segment periprostatic adipose tissue quite well. Cropped images with a 270 × 270-pixel matrix are more appropriate for use with the U-Net convolutional neural network based on contour features; reducing the resolution of the original image will lower the accuracy of the U-Net convolutional neural network. FCN and SegNet are not appropriate for identifying PPAT on T2 sequence MR images. Our method can automatically segment PPAT rapidly and accurately, laying a foundation for PPAT image analysis.
Keywords: Prostate cancer, Periprostatic adipose tissue, Deep learning, U-shaped fully convolutional neural network (U-Net), Contour feature
1. Introduction
Prostate cancer (PCa) is the most common cancer of the male genitourinary tract and remains the most common cancer and a significant cause of death in men worldwide, with approximately 1.6 million new cases and 366,000 deaths per year [1]. The prostate is surrounded by a unique adipose depot called periprostatic adipose tissue (PPAT), whose characteristics can contribute to high-grade or highly aggressive PCa [[2], [3], [4], [5], [6], [7]] and to shorter progression-free survival of patients with PCa under active surveillance [8]. In patients with PCa receiving androgen deprivation therapy (ADT) as primary treatment, PPAT features are an essential predictor of time to progression of castration-resistant PCa (CRPC) [9]. Adipokines secreted by PPAT may promote the aggressiveness and local spread of PCa [[10], [11], [12]]. PPAT is vital in the progression and aggressiveness of PCa. Therefore, having tools that can rapidly and automatically segment images of PPAT and then calculate its volume, area, and thickness and integrate them into the PCa risk assessment models can reduce the time, improve efficiency, and improve the understanding of the PCa.
Image segmentation is the primary part of medical image processing and subsequent analysis. Precise medical image segmentation is still challenging due to the diverse morphology of human organs and lesions and image background noise interference. Although convolutional neural network (CNNs) have significantly improved upon traditional image segmentation methods [13,14], they have disadvantages such as redundant computation and their inability to capture local and global features simultaneously. A U-shaped fully convolutional neural network (U-Net) has been proposed to compensate for the shortcomings of CNNs. U-Net currently has better results in the segmentation of prostate glands and lesions in magnetic resonance imaging (MRI) images [[15], [16], [17], [18]], but its algorithm still needs to be optimized to improve its recognition and segmentation.
The delineation of PPAT is not yet standardized. The distribution of PPAT is not consistent in all prostate directions, especially in the lack of large areas of adipose tissue on the posterior surface [19]. Some researchers [[20], [21], [22]] have measured and counted perivesical fat, perirectal fat, anal saphenous fat, and subcutaneous fat as PPAT. Perirectal fat is not part of PPAT. It has been found that the embryonic origin and anatomical characteristics of Denonvilliers’ fascia differ from the urogenital fascial organ layer that constitutes the perirectal fascia [23].
Most image analysis software used to measure periprostatic fat parameters accompanies the examination instrument, but some third-party medical image processing software is used, such as OsiriX MD [7], MIM [8], and ImageJ [24]. No image analysis software is dedicated to the prostate, and segmenting PPAT has a long learning curve and lacks the creation of an automated image analysis function module. No studies have used deep learning methods to recognize and segment PPAT. Further exploration of applicable algorithms and models is needed to achieve rapid and accurate automated recognition and segmentation of PPAT.
We defined the delineation range of PPAT on MR images and proposed an innovative, improved U-Net network based on contour feature learning to segment PPAT. By using manual annotation of T2-weighted (T2W) MR images of PCa with small sample amounts, data augmentation, and customized loss functions, the U-Net network emphasizes the learning of PPAT contour feature points. It then predicts the PPAT contour to achieve automatic segmentation of PPAT for further image analysis and integration into the clinical workflow.
2. Materials and methods
2.1. Patient selection
MR images of patients admitted to Haikou Hospital of Xiangya Medical College of Central South University from January 2013 to December 2016 who were diagnosed with PCa after the first biopsy for suspected PCa were selected for the study. This study was a retrospective cohort study approved by the Biomedical Ethics Committee of Haikou People's Hospital (reference 2023-(Ethical Review)-199).
2.2. Inclusion and exclusion criteria
Inclusion criteria:
-
①
prostate multiparametric (mp)-MRI followed by transrectal ultrasound-guided prostate biopsy (systematic and targeted biopsy) with complete pathology;
②MRI sequence that included transverse (axial) T2W images.
Exclusion criteria:
-
①
Repeat prostate biopsy;
-
②
MR image artifacts or poor display affecting image determination;
-
③
Previous transurethral surgery, rectal or anal surgery, endocrine therapy, chemotherapy or radiotherapy;
-
④
Pathological findings not of adenocarcinoma or no Gleason score;
-
⑤
Taking statins or hormonal drugs that affect lipid metabolism for more than three months prior to MRI;
-
⑥
Pelvic lipomatosis was considered based on the MRI.
2.3. MR image selection
MRI is the predominant imaging examination for staging and diagnostic evaluation of PCa, with favorable soft tissue resolution. Identification of PPAT on MR images is the method of choice because it is the most accurate and the most independent of investigator experience [10]. T2W is one of the two predominant imaging sequences on MRI, and it is the predominant sequence for assessing anatomical structures [25]. Since T2W MRI reveals the prostate anatomy, the best angle to view the prostate on MRI is the axis (transverse). Therefore, we chose the T2 sequence to label the prostate gland and surrounding fat. The pixel matrix of the T2W MRI was 512 × 512.
