Abstract
Campylobacter jejuni, a microaerophilic, gram-negative bacterium, is a common cause of gastrointestinal disease in humans. Heat shock proteins are a group of highly conserved, coregulated proteins that play important roles in enabling organisms to cope with physiological stresses. The primary aim of this study was to characterize the heat shock response of C. jejuni. Twenty-four proteins were preferentially synthesized by C. jejuni immediately following heat shock. Upon immunoscreening of Escherichia coli transformants harboring a Campylobacter genomic DNA library, one recombinant plasmid that encoded a heat shock protein was isolated. The recombinant plasmid, designated pMEK20, contained an open reading frame of 1,119 bp that was capable of encoding a protein of 372 amino acids with a calculated molecular mass of 41,436 Da. The deduced amino acid sequence of the open reading frame shared similarity with that of DnaJ, which belongs to the Hsp-40 family of molecular chaperones, from a number of bacteria. An E. coli dnaJ mutant was successfully complemented with the pMEK20 recombinant plasmid, as judged by the ability of bacteriophage λ to form plaques, indicating that the C. jejuni gene encoding the 41-kDa protein is a functional homolog of the dnaJ gene from E. coli. The ability of each of two C. jejuni dnaJ mutants to form colonies at 46°C was severely retarded, indicating that DnaJ plays an important role in C. jejuni thermotolerance. Experiments revealed that a C. jejuni DnaJ mutant was unable to colonize newly hatched Leghorn chickens, suggesting that heat shock proteins play a role in vivo.
Campylobacter jejuni, a microaerophilic, gram-negative bacterium, is a member of the epsilon subdivision of eubacteria. C. jejuni is recognized as a common cause of diarrheal disease in humans (9). Infection with C. jejuni is commonly acquired by eating undercooked chicken or drinking unpasteurized milk or contaminated water. Epidemiological studies have revealed that chickens serve as important reservoirs for C. jejuni and are generally colonized by C. jejuni early in life (7, 33). Chicken carcasses become extensively contaminated with C. jejuni during the slaughtering process, and the number of C. jejuni-contaminated carcasses is amplified by contaminated processing equipment (8). It is estimated that 50 to 90% of chicken carcasses are contaminated with C. jejuni by the time of sale. Given the importance of chickens as a major reservoir of C. jejuni and the fact that so many C. jejuni infections are acquired by eating contaminated poultry, studies are required to examine the role of bacterial heat shock proteins (Hsps) in C. jejuni colonization of the intestinal tracts of chickens.
Organisms respond to thermal stress by inducing the synthesis of a group of highly conserved proteins called Hsps (22, 23). Hsps are a group of coregulated proteins whose rate of synthesis varies with temperature but increases rapidly after a sudden increase in temperature or in response to other environmental stresses. In addition to playing important roles in thermotolerance and coping with other physiological stresses, Hsps serve vital roles in normal cell function by acting as chaperones to promote the folding of most cellular proteins and proteolysis of potentially deleterious, misfolded proteins. More specifically, in E. coli, DnaK, DnaJ, and GrpE aid in protein folding, bacteriophage λ replication (1, 32), replication of plasmids mini-P1 and mini-F (17, 35), host DNA replication (30), and the proteolysis of normally unstable proteins. Mutations in dnaJ and dnaK have similar phenotypes, a finding that is explained by the fact that the two proteins they encode work together.
Thermoregulation plays an important role in virulence gene expression in pathogenic bacteria, including Escherichia coli, Salmonella spp. Shigella spp., and Yersinia spp. Given the importance of the heat shock response in the pathogenesis of other enteric pathogens, the heat shock response may play a role in the pathogenesis of C. jejuni-mediated enteritis. The purpose of this study was to examine the heat shock response of C. jejuni to identify proteins whose synthesis is increased in response to thermal stress. Experiments were also performed to address the importance of the thermal stress response in allowing C. jejuni to grow at temperatures of 43°C and greater by generating a C. jejuni dnaJ mutant. Interestingly, the optimal growth temperature for C. jejuni in the laboratory has been reported to be 42°C (36), which approximates the core temperature of chickens (14).
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
C. jejuni F38011, M129, 78-27, 81116, and 33560 were cultured on Mueller-Hinton (MH) agar plates containing 5% citrated bovine blood at 37°C in Gas-Pak jars with CampyPak Plus packets (BBL Microbiology Systems, Becton Dickinson, Cockeysville, Md.). Each isolate was passaged every 24 to 48 h. C. jejuni F38011 and M129 were provided by K. Ryan (University Medical Center, University of Arizona, Tucson), 81116 was from L. S. Tompkins (Stanford University, Stanford, Calif.), 78-27 was from M. J. Blaser (Vanderbilt University, Nashville, Tenn.), and 33560 was from the American Type Culture Collection (Rockville, Md.). E. coli isolates XL1-Blue MRF, K-12 strain BMH71 (mutL), MF670, and BR3672 were cultured in Luria-Bertani broth (LB; 10 g of Bacto Tryptone, 5 g of yeast extract and 5 g of sodium chloride per liter) with aeration or on LB plates containing 15 g of Bacto Agar per liter in a 37°C incubator supplemented with the appropriate antibiotic. Culture media were supplemented with, per milliliter, 50 μg of ampicillin, 50 μg of kanamycin, or 15 μg of tetracycline, as needed.
Metabolic labeling of C. jejuni proteins.
