Abstract
Nonsense-mediated decay (NMD) is an RNA surveillance mechanism that degrades mRNAs containing premature termination (nonsense) codons. The second signal for this pathway in mammalian cells is an intron that must be at least ∼55 nucleotides downstream of the nonsense codon. Although the functional significance of this ‘–55 boundary rule’ is not known, it is widely thought to reflect the important role of an exon junction protein complex deposited just upstream of exon–exon junctions after RNA splicing. Here we report that a T-cell receptor (TCR)-β gene did not conform to this rule. Rather than a definitive boundary position, nonsense codons had a polar effect, such that nonsense codons distant from the terminal downstream intron triggered robust NMD and proximal nonsense codons caused modest NMD. We identified a region of the TCR-β gene that conferred this boundary-independent polar expression pattern on a heterologous gene. Collectively, our results suggest that TCR-β transcripts contain one or more sequence elements that elicit an unusual NMD response triggered by a novel second signal that ultimately causes boundary-independent polar regulation. TCR genes may have evolved this unique NMD response because they frequently acquire nonsense codons during normal development.
INTRODUCTION
Eukaryotic cells possess an RNA surveillance pathway known as nonsense-mediated decay (NMD) that reduces the abundance of mRNAs harboring nonsense codons. In mammalian cells, two signals are required for NMD: a nonsense codon and a downstream intron (Maquat, 1995; Li and Wilkinson, 1998; Hentze and Kulozik, 1999). It is perplexing that an intron is the second signal for NMD, as introns are spliced in the nucleus and, until recently, translation was thought to occur only in the cytoplasm. A possible solution to this paradox originally proposed by Cheng et al. (1994) and later modified by Carter et al. (1996) and Thermann et al. (1998) is that the second signal is not the intron but instead ‘a mark’ left after intron splicing. A protein complex that is deposited 20–24 nucleotides (nt) upstream of exon–exon junctions after RNA splicing may be this mark (Le Hir et al., 2000a). This exon junction (EJ) protein complex contains the general splicing activator RNPS1, the RNA export factor Aly/REF, the shuttling protein Y14, the nuclear matrix-localized serine–arginine-containing protein SRm160, the oncoprotein DEK and UPF proteins (Le Hir et al., 2000b, 2001; Shyu and Wilkinson, 2000; Kim et al., 2001a,b; Lykke-Andersen et al., 2001). Experiments in which individual members of the EJ complex were artificially tethered downstream of a 3′-terminal stop codon indicated that RNPS1, UPF proteins and possibly Y14 can trigger an NMD-like response (Lykke-Andersen et al., 2000, 2001).
Support for the notion that the EJ complex is a second signal for NMD is the ‘–55 boundary rule’, which states that only nonsense codons at least ∼55 nt upstream of the terminal intron can trigger NMD. This rule is based on the analysis of the effect of nonsense codons at different positions in triose phosphate isomerase (TPI), β-globin, mouse major urinary protein and gpx-1 transcripts (Nagy and Maquat, 1998). This rule is explained by the location of the EJ complex on spliced mRNAs: a translocating ribosome would displace an EJ complex upstream of the last exon–exon junction before the ribosome could reach a stop codon located <55 nt from the exon–exon junction. Without the EJ complex, there is no second signal for NMD and hence intron–proximal nonsense codons do not elicit NMD. This –55 boundary rule also explains why termination codons that allow for the production of normal full-length proteins usually do not elicit NMD: most are in the final exon and 98% of those that reside within upstream exons are <50 nt upstream of the 3′-most intron (Nagy and Maquat, 1998).
We previously showed that although mouse T-cell receptor (TCR)-β transcripts require at least one intron downstream of a stop codon to trigger NMD, a nonsense codon can be closer than 55 nt upstream of the IVS-JCβ intron (when situated in the 3′-most terminal position) and still elicit NMD (Carter et al., 1996). To gain further insight into this exception to the –55 boundary rule, we have examined whether it is a conserved feature of TCR-β introns and how it is mediated.
RESULTS
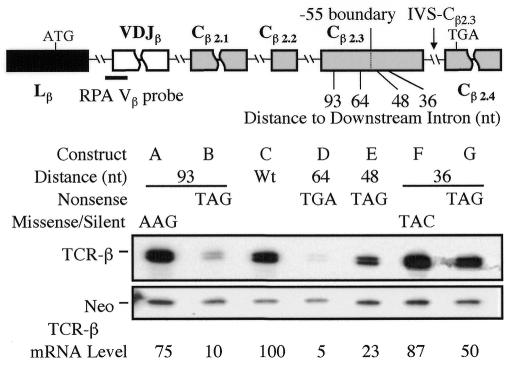
To test whether the –55 boundary rule applies to TCR genes, we introduced nonsense codons at various positions upstream of the terminal intron (IVS-Cβ2.3) in a functionally rearranged Vβ8.1Dβ2Jβ2.3Cβ2 TCR-β gene that we previously showed is strongly downregulated by NMD in transfected T cells and HeLa cells (Carter et al., 1995, 1996; Li et al., 1997). RNase protection analysis (RPA) demonstrated that nonsense codons both upstream (nt –64 and –93) and downstream (nt –48 and –36) of the nt –55 ‘boundary’ elicited NMD (Figure 1). In contrast, missense mutations did not affect mRNA levels significantly, demonstrating the specificity of the response. These results indicated that the –55 boundary rule does not apply to the intron IVS-Cβ2.3.