On the T2 sequence images, we can observe the boundary between PPAT and perirectal fat, and there is also a difference in the signal and density of the two types of fat. PPAT has a homogenous density and brighter signal, while the density of perirectal fat is granular and darker (see Fig. 1). We delineated the PPAT as anterior to the pubic symphysis, flanked by the medial foramina, posterior to Denonvilliers’ fascia (excluding the rectal mesenteric fat), superior to the bladder, and inferior to the urethral sphincter. This range is consistent with that of David [26].
Fig. 1.

Delineation of periprostatic adipose tissue. Note: Anteriorly is the pubic symphysis, flanked by the internal obturator muscle. Posteriorly is Denonvilliers’ fascia (excluding rectal mesenteric fat). The upper boundary is the bladder, and the lower boundary is the urethral sphincter. The yellow arrow points to the boundary between the perirectal fat and Denonvilliers’ fascia; the red arrow with the yellow arrow and the outline of the prostate is surrounded by the PPAT. PS: pubic symphysis; OM: internal omentum; P: prostate; R: rectum.
2.4. Labeling for the network
This method used an interpretable label to express the PPAT location and edge information. The label consisted of two parts: a mask generated from the PPAT annotated image and a gradient image. Our T2 sequence image dataset saved the images in RGB format. The manually labeled PPAT contour lines were labeled in yellow (see Appendix I for details), which was defined as the gold standard. These contours were obtained and filled by Python code, and the feature points representing the contour were randomly and uniformly selected on the contour to form a mask with only the contour features, which we used as Label-1 for U-Net. Then, the Laplacian and Sobel operators for image edge detection were used to obtain the gradient image as Label-2 for U-Net.
2.5. Data augmentation
U-Nets are data-driven networks that can be augmented to increase the amount of data when there is too little. Data augmentation usually increases the number of copies of an image by panning, rotating, flipping, scaling, and so on, so that the network can “learn” more image features, to reduce redundant information in the training dataset, and to increase the data usage and generalization ability of the network [27]. Data augmentation is only used for the training dataset and not for the validation dataset. In this study, we expanded the number of images in the training dataset from 400 to 1600 by positive and negative rotation and horizontal and vertical flipping.
2.6. Network training
The neural network of this method used the U-shaped fully convolutional network U-Net as the base network and modified its structure. Since the U-Net network, compared with other convolutional neural networks, can be trained with less data to obtain a model with good results, has fewer training parameters, and has a strong ability to learn edge information, this network was used to learn the contours of PPAT.
We normalized the dataset and limited the data range to eliminate the interference of individual abnormal samples with the network model. Additionally, the training method was set to shuffle mode to reduce network learning bias due to the interconnection of adjacent images. The dataset in this paper was divided into a training set and a test set. The training set was used to train the network parameters and obtain the network model; the test set was used to predict the results and evaluate the model. The training set was used with the label of the network to calculate the loss value, and iterative optimization was continuously done to shrink the loss value so that the network model gradually converged. We used 80% of the data (400 images) for training and the other 20% of the data (100 images) for testing. The pixel matrix of the T2W images was 512 × 512. To train the network to focus on learning to identify PPAT, we cropped the original T2W image, choosing an image matrix of 270 × 270 pixels for the first cropping and 256 × 256 pixels for the second cropping. We chose the 270 × 270 pixel size to ensure that the fatty tissue was cropped within the image field of view and that a fully convolutional network (FCN) could accommodate arbitrarily sized images. In contrast, a 256 × 256 was obtained by resizing the 512 × 512 image. The difference between the two was that the 270 × 270-pixel image was only cropped and not scaled down, while the 256 × 256 image was only scaled down and not cropped.
In this method, the network included two parts: input and labels, where the input was the original MRI T2W image, and labels included mask and gradient images. The network continuously obtained the edge information of the fat tissue in the T2W image by 4 × downsampling, 4 × upsampling to the original image size, and feature fusion with the downsampling counterparts to obtain more accurate feature information. It finally generated the outer contour line of the fat tissue. At this point, the current loss was calculated, and the Adam optimizer iteratively optimized the model to converge. The process is shown in Fig. 2.
Fig. 2.
Training flow diagram.
2.7. Customized loss function
During the training process of a network model, the loss function can reflect the gap between the model and the actual data well, and then the optimizer is used to optimize and update the weights of the parameters in each layer of the network. Since the commonly used loss functions, such as cross-entropy loss (CE) and mean square error (MSE), cannot predict the contour points of the tissue around the prostate well, we used a customized loss function, as described by Equation (1):
| (1) |
where Lmax represents the maximum distance between the predicted vital point and the actual value point, as defined in Equation (2);
| (2) |
where represents the mean distance between the predicted vital point and the actual value point, as outlined in Equation (3);
| (3) |
where represents the overlap index between polygon A enclosed by all predicted vital points and polygon B enclosed by the actual value points, as depicted in Equation (4);
| (4) |
with , as the empirically selected values.
In the calculation of the loss function, the calculation was performed twice. The first time was to calculate the position difference between the PPAT contour by the network and the edge of the T2W image adipose tissue mask; the second time was to calculate the difference between the PPAT contour by the network and the corresponding pixel of the gradient image. The two calculated loss functions can reflect the learning of the current model for the adipose tissue contour, and the optimization can be iterated continuously.