C. jejuni was harvested from MH blood agar plates in phosphate-buffered saline (PBS) and pelleted by centrifugation at 6,000 × g for 10 min at 4°C. The bacterial pellets were suspended in labeling medium (Eagle’s minimal essential medium minus methionine [ICN Biomedicals, Inc., Aurora, Ohio] supplemented with 1% dialyzed bovine serum) to an optical density of 0.3 at 540 nm. The bacterial suspensions (3 ml of each sample) were equilibrated at 37°C for 10 min and then incubated between 43 and 46°C for 1, 3, 5, or 10 min to induce a heat shock response. [35S]methionine (25 μCi/ml) was added to all of the cultures, which were then incubated for an additional 15 min, to identify the proteins whose synthesis was increased or enhanced following the heat shock. The labeling period was terminated by a chase with unlabeled methionine (1 mM, final concentration) and placement of the cultures on ice. Bacterial cells were pelleted by centrifugation at 6,000 × g at 4°C, washed twice in PBS, and suspended in 50 μl of water. All samples were stored at −20°C. C. jejuni cocultured with INT 407 epithelial cells was also labeled with [35S]methionine as previously described (19).
One- and two-dimensional gel electrophoresis.
One-dimensional gel electrophoresis was performed by mixing the same volume of each bacterial suspension with double-strength electrophoresis sample buffer. Samples were placed in boiling water for 5 min and allowed to cool to room temperature. Proteins were resolved in sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gels by using the discontinuous buffer system described by Laemmli (21). Two-dimensional gel electrophoresis was performed as described elsewhere (26). Labeled bacterial proteins were visualized by autoradiography. Analyses of one- and two-dimensional autoradiographs were performed by using the Multi-Analyst and Melanie II software packages (Bio-Rad Laboratories, Richmond, Calif.).
Construction and screening of the C. jejuni genomic DNA library.
Chromosomal DNA was isolated from C. jejuni F38011 as previously described (24). C. jejuni chromosomal DNA partially digested with Sau3AI was ligated into the unique BamHI site of pBluescriptII SK+ (pBSKII+) with T4 DNA ligase (Boehringer Mannheim, Indianapolis, Ind.) in an overnight incubation at 14°C under the conditions described by the supplier. To prevent autoligation of the vector, the linearized pBSKII+ plasmid vector was treated with calf intestinal alkaline phosphatase (Promega, Madison, Wis.) prior to the ligation reaction. The ligated products were precipitated by addition of 0.5 volume of 7.5 M ammonium acetate and 2.5 volumes of 100% ethanol. The precipitated DNA was washed once with 70% ethanol, air dried, and suspended in water. The DNA was then transformed into E. coli XL1-Blue by electroporation with a Gene Pulser (Bio-Rad Laboratories).
E. coli transformants were transferred to nitrocellulose membranes and screened with an antiserum prepared against whole-cell C. jejuni cultured with epithelial cell monolayers as described previously (31). Briefly, membranes with bacterial colonies were placed on chloroform-saturated Whatman paper for 15 min, air dried, incubated in a bacterial cell lysis buffer, and rinsed. Membranes were then incubated for 18 h at 4°C with a 1:250 dilution of the rabbit anti-C. jejuni serum in PBS (pH 7.4)–0.01% Tween 20 containing 20% bovine serum and an E. coli extract (Protoblot; Promega) to reduce the background. Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Organon Teknika Corp., West Chester, Pa.) as the secondary antibody and 4-chloro-1-naphthol (Sigma Chemical Co., St. Louis, Mo.) as the chromogenic substrate.
DNA sequencing.
DNA sequencing was performed with a double-stranded DNA cycle-sequencing kit in accordance with the supplier’s (Life Technologies Inc., Gaithersburg, Md.) instructions. Sequencing primers were synthesized by Ransom Hill Bioscience, Inc. (Ramona, Calif.). Samples were heated to 95°C for 5 min prior to electrophoresis in 8% polyacrylamide–8 M urea sequencing gels in TBE (0.089 M Tris base, 0.089 M boric acid, 0.002 M EDTA [pH 8.0]). After electrophoresis, the gels were transferred to 3MM paper (Whatman), dried, and analyzed by autoradiography.
Analysis of recombinant plasmid-encoded proteins.
In vitro transcription-translation analyses of purified recombinant plasmids were performed with an E. coli S30-coupled Transcription Translation System as described by the supplier (Promega). The translated products were labeled with [35S]methionine (New England Nuclear [NEN], Boston, Mass.) and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE) with the buffer system described by Laemmli (21). Labeled proteins in dried gels were detected by autoradiography.
Bacteriophage infections.
E. coli isolates were cultured in LB broth containing the appropriate antibiotic at 37°C with shaking for 24 h. Overnight bacterial cultures (200 μl) were added to 10 ml of LB broth supplemented with 0.2% maltose and 10 mM MgSO4. Following a 3-h incubation at 37°C with shaking, the bacteria were pelleted by centrifugation at 6,000 × g. The bacterial pellets were suspended in 10 mM MgSO4 to an optical density of 0.5 at 600 nm. Each bacterial suspension (600 μl) was then mixed with 8.5 ml of melted NZY top agar (10 g of NZ amine [casein hydrolysate], 5 g of yeast extract, 5 g of sodium chloride, 2 g of MgSO4 · 7H2O, and 7.5 g of agar per liter) and spread onto a 150-mm-diameter plate of NZY agar. Bacteriophage infections were performed as described elsewhere (Stratagene, La Jolla, Calif.). Alternatively, phage stocks (20 μl) were spotted onto the surface of NZY top agar and the plates were incubated at 37°C for 24 h. As E. coli dnaJ mutants are unable to support bacteriophage λ replication, complementation of an E. coli dnaJ mutant with a recombinant plasmid was judged by the ability of bacteriophage λ to form plaques.
Southern hybridization.
Southern hybridization of C. jejuni chromosomal DNA with a 558-bp fragment that was amplified by PCR from plasmid pMEK20 was performed under conditions described elsewhere (24). The gel-purified DNA was nick translated with [α-32P]dCTP (NEN) by using a nick translation kit from Promega. C. jejuni chromosomal DNA was digested with restriction endonucleases under the conditions described by the supplier (New England Biolabs, Beverly, Mass.), fractionated in 0.8% agarose gels (16 h at 25 V) in TBE buffer, and transferred to GeneScreen hybridization transfer membranes (NEN) by vacuum blotting with a VacuGene vacuum blotting apparatus in accordance with the manufacturer’s (Pharmacia) recommendations. The DNA was fixed to the membrane by UV cross-linking using a GS Gene Linker UV chamber (Bio-Rad).