Fig. 1. Boundary-independent polar NMD regulation. The upper panel shows the positions of nonsense and missense mutations introduced in the Cβ2.3 exon [the numbers refer to the distance (in nt) between the end of the mutated codon and the exon–intron boundary; the size of the Cβ2.3 exon is 107 nt]. The lower panel shows RPA of total cellular RNA (10 µg) from HeLa cells transiently transfected with the constructs indicated. The TCR-β mRNA band protected by the TCR-β Vβ probe (position shown) is ∼72 nt, which is the expected size based on the positions of the splice sites in TCR-β mRNA. TCR-β mRNA levels were determined by normalizing against the levels of neomycin (neo) mRNA, which was expressed from the same plasmids as was TCR-β mRNA. Similar results were obtained for each construct in at least three independent transfection experiments.
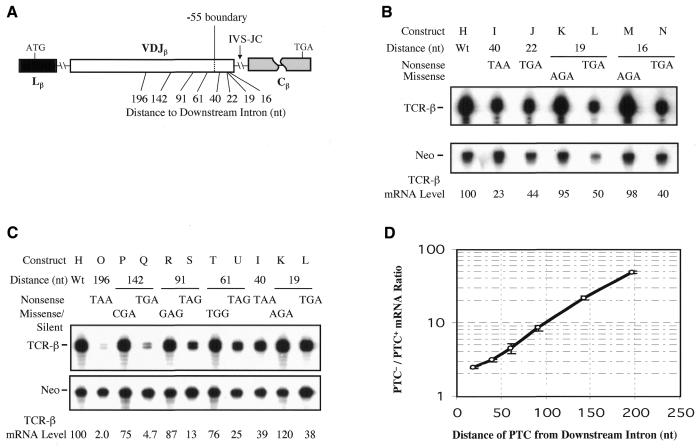
To test whether the failure to observe the –55 boundary rule is a conserved feature of TCR introns, we introduced nonsense codons upstream of another intron, IVS-JCβ. We made IVS-JCβ the terminal intron by deleting the downstream Cβ introns (Figure 2A). We found that nonsense codons at various positions downstream of the –55 nt boundary (40, 21, 19 and 16 nt upstream of IVS-JCβ) caused TCR-β mRNA downregulation, whereas missense mutations did not (Figure 2B). We conclude that at least two introns in TCR-β gene do not abide by the –55 boundary rule.
Fig. 2. Boundary-independent polar NMD regulation is a conserved feature of TCR-β introns. (A) The position of nonsense and missense mutations introduced in the VDJ exon (its total length is 354 nt). (B and C) RPA performed and quantitated as in Figure 1. (D) The effect of premature termination codon (PTC) position on the degree of NMD (PTC–/PTC+ mRNA ratio), based on at least three experiments for each construct, some of which are shown in (B) and (C); error bars indicate standard deviations.
Another novel feature of the TCR-β NMD response that we observed was that nonsense codons elicited a polar effect, such that more 5′ nonsense codons caused stronger NMD than 3′ nonsense codons did. This was found for nonsense codons upstream of both IVS-Cβ2.3 (Figure 1) and IVS-JCβ (Figure 2C and D). For IVS-JCβ, we observed a strong polar effect, such that the 5′-most nonsense codon (nt –196) elicited a dramatic (∼50-fold) reduction in mRNA levels, intermediate position nonsense codons (nt –91 to –142) triggered less of an mRNA decrease (8- to 21-fold) and 3′ nonsense codons (nt –16 to –61) caused only modest NMD (2- to 4-fold). Because intron–proximal nonsense codons elicited modest and somewhat variable degrees of NMD, we cannot be sure that the polarity extended to this region, but clearly there was a polar effect over much of the rest of the VDJ exon.
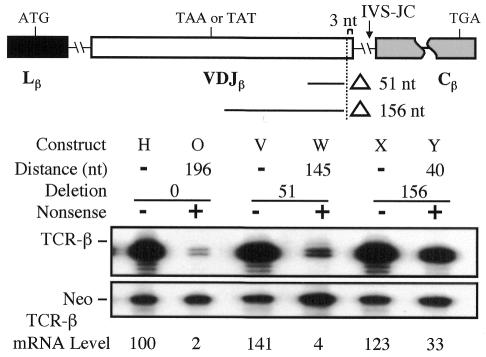
As an independent test of whether TCR-β transcripts are both polar regulated and fail to observe the –55 boundary rule, we performed deletion analysis (Figure 3). We deleted either 51 or 156 nt from the VDJ exon, which moved a TAA nonsense mutation originally in the middle of the exon (nt –196) closer to the downstream intron (nt –145 and –40). This decreased the degree of NMD, consistent with a polar mechanism. Importantly, the deletion that brought the nonsense codon to nt –40 still allowed NMD, providing further evidence that TCR-β transcripts do not obey the –55 boundary rule. These results indicated that both the polar effect and the failure to observe the –55 boundary rule are independent of nonsense codon context.