2.8. Generating contours
For the model prediction results, further processing was needed to obtain the contours of the PPAT. First, the model-predicted PPAT contour was filtered out using an erosion operation to remove inconspicuous areas at the edges. The entire periprostatic adipose target contour was highlighted using binarization to visually evaluate the predicted PPAT contour. The predicted PPAT was then used to generate the corresponding PPAT contour. Finally, the final PPAT contours were detected and generated using the convex hull method of the Open Source Computer Vision Library (OPENCV) to complete the automatic annotation of PPAT contours on T2W images. The workflow is shown in Fig. 3.
Fig. 3.
An illustration of contour generation pipeline.
2.9. Quantitative evaluation of segmentation
Three evaluation parameters commonly used in medical image segmentation were used to quantitatively evaluate the ability of different methods to predict the PPAT contour segmentation of the T2W images: the Dice similarity coefficient (DSC), Hausdorff distance (HD), and intersection over union (IoU).
DSC refers to the degree of agreement between the model’s predicted fat contour and the doctor's manual annotation. It is calculated as Equation (5):
| (5) |
where A and B represent the point sets contained in the two fat contour regions. The closer the DSC value is to 1, the closer the point sets of the two groups are.
-
⑵
HD calculates the maximum difference between the two sets of contour point sets and represents the maximum degree of mismatch between the segmentation result and the two point sets on the fat contour region of the valid label. The formula is as Equation (6):
| (6) |
where h(A, B) and h(B, A) are called the one-way Hausdorff distances from set A to set B and from set B to set A, respectively, which are defined as Equations (7), (8).
| (7) |
| (8) |
A smaller value of HD suggests that the maximum difference between the two sets is minor, indicating higher effectiveness of the segmentation.
-
⑶
IoU is the union of two point set intersection ratios, representing the degree of similarity between them. The larger the value, the better the effect. See Equation (9) for the definition. IoU is an evaluation parameter commonly used in target detection. The similarity or overlap of two samples can be calculated by the ratio between the predicted border and the actual border [28]:
| (9) |
2.10. Comparison with other CNNs
Like the U-Net network, other fully convolutional networks, such as the standard fully convolutional network (FCN) and semantic segmentation convolutional network (SegNet), are also constructed by encoding and decoding. The difference is that the latter networks adopt the VGG16 network to extract feature information. Our proposed approach, based on the improved U-Net network with contour features, is compared on image segmentation capabilities against FCN, SegNet, and U-Net.
2.11. Experimental hardware and software environment
The experiment was run on the Windows 10 operating system. The graphics card was the NVIDIA GeForce GTX 1050Ti, the deep learning framework was PyTorch 1.2, and the experimental environment was Python 3.7. Network training was set to 160 training rounds (epochs), and the initial learning rate was set to 0.0005 and gradually decreased with epoch number. The training batch was set to 1. Since the optimizer can accelerate the training speed of convolutional neural networks, we sampled the adaptive moment estimation (Adam) optimizer to update the weights of each layer of the network and the customized loss function. The general flow chart is shown in Fig. 4.
Fig. 4.
The core steps of the proposed solution.
2.12. Statistical analysis
SPSS 20.0 was used for all statistical analyses. Quantitative data conforming to a normal distribution are expressed as mean and standard deviation. The T test was used for statistical analysis between the two groups, and one-way analysis of variance was used for statistical analysis of three groups. The least significant difference test was used as the ANOVA post hoc test for data with homogeneity of variance, and the Games-Howell test was used for data with heterogeneity of variance. Enumeration data are expressed as ratio (%) and were analyzed by Fisher's exact probability method. P < 0.05 indicated statistical significance.
3. Results
3.1. Graph of the iterative loss function
In deep learning, by minimizing the loss function, the model reaches the convergence state and reduces the error of the model's predicted value. As seen from the loss function curve (Fig. 5), the loss function gradually decreases and eventually stabilizes with the epoch increase, indicating that the model has converged to the optimal state. The epoch number is more than 160 times, or the change of the training loss is less than a given threshold (less than 300) considered converged.
Fig. 5.
The training loss function curve.
3.2. Gradient image labels
Gradient image labels were divided into two categories, one for images obtained with the Laplace operator (shown in Fig. 6) and the other for images obtained with the Sobel operator (see Fig. 7).
Fig. 6.
An example of result image by using the Laplacian operator. Notes: A Original image, B Laplacian extracted image.
Fig. 7.
An example of result image by using the Sobel operator. Notes: A Original diagram, B vertical gradient image, C horizontal gradient image.
3.3. Generated contour
Fig. 8 shows the original T2 sequence from the base to the apex of the prostate, the manually annotated images, the contour of adipose tissue around the prostate predicted by the proposed method, and the final adipose tissue contour generated. The contour of adipose tissue generated is red.
Fig. 8.
The results of contour prediction and generation across four prostate levels, from the base (row 1) to the apex (row 4), each depicted at a resolution of 270 × 270. Note: In Manually Segmentation, yellow is the contour of PPAT; red is the prostate contour; blue is the contour of PCa lesions; green is the contour of the PCa transition zone; in Generated Contours, red outlines indicate adipose tissue contours.