Isolation of two isogenic C. jejuni dnaJ mutants.
The dnaJ gene in C. jejuni F38011 was disrupted by homologous recombination as described elsewhere (20). Briefly, a 558-bp internal fragment of the dnaJ gene from C. jejuni F38011 was amplified by PCR and cloned into the pCRII cloning vector (TA Cloning System; Invitrogen, San Diego, Calif.). The cloned insert was excised by restriction endonuclease digestion with EcoRI, gel purified, and ligated into the pBSKII+ vector containing a Campylobacter kanamycin resistance gene. The pBSKII+ vector was digested with EcoRI and treated with calf intestinal alkaline phosphatase prior to ligation. The resultant plasmid was introduced into C. jejuni F38011 by electroporation and acted as a suicide vector for delivery of the internal fragment of the dnaJ gene into the C. jejuni chromosomal gene through allelic exchange. Two potential C. jejuni dnaJ insertional mutants were identified by the acquisition of kanamycin resistance. Both of the C. jejuni DnaJ null mutants were confirmed by Southern hybridization analyses.
Plating efficiency.
C. jejuni was harvested from MH blood agar plates in PBS, pelleted by centrifugation at 6,000 × g, and suspended in PBS to an optical density of 0.2 at 540 nm. Serial dilutions of each bacterial suspension were plated on MH blood agar plates, and the resultant colonies were counted after a 3-day incubation period at the appropriate temperature.
In vivo colonization studies.
Day-of-hatch Leghorn chickens (Hy-Line W-36) were obtained from a commercial hatchery (Hy-Line International, Bryan, Tex.) and placed in electrically heated commercial brooder batteries located within a biological hazard isolation unit. Five chickens were euthanized on arrival at the laboratory and determined to be free of Campylobacter isolates by aseptic removal and culturing of the cecal contents on Campy-Cefex agar plates (34). Chickens were provided water and a balanced, unmedicated corn-soybean ration ad libitum. Food and water were withheld for 2 to 3 h prior to challenge but given ad libitum immediately after challenge. For challenge, C. jejuni was cultured in a microaerophilic atmosphere on Campy-Cefex agar plates for 48 h at 42°C and then harvested in PBS. Previous experiments revealed that the heat shock response of C. jejuni is exhibited at 43°C and higher temperatures. Each chicken was given 1 ml of a suspension containing approximately 104 to 106 viable C. jejuni bacteria by oral gavage. The number of viable C. jejuni bacteria in each suspension was determined by direct plate counting. Seven days after challenge, the chicks were humanely euthanized and the cecal contents were collected aseptically. The cecal contents were serially diluted and 0.1-ml samples of each dilution were spread onto the surfaces of Campy-Cefex agar plates to give final dilutions of 10−2 to 10−6. The number of viable C. jejuni bacteria in each sample was determined by counting the resultant colonies on the Campy-Cefex agar plates following 2 to 3 days of incubation under the conditions described above.
Nucleotide sequence accession number.
The GenBank accession number of the sequence shown in Fig. 4 is AF052661.
FIG. 4.
Nucleotide and deduced amino acid sequences of the C. jejuni dnaJ gene. The deduced amino acid sequence is indicated below the nucleotide sequence in single-letter code. One putative ς32 (−35 [CTTGTAA] and −10 [CTTTAA]) and two putative ς70 promoter elements are indicated by the rectangles and ovals, respectively. The proposed ribosome-binding site (AGGA) is overlined. The two in-frame translational stop codons, both of which are ochre codons, are underlined. A possible terminator is indicated by converging arrows over an inverted repeat. The circled cysteines and glycines make up the cysteine-rich repeat motif (Cys-X-X-Cys-X-Gly-X-Gly) characteristic of DnaJ proteins.
RESULTS
Identification of C. jejuni Hsps.
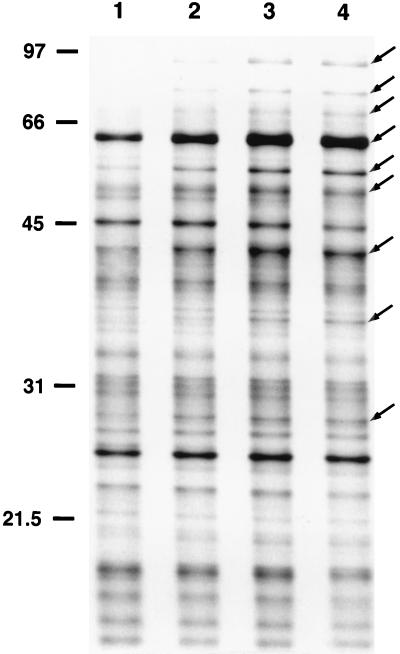
Hsps are defined as those proteins whose synthesis is dramatically induced at high temperatures. To identify C. jejuni Hsps, bacteria were pulse-labeled with [35S]methionine at 37°C or at elevated temperatures (43 or 46°C) and proteins were separated by one-dimensional gel electrophoresis. A typical autoradiograph showing the heat shock response of C. jejuni is presented in Fig. 1. C. jejuni Hsps were identified by computer analysis of the autoradiographs. Nine C. jejuni Hsps were detected when the bacteria were shifted from 37 to 43°C, as judged by [35S]methionine labeling coupled with SDS-PAGE analysis (Fig. 1). The molecular masses of these proteins ranged from 28 to 93 kDa. No differences were observed in the protein synthetic profiles of bacteria shifted from 37 to 43 or 46°C (data not shown). In addition, no differences were noted in the heat shock responses of C. jejuni bacteria which were harvested after a 24-h incubation period from MH broth cultures or from MH blood agar plates (data not shown). Therefore, additional assays were conducted with C. jejuni harvested from MH blood agar plates after a 24-h incubation period.