3.4. Comparison of predicted contours with hand-annotated contours
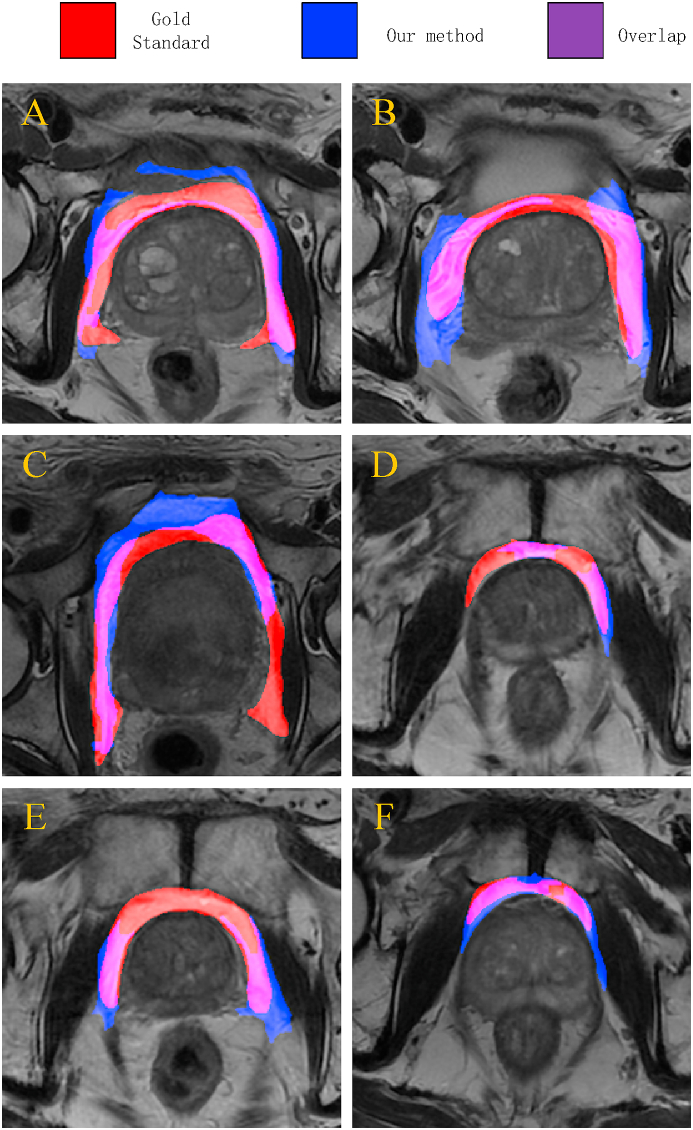
Fig. 9 shows the superposition of the predicted adipose tissue contour and the hand-annotated contour (gold standard). The predicted adipose tissue is blue, the hand-marked adipose tissue is red, and the overlap is purple. As seen from this figure, our method segmented and labeled the PPAT well, but there were still some differences from the gold standard of manual labeling.
Fig. 9.
A visualization of overlay maps between the predicted contour and gold standard for six patients. Note: Blue represents the predicted contours of PPAT, red indicates manual segmentation, and purple shows the overlap between the two.
3.5. Quantitative evaluation and comparison of image segmentation
Before training, for the network to focus on learning to identify adipose tissue to improve the accuracy of network learning, cropping of T2 sequence images is needed. We altered the images into two sizes (270 × 270- and 256 × 256-pixel matrices) and then compared the segmentation ability on the two sizes in the network model. Additionally, our proposed network was compared with the image segmentation results of the FCN, U-Net, and SegNet networks by three evaluation metrics, DSC, HD, and IoU.
3.6. Cropped image (270 × 270 pixels)
Before training, to improve the accuracy of the results and to make the network model focus on learning the PPAT of the MR images, the images needed to be cropped, and the size of the first cropped image was 270 × 270 pixels. The quantitative evaluation values of different segmentation methods on this dataset are given in Table 1. Table 1 shows that our new proposed U-Net network based on feature learning outperformed the other three networks in all three evaluation parameters, DSC, HD, and IoU.
Table 1.
Quantitative evaluation of image segmentation (270 × 270).
| Method | DSC/% | HD/mm | IoU/% | P value |
|---|---|---|---|---|
| FCN | 9.4 | 76.0 | 5.4 | 0.001 |
| U-Net | 51.1 | 51.3 | 37.2 | |
| SegNet | 15.2 | 87.0 | 8.2 | |
| Ours | 70.1 | 27.0 | 56.1 |
3.7. Downscaled image (256 × 256 pixels)
The size of the downscaled image was 256 × 256 pixels. Table 2 gives this dataset's quantitative evaluation values of different segmentation methods. Table 2 shows that our proposed U-Net network based on contour feature learning and the standard U-Net network was significantly better than the other two networks. The standard U-Net network slightly outperformed our network in the DSC and IoU, but our proposed network model had better evaluation parameter values in the HD.
Table 2.
Quantitative evaluation of image segmentation (256 × 256).
| Method | DSC/% | HD/mm | IoU/% | P value |
|---|---|---|---|---|
| FCN | 37.9 | 44.2 | 25.6 | 0.002 |
| U-Net | 70.1 | 29.9 | 55.6 | |
| SegNet | 17.0 | 63.2 | 10.1 | |
| Ours | 68.7 | 26.7 | 54.1 |
3.8. Comparison with other convolutional networks
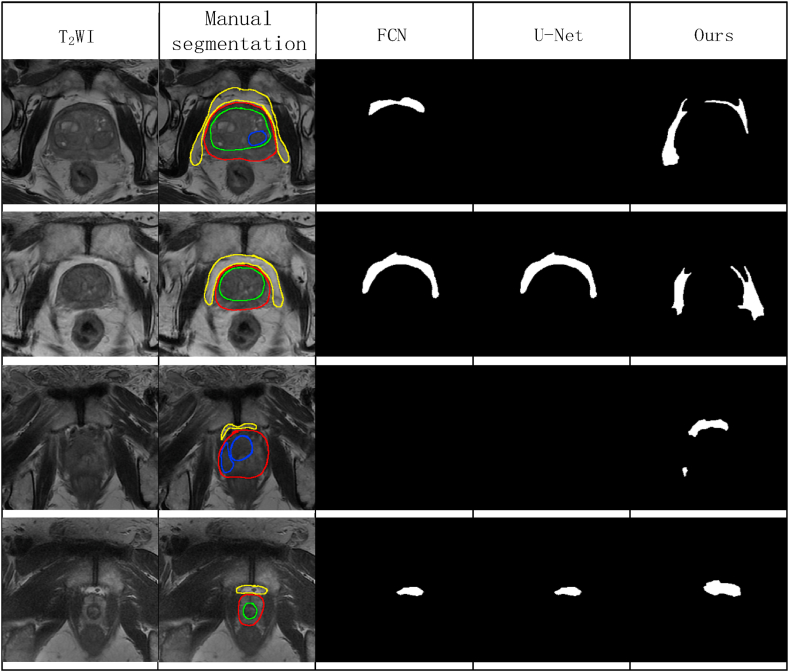
Fig. 10 shows compares the adipose tissue contours from the segmentation by our proposed improved convolutional network with the results of other convolutional networks. Since our proposed improved U-Net network had the best segmentation evaluation value on the cropped image of 270 × 270 pixels, we choose the cropped image of 270 × 270 pixels for segmentation prediction. Since the evaluation parameters of SegNet were the worst, we did not include SegNet for comparison. Each row of the figure represents images of different levels of the prostate (from the base to the apex of the prostate). This figure shows that our U-Net predicted PPAT at all levels (each row had a predicted segmentation map of fat), while FCN and U-Net did not predict PPAT at all levels. In the third row, neither FCN nor U-Net predicted the segmentation of PPAT; in the fourth first row, U-Net could not (predict the segmentation of PPAT).
Fig. 10.
The visual results of FCN, U-Net, and our method in PPAT image segmentation across four prostate levels, from the base (row 1) to the apex (row 4). Note: FCN, fully convolutional neural network; U-Net, U-shaped convolutional neural network; Ours, our method.
4. Discussion
4.1. Label selection and network training
AI has penetrated into our daily lives, playing novel roles in industry, healthcare, transportation, education, and many more areas that are close to the general public. AI is believed to be one of the major drives to change socio-economical lives. In another aspect, AI contributes to the advancement of state-of-the-art technologies in many fields of study, as helpful tools for groundbreaking research [29]. A medical image represents the internal structure and function of the anatomical region, and image segmentation is the basis of subsequent analysis. Before the emergence of deep learning, segmentation of medical images was usually performed using traditional image processing and machine learning methods, such as the K-nearest neighbor (KNN), Gaussian mixture model (GMM), support vector machine (SVM), and other algorithms. However, the above methods are based on shallow feature analysis of the image, so they often learn only local features during feature learning, and manual assistance is often needed. Deep learning, giving it good robustness [30] and the ability to process the original data directly. Compared with traditional image segmentation methods, it performs significantly better at medical image segmentation [14].
The CNN, a representative algorithm of deep learning, is the central algorithmic model in computer vision, and its application to the analysis of medical images has obvious advantages [31].
To solve the problem that the CNN is slow in calculation and to take into account both local and global features, Ronneberger et al. proposed the U-Net convolutional neural network [32] based on the FCN, which was so named because its structure resembles the letter U. The advantages of U-Net are that it is fast, spatially consistent, and can accommodate images of any size. The U-Net model is small, can adapt to a small training set, and has a strong learning ability for edge information [32], so it is very suitable for small-sample medical image segmentation under a lack of data.
Medical images often have unclear tissue boundaries and complex pixel gradients, and only higher-resolution information can be used for accurate segmentation. U-Net extracts feature information through downsampling and dimension reduction, restoring resolution to the maximum extent through upsampling. It effectively improves segmentation accuracy and achieves good positioning accuracy through the network structure containing the coding–decoding structure of the jump connection [15]. U-Net combines low-resolution information (the basis for identifying object categories) with high-resolution information (the basis for accurate segmentation and localization), which is suitable for medical image recognition and segmentation. The U-Net can accurately detect and segment prostate glands and lesions on MR images [[15], [16], [17]]. The U-Net trained with T2 and diffusion-weighted images performed similarly to that evaluated by clinical prostate imaging reports and data systems [18]. While we collected a total of 500 labeled images from 102 patients, the overall sample size was not large, so the we chose U-Net to identify segmented PPAT.
There is still no research on the segmentation of PPAT using any deep learning methods, and we need to explore applicable algorithms and models to achieve fast and accurate automated segmentation of PPAT. Given the irregular boundaries of the PPAT, its signal differentiation from the surrounding adjacent muscles, bones, and fascia is significant in T2W images of MRI.