FIG. 1.
Autoradiograph of a one-dimensional gel of proteins synthesized by C. jejuni M129. C. jejuni bacteria were preincubated at 37°C for 10 min and then either maintained at 37°C for 10 min (lane 1) or shifted to 43°C for 1, 3, or 5 min (lanes 2 to 4, respectively) to identify proteins whose synthesis is increased following a rapid temperature upshift. Proteins were labeled with [35S]methionine for 15 min at either 37°C (lane 1) or 43°C (lanes 2 to 4). C. jejuni Hsps ranging in molecular mass from 28 to 93 kDa are indicated on the right (arrows). The positions of molecular mass standards (sizes are in kilodaltons) are indicated on the left.
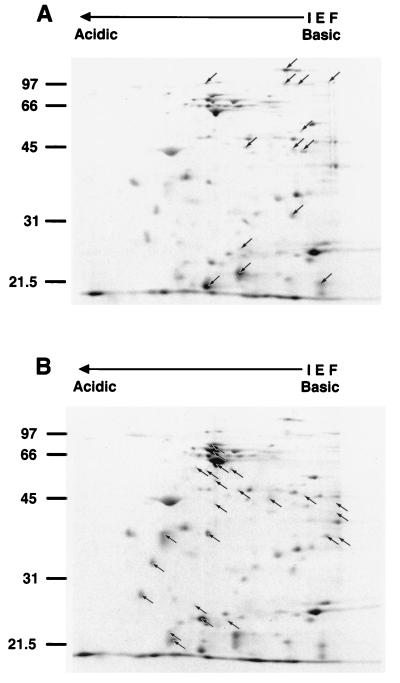
To further characterize the Hsps synthesized by C. jejuni, bacteria cultured at 37°C were pulse-labeled with [35S]methionine for 15 min at either 37, 43, or 46°C and analyzed by two-dimensional gel electrophoresis coupled with autoradiography. A typical autoradiograph showing the heat shock response of C. jejuni is presented in Fig. 2. Twenty-four C. jejuni Hsps were identified (Fig. 2B). The molecular masses of these Hsps ranged from 22 to 77.4 kDa (Table 1). As observed with other pathogens (3, 12, 15), many of the C. jejuni Hsps were detectable prior to the temperature upshift but were preferentially synthesized immediately following heat shock. The apparent molecular masses of the major Hsps detected by two-dimensional gel electrophoresis corresponded to those identified by one-dimensional gel electrophoresis. Again, the heat shock response of C. jejuni was found to be essentially the same regardless of whether the temperature was elevated from 37 to 43°C or from 37 to 46°C (data not shown). Several C. jejuni proteins were also identified whose synthesis decreased following the sudden temperature upshift (Fig. 2A). The molecular masses of these proteins ranged from 21.2 to 124 kDa (Table 1). These findings suggest that the heat shock response of C. jejuni, with respect to the number and apparent molecular masses of proteins whose synthesis is increased or decreased, resembles that of other, more extensively characterized enteric pathogens (15).
FIG. 2.
Autoradiograph of a two-dimensional gel of proteins synthesized by C. jejuni F38011. C. jejuni bacteria were preincubated at 37°C for 10 min and then either maintained at 37°C for 10 min or shifted to 46°C for 10 min to identify proteins whose synthesis is either increased or decreased following a temperature upshift. Proteins were labeled with [35S]methionine for 15 min at either 37°C (A) or 46°C (B) following the temperature upshift. Fourteen proteins were identified whose synthesis was decreased following the temperature upshift (A, arrows), and 24 proteins were identified whose synthesis was increased following the temperature upshift (B, arrows). The positions of molecular mass standards (sizes are in kilodaltons) are indicated on the left. IEF, isoelectric focusing.
TABLE 1.
Altered protein synthetic profile of C. jejuni F38011 in response to heat shock
| Protein type | Molecular masses (kDa) |
|---|---|
| Induceda | 77.4, 73.5, 61, 58.2, 57, 56, 51.9, 48.2, 45.7, 44.2, 43, 42.6, 41.2, 37.5, 37.3, 37, 36.8, 33, 28.3, 26.5, 24.8, 24.5, 23, 22 |
| Suppressedb | 124, 98.6, 97.3, 96, 96, 52, 45, 44.2, 43.8, 32, 25.5, 22.5, 21.5, 21.2 |
Indicates the apparent molecular masses of the 24 proteins for which synthesis was increased in response to a sudden upshift in temperature.
Indicates the apparent molecular masses of the 14 proteins for which synthesis was decreased in response to a sudden upshift in temperature.
Characterization of recombinant plasmid pMEK20.
Several immunoreactive colonies were identified by screening a C. jejuni genomic DNA plasmid library with antiserum prepared against whole C. jejuni cells incubated with epithelial cells (Cj+INTs). In the course of sequencing several of the recombinant plasmids, one was chosen for further characterization based on the putative identification of a gene whose deduced amino acid sequence shared similarity with those of proteins that belong to the Hsp-40 family of molecular chaperones. The identification of a gene coding for a putative Hsp provided the opportunity to gain a better understanding of the importance of a thermal stress response in allowing C. jejuni to grow at elevated temperatures. The question of whether an Hsp plays a role in enabling C. jejuni to colonize the intestinal tracts of chickens could also be addressed by constructing a null mutation in a gene encoding an Hsp.
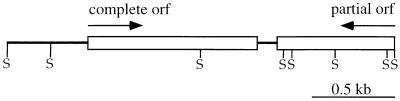
Sequencing of the entire 2.4-kb insert contained within recombinant plasmid pMEK20 revealed the presence of one partial open reading frame (ORF) of 695 bp that was continuous and in frame with the β-galactosidase-encoding gene within the pBSKII vector and a second complete ORF 1,119 bp long (Fig. 3). The deduced amino acid sequence of the complete ORF shared 56.74 to 66.76% similarity with DnaJ proteins from a number of bacteria (mean percentage ± standard deviation, 60.05 ± 3.2 [n = 10]). To determine whether the Cj+INTs serum contained antibodies reactive to DnaJ from C. jejuni, whole-cell lysates of E. coli XL1-Blue harboring the pBSKII+ parental or pMEK20 recombinant plasmid were subjected to SDS-PAGE coupled with immunoblot analysis. Although an immunoreactive band of 33 kDa was observed with the Cj+INTs serum, the predicted molecular mass of DnaJ was estimated to be approximately 42 kDa (data not shown). Based on this finding, the immunoreactivity of the E. coli transformant harboring recombinant plasmid pMEK20 was hypothesized to be a result of the expression of the partial ORF from the lacZ promoter contained within the pBSKII+ cloning vector. To identify the 5′ end of the gene coding for the partial ORF, a C. jejuni genomic DNA λ phage library was screened with oligonucleotide primers. This strategy resulted in the isolation of two reactive plaques whose DNA inserts were subsequently sequenced. Both inserts contained fragments of C. jejuni DNA that overlapped the insert contained within recombinant plasmid pMEK20. We identified a gene of 1,263 nucleotides that is capable of coding for a protein of 420 amino acids with an Mr of 45,267 (data not shown). A search of the PIR and SwissProt databases with the deduced 420-amino-acid sequence revealed that it exhibited 96.7% identity with the major outer membrane protein (MOMP) from C. jejuni 2483. Each of the antisera, which were generated in rabbits against either whole-cell or outer membrane protein extracts from C. jejuni, contains antibodies that react with the 43-kDa MOMP from C. jejuni. The molecular characterization of the gene encoding the MOMP from C. jejuni will be described elsewhere, as it is not the focus of this report.
FIG. 3.
Physical map of pMEK20. The 2,237-bp insert contains one complete ORF of 1,119 bp encoding a protein that shares similarity with DnaJ protein from other organisms and one partial ORF of 692 bp. The partial ORF represents the 3′ end of the gene that encodes the MOMP from C. jejuni. The arrows indicate the directions in which the genes are transcribed. S, Sau3AI restriction endonuclease site.
Molecular characterization of the gene coding for DnaJ from C. jejuni.
The complete, 1,119-bp ORF contained within recombinant plasmid pMEK20 is capable of encoding a protein of 372 amino acids with a calculated molecular mass of 41,436 Da (Fig. 4). The ORF begins with a valine (GTG) initiation codon and is terminated by two in-frame stop (TAA) codons. A putative ribosome-binding site is located 5 nucleotides upstream of the proposed valine initiation codon. Also identified upstream of the proposed valine initiation codon and ribosome-binding site were putative ς32 and ς70 promoter elements. Downstream of the two in-frame stop codons is a putative terminator sequence that is capable of forming a 9-bp inverted repeat. The G+C content of the gene encoding the 41-kDa protein is 34 mol%, which is consistent with the base composition of Campylobacter DNA (28).
The deduced amino acid sequence of the gene capable of encoding the 41-kDa protein was compared to sequences in the PIR and SwissProt databases. The deduced amino acid sequence of the 41-kDa protein exhibited similarity with DnaJ proteins from a number of bacteria, including E. coli (60.8%), Salmonella typhimurium (59.6%), and Clostridium acetobutylicum (58.9%) (Fig. 5). The deduced amino acid sequence of the C. jejuni 41-kDa protein also contains four cysteine-rich repeats (Cys-X-X-Cys-X-Gly-X-Gly) located between Cys-147 and Gly-206 (Fig. 4 and 5). These four cysteine-rich repeats are characteristic of DnaJ proteins found in other bacteria (2).
FIG. 5.
Alignment of the deduced amino acid sequence of DnaJ from C. jejuni (dnaj_cj) with the DnaJ proteins from E. coli (dnaj_ecoli), S. typhimurium (dnaj_salty), and C. acetobutylicum (dnaj_cloab). Gaps, indicated by dashes, were introduced to obtain maximal alignment. Amino acids are indicated by single-letter codes and are numbered from the first valine or methionine residue. Identical or conserved amino acid residues are boxed. Comparison of the C. jejuni DnaJ amino acid sequence to the DnaJ proteins from E. coli, S. typhimurium, and C. acetobutylicum yielded amino acid sequence similarity values of 60.8, 58.9, and 59.6%, respectively.
In vitro transcription-translation analysis of recombinant plasmid pMEK20.
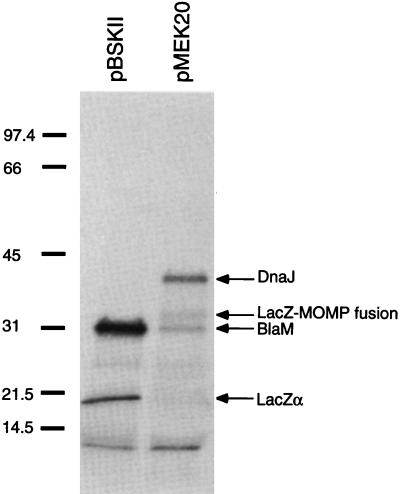
The products synthesized by pMEK20 were labeled with [35S]methionine by using the in vitro E. coli cell-free transcription-translation (S30) system (Fig. 6). Aliquots of the in vitro assay were separated by SDS–12.5% PAGE, and labeled proteins were detected by autoradiography. Examination of the autoradiograph revealed that pMEK20 synthesized two products with relative molecular masses of 41 and 33 kDa and a third product of 31 kDa, which was also synthesized by the pBSKII+ parental plasmid. The 41-kDa band likely represents the DnaJ protein, the 32-kDa band represents a LacZ-MOMP fusion protein, and the 31-kDa band represents the BlaM product encoded by the pBSKII vector. Based on this analysis, the gene coding for the 41-kDa product is hypothesized to be expressed from an endogenous promoter, given that the gene is located in the opposite orientation from the lacZ promoter of the pBSKII+ cloning vector. In support of this hypothesis, a ribosome-binding site and putative promoter elements were identified upstream of the gene’s proposed valine initiation codon.