A contour is a curve composed of a series of connected points, representing the basic shape of the object. The contour is continuous, while the edge is not all continuous. The border of PPAT is its contour, and the feature points on the contour are its pixels. Because of the large amount of contour data in the image, it is worth studying how to find the feature points that reflect the contour of fat around the prostate from these feature points. Only the characteristic points of the contour of adipose tissue around the prostate can be identified and segmented. Therefore, the pixel difference of images between adjacent tissues can be considered for achieving segmentation. Fat contour is linear. To improve the efficiency of network learning and reduce redundancy, we used the feature points (also known as control points) method of image contour detection [33] to convert the hand-labeled linear fat tissue contour into the feature points of the contour as the learning label-1 of the network and input them into the U-Net network for learning. The feature points are pixels, and there is a large amount of contour data in the image. Therefore, it is worth studying how to find the feature points that reflect the main shape features of the contour from these data points. Fig. 5 shows the heatmap of the contour line converted to feature points, and it can be seen that the feature points are more evenly distributed on the contour line.
The Laplacian and Sobel operators are standard algorithms for image gradient detection. The Sobel operator is a first-order differential operator that detects edges from different directions and strengthens the weight of pixels in four directions above and below the center pixel. As a result, the edges in the image are relatively bright, yielding a more intuitive visual effect and playing a particular role in suppressing isolated noise [34]. The Laplacian operator is a second-order differential operator with the property of rotation invariability, that is, isotropy, so it has the same effect in all directions. This makes the deviation error small, but it also makes the deviation direction unpredictable [34]. These two operators have complementary properties. Therefore, we used the images extracted by these two operators as Label-2 input into our modified U-Net to allow the network to better learn the contour features of the PPAT. Fig. 6, Fig. 7 show that we successfully extracted the images of these respective operators.
In the training of neural networks, two essential tools to improve their performance are the loss function and optimizer. By minimizing the loss function, the network model achieves convergence (the loss value tends to be stable) and reduces the error of the model's predicted value. According to the graph of our iterative loss function (Fig. 4), it can be seen that with more training batches, the loss function value gradually decreases and tends to be stable, that is, the convergence state. This result reflects that the prediction ability of our model is good. An optimizer is used to update and calculate the network parameters that affect model training and model output to minimize the loss function, and its primary function is to accelerate the training speed of the convolutional neural network. It dynamically adjusts each parameter's learning rate by using the gradient's first-order and second-order moment estimation [35]. The main advantage of Adam is that after bias correction, the learning rate of each iteration has a specific range, which makes the parameters relatively stable [36].
4.2. Evaluation and comparison of convolutional networks segmentation
Compared with the U-Net network, other fully convolutional networks, such as FCN and SegNet, are also constructed as encoding and decoding. The difference is that SegNet adopts the VGG16 network to extract feature information. Furthermore, the three feature fusion methods are different: (1) the FCN network feature fusion method is added, that is, the feature map after upsampling and the feature map after downsampling are added, the number of features is unchanged, but the feature value is changed; (2) the feature fusion method of the U-Net network is Concat, that is, the feature map after upsampling is trimmed and stacked with the feature map after downsampling so that the feature information is thicker, the number of features is increased, and the feature value is unchanged. (3) SegNet [37] was initially designed to deal with the task of street view understanding. The most significant advantage of this network is that the subscripts of the location information of the feature values are saved in the highest pooling layer of the downsampling, and these subscripts restore the position of the feature values during upsampling to improve the prediction accuracy. The add method increases the amount of information in each dimension of the features describing the image under the condition that the dimensions are unchanged. The Concat method is the combination of the number of channels. That is, the feature dimensions describing the image are increased, while the amount of information in each dimension is unchanged.
Before training, to make the network focus on learning the PPAT and improve the accuracy of the network, T2 sequence images needed to be made smaller. We cropped the image into the 270 × 270 size and downscaled them to 256 × 256 and then compared the advantages and disadvantages of the two sizes in the network. At the same time, we compared the image segmentation results of our proposed network with those of the FCN, U-Net, and SegNet networks by DSC, HD, and IoU.
Table 1 shows that our proposed U-Net network based on feature point learning outperforms the other three networks in all evaluation metrics, DSC, HD, and IoU, at the 270 × 270-pixel size. Table 2 shows that our proposed feature point learning-based U-Net network and the standard U-Net network significantly outperform the other two networks in all three evaluation metrics, DSC, HD, and IoU, in the 256 × 256-pixel matrices. The standard U-Net slightly outperforms our network in the DSC and IoU metrics, but in the HD metric, our proposed improved network is better. This also means that our U-Net network based on the contour features is better than FCN, U-Net, and SegNet for the recognition and segmentation efficacy of PPAT on T2 sequence images of MRI.
By comparing the three evaluation indexes at 270 × 270 and 256 × 256, we see that the two metrics, DSC and IoU, are better at 270 × 270 pixels than at 256 × 256. Moreover, HD on the 256 × 256 size is only 0.3 mm more than at the 270 × 270 size. Therefore, the cropped image of 270 × 270 pixels is more suitable for our proposed learning model of segmenting the adipose tissue around the prostate. The 270 × 270-pixel image was cropped only without deflation, while the 256 × 256-pixel image was reduced only without cropping. This shows that the deflation of the original image affects the recognition accuracy of the convolutional neural network. Based on the above results, we compared the prediction results and the final generated PPAT contour map output from the network model with the manually labeled map (Fig. 8) and visualized and compared the two by superimposing the images (Fig. 9).
In summary, the neural network model we constructed can segment PPAT well, but there is still a gap between it and the gold standard of manual labeling. Since all three evaluation metrics of the SegNet network were the worst, we did not include them in the comparison. Fig. 10 show that our proposed U-Net network based on contour feature points predicted the complete PPAT on T2 sequence images at different levels, while FCN and U-Net could not predict the full range of adipose tissue contours.