FIG. 6.
In vitro transcription-translation analysis of recombinant plasmid pMEK. Translated products were labeled with [35S]methionine and separated by SDS–12.5% PAGE, and products were visualized by autoradiography. pBluescriptII SK+ (pBSKII) is the parental plasmid, and pMEK20 (dnaJ+) is a recombinant plasmid. The products translated by one or both of the plasmids are given on the right. The positions of molecular mass standards (sizes are in kilodaltons) are indicated on the left.
Complementation of an E. coli dnaJ mutant.
E. coli dnaJ mutants are unable to support plaque formation by bacteriophage λ at any temperature (32). To determine whether the C. jejuni gene encoding the 41-kDa protein could complement an E. coli dnaJ mutant, bacteriophage λ infection studies were performed with an E. coli dnaJ deletion mutant. In contrast to transformants harboring the parental pBSKII plasmid, plaques were observed following infection of the E. coli dnaJ mutant harboring the pMEK20 recombinant plasmid (data not shown). Based on this finding and the additional data presented above, the C. jejuni gene encoding the 41-kDa protein was designated dnaJ.
C. jejuni dnaJ gene copy number.
Southern hybridization analysis of chromosomal DNAs from four different C. jejuni isolates (F38011, 78-27, 8116, and 33560) was performed with a gel-purified 558-bp fragment that lies internal to the dnaJ gene within pMEK20 (data not shown). The size and number of bands observed corresponded to the physical map of the insert in pMEK20, regardless of the enzyme employed for digestion. These findings suggest that there is a single copy of the dnaJ gene in the genome of C. jejuni isolates.
Phenotypic properties of C. jejuni dnaJ mutants.
Although E. coli DnaJ mutants are viable and capable of forming colonies on agar plates at 37°C, DnaJ is required for bacterial survival at higher temperatures, as evidenced by the inability of dnaJ insertion mutants to form colonies at 43°C. To determine whether DnaJ is required for the growth of C. jejuni at higher temperatures, two C. jejuni dnaJ mutants that contained insertional disruptions of the dnaJ gene were isolated. Disruption of the dnaJ gene in the two C. jejuni isolates, designated dnaJ mutants A and B, was confirmed via Southern hybridization analysis (data not shown). No significant differences were noted between the abilities of the C. jejuni parent and DnaJ mutants to form colonies at temperatures of less than 45.0°C. However, the ability of both of the DnaJ mutants to form colonies at 46°C was severely diminished (Table 2), indicating that DnaJ plays an important role in the C. jejuni thermal stress response.
TABLE 2.
Plating efficiency of C. jejuni strains
| Strain | Mean no. of viable bacteriaa ± SD
|
Plating efficiencyb (%) | ||
|---|---|---|---|---|
| 37°C | 42°C | 46°C | ||
| F38011 | (7.5 ± 2.6) × 108 | (6.0 ± 2.3) × 108 | (5.8 ± 0.8) × 108 | 77.3 |
| F38011 dnaJ mutant A | (1.1 ± 0.8) × 109 | (2.5 ± 2.1) × 109 | (3.7 ± 3.2) × 103 | 0.0003 |
| F38011 dnaJ mutant B | (7.8 ± 3.2) × 108 | (1.5 ± 0.5) × 109 | (3.8 ± 2.6) × 104 | 0.005 |
Data generated from a typical experiment. Serial dilutions of each bacterial suspension were plated on MH blood agar plates, and the resultant colonies were counted after 3 days of incubation at the appropriate temperature (five plates at each temperature). The assay was repeated three times to ensure the reproducibility of the results.
Plating efficiency was determined by dividing the number of CFU at 46°C by the number of CFU at 37°C and multiplying the result by 100.
In vivo colonization of chickens.
As Hsps play an important role in enabling bacteria to cope with physiological stresses, we hypothesized that synthesis of DnaJ is required for the successful colonization of chickens by C. jejuni. To test our hypothesis, newly hatched Leghorn chickens were infected with wild-type C. jejuni and one of the DnaJ isogenic mutants (Table 3). C. jejuni colonization of the chickens was assessed by plating serial dilutions of the cecal contents 1 week after infection. C. jejuni bacteria were recovered from every chicken infected with F38011, the parental isolate. In contrast, C. jejuni bacteria were not recovered in the cecal contents of any of the chickens infected with the DnaJ mutant, suggesting that C. jejuni must adapt to this in vivo environment by upregulating Hsps.
TABLE 3.
In vivo colonization of Leghorn chickens
| Trial and strain | Infective dose (no. of bacteria) | No. of chickens | Mean no. of viable C. jejuni bacteria recovered/ml of cecal contents ± SD | Range |
|---|---|---|---|---|
| 1 | ||||
| F38011 | 104 | 15 | (1.34 ± 1.30) × 108 | 8.4 × 106–5.0 × 108 |
| F38011 | 106 | 15 | (9.46 ± 7.44) × 107 | 1.4 × 107–2.4 × 108 |
| F38011 dnaJ mutant A | 104 | 15 | NR | |
| F38011 dnaJ mutant A | 106 | 15 | NR | |
| 2 | ||||
| F38011 | 2.2 × 105 | 10 | (1.24 ± 0.78) × 108 | 1.1 × 107–2.4 × 108 |
| F38011 dnaJ mutant A | 2.6 × 105 | 10 | NR |
NR, none recovered.