4.3. Limitations
The patient data came from a single center, and there was a small sample size of imaging data. Our modified U-Net convolutional neural network based on contour line features was validated using its dataset, and it had problems such as unclear segmentation edges and missing spatial information. External validation was not possible because there is no publicly available database of labeled PPAT. The final predicted contour in this paper was generated based on the convex hull approach, so the effect may be less satisfactory for the few cases where the prostate contour has a concave shape.
5. Conclusions
In this paper, a novel method of segmentation of PPAT by the U-Net network based on contour feature points is proposed to achieve fast and relatively accurate automatic recognition and segmentation of PPAT. This study shows that the cropped image of 270 × 270 pixels is more suitable for training the U-Net convolutional neural network than 256 × 256, as the deflation of the original image affects the recognition accuracy of the CNN on the image. Furthermore, the FCN and SegNet are not suitable for predicting PPAT on T2W MR images. This study verifies the feasibility of our modified U-Net, but it still needs optimization. Whether this neural network can perform stably on images from different MRI devices and medical centers still needs to be validated by expanding the sample size and the number of centers.
Ethics declarations
This study was reviewed and approved by the Biomedical Ethics Committee of Haikou People's Hospital, with the approval number: 2023-(Ethical Review)-199.
Informed consent
As this study was retrospective, informed consent from patients was not required.
Funding
This work was supported by fundings from the National Natural Science Foundation of China [Grant #: 82260362], Key R&D Projects of Hainan Province [Grant No. ZDYF2017084] and the Major Science and Technology Project of Haikou [Grant #: 2020-009].
Data availability statement
The data are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Gang Wang: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Jinyue Hu: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Yu Zhang: Writing – review & editing, Writing – original draft, Methodology, Data curation. Zhaolin Xiao: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation. Mengxing Huang: Writing – review & editing, Writing – original draft, Methodology. Zhanping He: Writing – review & editing, Writing – original draft, Resources. Jing Chen: Writing – review & editing, Writing – original draft, Resources, Project administration, Methodology, Funding acquisition, Conceptualization. Zhiming Bai: Writing – review & editing, Writing – original draft, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25030.
Contributor Information
Jing Chen, Email: jingchen_haiko@163.com.
Zhiming Bai, Email: Drzmbai@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Roermund J.G.H., Hinnen K.A., Tolman C.J., Bol G.H., Witjes J.A., Bosch J.L.H.R., et al. Periprostatic fat correlates with tumour aggressiveness in prostate cancer patients. BJU Int. 2011;107:1775–1779. doi: 10.1111/j.1464-410X.2010.09811.x. [DOI] [PubMed] [Google Scholar]
- 3.Woo S., Cho J.Y., Kim S.Y., Kim S.H. Periprostatic fat thickness on MRI: correlation with Gleason score in prostate cancer. AJR Am. J. Roentgenol. 2015;204:W43–W47. doi: 10.2214/AJR.14.12689. [DOI] [PubMed] [Google Scholar]
- 4.Tan W.P., Lin C., Chen M., Deane L.A. Periprostatic fat: a risk factor for prostate cancer? Urology. 2016;98:107–112. doi: 10.1016/j.urology.2016.07.042. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q., Sun L., Qi J., Yang Z., Huang T., Huo R. Periprostatic adiposity measured on magnetic resonance imaging correlates with prostate cancer aggressiveness. Urol. J. 2014;11:1793–1799. [PubMed] [Google Scholar]
- 6.Cao Y., Cao M., Chen Y., Yu W., Fan Y., Liu Q., et al. The combination of prostate imaging reporting and data system version 2 (PI-RADS v2) and periprostatic fat thickness on multi-parametric MRI to predict the presence of prostate cancer. Oncotarget. 2017;8:44040–44049. doi: 10.18632/oncotarget.17182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahran N., Szewczyk-Bieda M., Wei C., Vinnicombe S., Nabi G. Normalized periprostatic fat MRI measurements can predict prostate cancer aggressiveness in men undergoing radical prostatectomy for clinically localised disease. Sci. Rep. 2017;7:4630. doi: 10.1038/s41598-017-04951-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregg J.R., Surasi D.S., Childs A., Moll N., Ward J.F., Kim J., et al. The association of periprostatic fat and grade group progression in men with localized prostate cancer on active surveillance. J. Urol. 2021;205:122–128. doi: 10.1097/JU.0000000000001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H., Chen S., Li W., Bai P., Wu X., Xing J. Periprostatic fat thickness on MRI is an independent predictor of time to castration-resistant prostate cancer in Chinese patients with newly diagnosed prostate cancer treated with androgen deprivation therapy. Clin. Genitourin. Cancer. 2019;17:e1036–e1047. doi: 10.1016/j.clgc.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Nassar Z.D., Aref A.T., Miladinovic D., Mah C.Y., Raj G.V., Hoy A.J., et al. Peri-prostatic adipose tissue: the metabolic microenvironment of prostate cancer. BJU Int. 2018;121(Suppl 3):9–21. doi: 10.1111/bju.14173. [DOI] [PubMed] [Google Scholar]
- 11.Gucalp A., Iyengar N.M., Zhou X.K., Giri D.D., Falcone D.J., Wang H., et al. Periprostatic adipose inflammation is associated with high-grade prostate cancer. Prostate Cancer Prostatic Dis. 2017;20:418–423. doi: 10.1038/pcan.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahran N., Szewczyk-Bieda M., Vinnicombe S., Fleming S., Nabi G. Periprostatic fat adipokine expression is correlated with prostate cancer aggressiveness in men undergoing radical prostatectomy for clinically localized disease. BJU Int. 2019;123:985–994. doi: 10.1111/bju.14469. [DOI] [PubMed] [Google Scholar]
- 13.Yi X., Walia E., Babyn P. Generative adversarial network in medical imaging: a review. Med. Image Anal. 2019;58 doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 14.Tajbakhsh N., Jeyaseelan L., Li Q., Chiang J.N., Wu Z., Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med. Image Anal. 2020;63 doi: 10.1016/j.media.2020.101693. [DOI] [PubMed] [Google Scholar]
- 15.Zabihollahy F., Schieda N., Krishna Jeyaraj S., Ukwatta E. Automated segmentation of prostate zonal anatomy on T2-weighted (T2W) and apparent diffusion coefficient (ADC) map MR images using U-Nets. Med. Phys. 2019;46:3078–3090. doi: 10.1002/mp.13550. [DOI] [PubMed] [Google Scholar]
- 16.Wong T., Schieda N., Sathiadoss P., Haroon M., Abreu-Gomez J., Ukwatta E. Fully automated detection of prostate transition zone tumors on T2-weighted and apparent diffusion coefficient (ADC) map MR images using U-Net ensemble. Med. Phys. 2021;48:6889–6900. doi: 10.1002/mp.15181. [DOI] [PubMed] [Google Scholar]
- 17.Sunoqrot M.R.S., Selnæs K.M., Sandsmark E., Langørgen S., Bertilsson H., Bathen T.F., et al. The reproducibility of deep learning-based segmentation of the prostate gland and zones on T2-weighted MR images. Diagnostics. 2021;11:1690. doi: 10.3390/diagnostics11091690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schelb P., Schelb P., Kohl S.A.A., Radtke J.P., Wiesenfarth M., Kickingereder P., et al. Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology. 2019;293:607–617. doi: 10.1148/radiol.2019190938. [DOI] [PubMed] [Google Scholar]
- 19.Ioannides C.G., Whiteside T.L. T cell recognition of human tumors: implications for molecular immunotherapy of cancer. Clin. Immunol. Immunopathol. 1993;66:91–106. doi: 10.1006/clin.1993.1012. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., Sun K., Zheng R., Zeng H., Wang S., Chen R., et al. Cancer incidence and mortality in China, 2015. Journal of the National Cancer Center. 2021;1:2–11. doi: 10.1016/j.jncc.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Y., Zhou X., Yang J., Shi H., Li H., Zhao X., et al. The role of tumor-stroma interactions in drug resistance within tumor microenvironment. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.637675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrova V., Annicchiarico-Petruzzelli M., Melino G., Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10. doi: 10.1038/s41389-017-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whiteside T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai T.-S., Hu L.-T., Ma W.-G., Chen X., Luo M., Jin L., et al. Peri-prostatic adipose tissue measurements using MRI predict prostate cancer aggressiveness in men undergoing radical prostatectomy. J. Endocrinol. Invest. 2021;44:287–296. doi: 10.1007/s40618-020-01294-6. [DOI] [PubMed] [Google Scholar]
- 25.王刚 禹刚, 陈晶 杨光, 徐海霞 王国任, et al. 简版前列腺影像报告和数据系统评分的双参数磁共振成像在初次前列腺活检中的诊断价值. 现代泌尿外科杂志. 2020;25:969–974+978. [Google Scholar]
- 26.Estève D., Roumiguié M., Manceau C., Milhas D., Muller C. Periprostatic adipose tissue: a heavy player in prostate cancer progression. Curr. Opin. Endocr. Metab. Res. 2020;10:29–35. doi: 10.1016/j.coemr.2020.02.007. [DOI] [Google Scholar]
- 27.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6:60. doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman M.A., Wang Y. In: Bebis G., Boyle R., Parvin B., Koracin D., Porikli F., Skaff S., et al., editors. vol. 10072. Springer International Publishing; Cham: 2016. Optimizing intersection-over-union in deep neural networks for image segmentation; pp. 234–244. (Advances in Visual Computing). [DOI] [Google Scholar]
- 29.Jiang Y., Li X., Luo H., Yin S., Kaynak O. Quo vadis artificial intelligence? Discov. Artif. Intell. 2022;2:4. doi: 10.1007/s44163-022-00022-8. [DOI] [Google Scholar]
- 30.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A., et al. Artificial intelligence in cancer imaging: clinical challenges and applications. Ca - Cancer J. Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Ronneberger O., Fischer P., Brox T. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Springer International Publishing; Cham: 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- 33.Carmona-Poyato A., Fernández-García N.L., Medina-Carnicer R., Madrid-Cuevas F.J. Dominant point detection: a new proposal. Image Vis Comput. 2005;23:1226–1236. doi: 10.1016/j.imavis.2005.07.025. [DOI] [Google Scholar]
- 34.雷丽珍. 数字图像边缘检测方法的探讨. 测绘通报. 2006 40–42. [Google Scholar]
- 35.Bottou L., Curtis F.E., Nocedal J. Optimization methods for large-scale machine learning. SIAM Rev. 2018;60:223–311. doi: 10.1137/16M1080173. [DOI] [Google Scholar]
- 36.Kingma D.P., Ba J. 2017. Adam: A Method for Stochastic Optimization. [DOI] [Google Scholar]
- 37.Badrinarayanan V., Kendall A., Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the corresponding author on reasonable request.