DISCUSSION
Both eukaryotic and prokaryotic organisms respond to heat and other environmental stresses by inducing the synthesis of a specific subset of proteins called Hsps (22, 23). In this study, the heat shock response of C. jejuni was examined to identify the Hsps of C. jejuni. The heat shock response of C. jejuni resembles that of other bacteria with respect to the relative number and molecular masses of the Hsps. Twenty-four C. jejuni Hsps were identified by performing a typical heat shock experiment and then analyzing the [35S]methionine protein synthetic profile by two-dimensional gel electrophoresis. As with other organisms, many of the C. jejuni Hsps were synthesized prior to heat shock. A number of C. jejuni proteins were also identified whose synthesis was reduced in response to heat shock.
Upon screening of E. coli transformants harboring a C. jejuni genomic DNA library with an antiserum prepared against C. jejuni cocultivated with INT 407 host cells, many immunoreactive clones were identified. One of the transformants harbored recombinant plasmid pMEK20, which contained a 2,237-bp insert of C. jejuni chromosomal DNA. Within the 2.2-kb insert, one complete ORF was found that is capable of encoding a protein with a calculated molecular mass of 41,436 Da. The deduced amino acid sequence of this ORF exhibited similarity to DnaJ proteins from a number of bacteria, including E. coli and S. typhimurium, and contained four cysteine-rich repeats (Cys-X-X-Cys-X-Gly-X-Gly). These cysteine-rich repeats are characteristic of DnaJ proteins from other bacteria, including E. coli (5, 27), S. typhimurium, C. acetobutylicum (6, 25), and Borrelia burgdorferi (2). An E. coli dnaJ mutant was successfully complemented with recombinant plasmid pMEK20 as judged by the ability to support bacteriophage λ replication and plaque formation. These data indicate that the C. jejuni gene encoding the 41-kDa protein is a functional homolog of the E. coli dnaJ gene and was therefore designated dnaJ.
As has been shown with some proteins in E. coli (13), the C. jejuni DnaJ protein is likely initiated with a valine (GTG) initiation codon. In support of this hypothesis, a putative ribosome-binding site is located immediately upstream of the proposed valine initiation codon. In addition, comparison of the C. jejuni DnaJ deduced amino acid sequence with DnaJ proteins from other bacteria revealed a stretch of identical amino acids prior to the first methionine codon in the C. jejuni DnaJ protein. Inspection of the sequence upstream of the proposed valine initiation codon revealed the presence of several possible promoter elements. The sequence CTTGTAA shares six of seven nucleotides with the −35 consensus sequence (CTTGAAA) of E. coli ς32 promoters (15) and four of six nucleotides with the −35 consensus sequence (TTGATA) of E. coli ς70 promoters. Overlapping ς32 and ς70 promoter elements have been identified in E. coli (15). A second putative ς70 promoter was located downstream of the ς32 promoter, whose TTGATT sequence shares five of six nucleotides with the −35 consensus sequence of E. coli ς70 promoters. The dnaKJ operon of Caulobacter crescentus is under the control of two functional promoters, with the ς32 promoter residing 5′ of the ς70 promoter (4). The groEL, groES, and grpE heat shock genes of E. coli are also under the control of ς32 and ς70 promoters, where the latter is used to ensure basal levels of their expression (11). It is unknown whether the dnaJ gene is transcribed from its own promoter in C. jejuni, as Northern hybridization or primer extension analyses were not performed. The dnaJ gene, however, has been shown to be transcribed in B. burgdorferi alone or as part of a larger transcript containing a dnaK homolog (2). The role and significance of the multiple promoter elements in the transcription of the dnaJ gene in C. jejuni are unknown.
Domestic poultry, which is frequently colonized by Campylobacter, represents one of the most common sources of infection with C. jejuni in the United States and other countries. We hypothesized that Hsps play an important role, albeit indirectly, in the colonization of chickens by C. jejuni by enabling the pathogen to adapt from a free-living environment to a new, and possibly stressful, environment. Specifically, it was theorized that C. jejuni is subjected to various stresses upon encountering an avian host, including changes in temperature and pH upon passage through the stomach to the cecum. Previous investigators have noted a decrease in the viability of dnaK (18) and dnaJ (29, 32) mutants exposed to acidic environments and high temperatures. Confronting these environmental stresses might result in an increase in the synthesis of a number of proteins, resulting in an increase in unfolded polypeptides within the bacterial cell. An increase in the synthesis of Hsps would be necessary for the proper folding of these polypeptides and, therefore, crucial for cellular survival. This hypothesis was tested by constructing a null mutation in the gene coding for DnaJ in C. jejuni. This approach was feasible, as C. jejuni DnaJ mutants could be generated via homologous recombination using a suicide vector harboring an internal fragment of the gene of interest and an antibiotic resistance gene. A C. jejuni DnaJ mutant was incapable of colonizing chickens. This finding represents the first in vivo experimental evidence that an Hsp plays an important role in enabling C. jejuni to colonize chickens. Given that previous studies with other enteric pathogens, such as Salmonella (10, 16), have demonstrated a role of Hsps in bacterium-host cell interactions, it would also be of interest to examine the pathogenic potential of a C. jejuni DnaJ mutant in an in vivo infection model.
ACKNOWLEDGMENTS
We thank Kit Tilly for providing E. coli isolates MF670 and BR3672 and for helpful discussions. We also thank Steve Garvis and Sean Gray for reading the manuscript and assistance with the preparation of figures. Finally, we thank Louis Mallavia and Ray Larsen for critically reviewing the manuscript.
This work was supported by grants from the NIH (1R01 DK50567-01A1) and the USDA National Research Initiative Competitive Grants Program (USDA/NRICGP, no. 9601496) awarded to M.E.K. B. Kim was supported by the Howard Hughes Fellowship Program.
REFERENCES
- 1.Ang D, Georgopoloulos C. The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J Bacteriol. 1989;171:2748–2755. doi: 10.1128/jb.171.5.2748-2755.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzola J, Luft B J, Gorgone G, Peltz G. Characterization of a Borrelia burgdorferi dnaJ homolog. Infect Immun. 1992;60:4965–4968. doi: 10.1128/iai.60.11.4965-4968.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnosti D N, Singer V L, Chamberlin M J. Characterization of heat shock in Bacillus subtilis. J Bacteriol. 1986;168:1243–1249. doi: 10.1128/jb.168.3.1243-1249.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avedissian M, Lessing D, Gober J W, Shapiro L, Gomes S L. Regulation of the Caulobacter crescentus dnaKJ operon. J Bacteriol. 1995;177:3479–3484. doi: 10.1128/jb.177.12.3479-3484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell J C A, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene: a gene that encodes a heat shock protein. J Biol Chem. 1986;261:1782–1785. [PubMed] [Google Scholar]
- 6.Behrens S, Narberhaus F, Bahl H. Cloning, nucleotide sequence and structural analysis of the Clostridium acetobutylicum dnaJ gene. FEMS Microbiol Lett. 1993;114:53–60. doi: 10.1016/0378-1097(93)90141-n. [DOI] [PubMed] [Google Scholar]
- 7.Berndtson E, Danielsson-Tham M L, Engvall A. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int J Food Microbiol. 1996;32:35–47. doi: 10.1016/0168-1605(96)01102-6. [DOI] [PubMed] [Google Scholar]
- 8.Berndtson E, Tivemo M, Engvall A. Distribution and numbers of Campylobacter in newly slaughtered broiler chickens and hens. Int J Food Microbiol. 1992;15:45–50. doi: 10.1016/0168-1605(92)90134-o. [DOI] [PubMed] [Google Scholar]
- 9.Blaser M J, Wells J G, Feldman R A, Pollard R A, Allen J R. Campylobacter enteritis in the United States. A multicenter study. Ann Intern Med. 1983;98:360–365. doi: 10.7326/0003-4819-98-3-360. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier N A, Hefron F. Induction of Salmonella stress proteins upon infection of macrophages. Science. 1990;248:730–732. doi: 10.1126/science.1970672. [DOI] [PubMed] [Google Scholar]
- 11.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 12.Carreiro M M, Laux D C, Nelson D R. Characterization of the heat shock response and identification of heat shock protein antigens of Borrelia burgdorferi. Infect Immun. 1990;58:2186–2191. doi: 10.1128/iai.58.7.2186-2191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draper D E. Translation initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 902–908. [Google Scholar]
- 14.Eddy T J, Gallup G G., Jr Thermal correlates of tonic immobility and social isolation in chickens. Physiol Behav. 1990;47:641–646. doi: 10.1016/0031-9384(90)90071-b. [DOI] [PubMed] [Google Scholar]
- 15.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 16.Hassett D J, Cohen M S. Bacterial adaption to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki Y, Wada C, Yura T. Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol Gen Genet. 1990;220:277–282. doi: 10.1007/BF00260494. [DOI] [PubMed] [Google Scholar]
- 18.Kohler S, Teyssier J, Cloeckaert A, Rouot B, Liautard J-P. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol Microbiol. 1996;20:701–712. doi: 10.1111/j.1365-2958.1996.tb02510.x. [DOI] [PubMed] [Google Scholar]
- 19.Konkel M E, Cieplak W., Jr Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect Immun. 1992;60:4945–4949. doi: 10.1128/iai.60.11.4945-4949.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konkel M E, Garvis S G, Tipton S, Anderson D E, Jr, Cieplak W., Jr Identification and molecular cloning of a gene encoding a fibronectin binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 23.Lindquist S, Craig E A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 24.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narberhaus F, Giebeler K, Bahl H. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J Bacteriol. 1992;174:3290–3299. doi: 10.1128/jb.174.10.3290-3299.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 27.Ohki M, Tamura F, Nishimura S, Uchida H. Nucleotide sequence of the Escherichia coli dnaJ gene and purification of the gene product. J Biol Chem. 1986;261:1778–1781. [PubMed] [Google Scholar]
- 28.Owen R J, Leaper S. Base composition, size and nucleotide sequence similarities of genome deoxyribonucleic acids from species of the genus. Campylobacter. FEMS Microbiol Lett. 1981;12:395–400. [Google Scholar]
- 29.Saito H, Uchida H. Organization and expression of the dnaJ and dnaK genes of Escherichia coli K12. Mol Gen Genet. 1978;164:1–8. doi: 10.1007/BF00267592. [DOI] [PubMed] [Google Scholar]
- 30.Sakakibara Y. The dnaK gene of Escherichia coli functions in the initiation of chromosome replication. J Bacteriol. 1988;170:972–979. doi: 10.1128/jb.170.2.972-979.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwan T G, Schrumpf M E, Hinnebusch B J, Anderson D E, Konkel M E. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J Clin Microbiol. 1996;34:2483–2492. doi: 10.1128/jcm.34.10.2483-2492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sell S M, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanker S, Lee A, Sorrell T C. Horizontal transmission of Campylobacter jejuni amongst broiler chicks: experimental studies. Epidemiol Infect. 1990;104:101–110. doi: 10.1017/s0950268800054571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stern N J, Wojton B, Kwiatek K. A differential-selective medium and dry ice-generated atmosphere for recovery of Campylobacter jejuni. J Food Prot. 1992;55:514–517. doi: 10.4315/0362-028X-55.7.514. [DOI] [PubMed] [Google Scholar]
- 35.Tilly K, Yarmolinsky M. Participation of Escherichia coli heat shock proteins DnaJ, DnaK, and GrpE in P1 plasmid replication. J Bacteriol. 1989;171:6025–6029. doi: 10.1128/jb.171.11.6025-6029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker R I, Caldwell M B, Lee E C, Guerry P, Trust T J, Ruiz-Palacios G M. Pathophysiology of Campylobacter enteritis. Microbiol Rev. 1986;50:81–94. doi: 10.1128/mr.50.1.81-94.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]