Fig. 3. Boundary-independent polar NMD regulation is independent of nonsense codon context. The upper panel shows the deletions introduced in the VDJ exon. The lower panel shows RPA performed and quantitated as in Figure 1. Similar results were obtained in at least two independent transfection experiments.
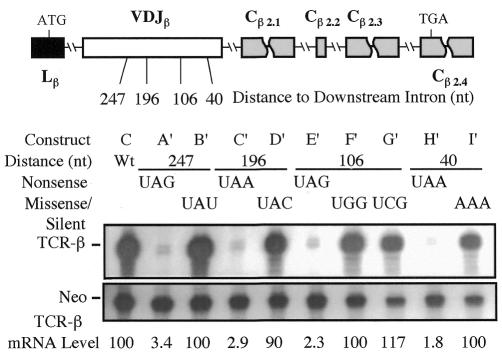
The polar effect we have defined in the preceding experiments is dictated by the distance of a nonsense codon to introns situated in a 3′-terminal position. Because the IVS-JCβ intron is normally not in a 3′-terminal position but rather in an internal position, we elected to ask whether it engendered a polar effect from this internal position. This allowed us to determine whether IVS-JCβ has a polar effect per se or only when in a 3′-terminal position. To test this, we introduced nonsense and corresponding missense or silent mutations in a VDJ exon that had several introns downstream (Figure 4). We found that nonsense codons 247, 196, 106 and 40 nt upstream of the adjacent intron all strongly downregulated mRNA levels (by 30- to 50-fold). This uniform degree of downregulation contrasted with the strong polar effect observed when IVS-JCβ was the terminal intron (Figure 2C). We conclude that the distance between a nonsense codon and the 3′-most terminal intron directs the polar effect.
Fig. 4. The polar effect is dictated by the distance to the 3′-most intron. The upper panel shows the positions of nonsense and missense mutations introduced in the VDJ exon. The lower panel shows RPA performed and quantitated as in Figure 1. Similar results were obtained in at least three independent transfection experiments.
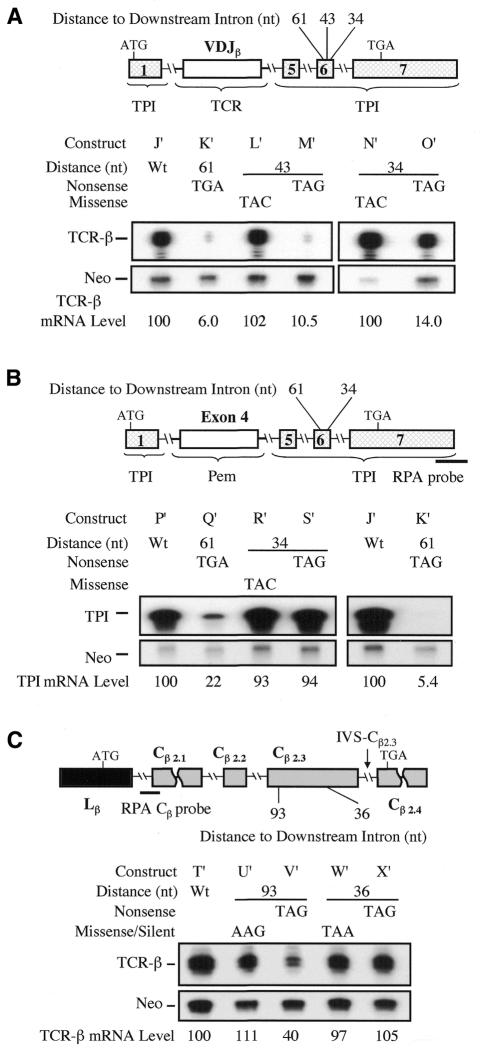
We next determined whether boundary-independent polar regulation is an intrinsic property of TCR or can be conferred on another gene. As a recipient heterologous gene, we chose the TPI gene, which normally observes the –55 boundary rule (Cheng et al., 1994; Zhang et al., 1998). We found that when the TCR VDJ exon and adjacent intron sequences were inserted into the TPI gene, this was sufficient to allow nonsense codons in the penultimate TPI exon to break the –55 boundary rule (Figure 5A). In particular, we found that the nonsense codon either 43 or 34 nt upstream of the 3′-terminal TPI intron elicited strong NMD. Missense mutations at the same positions did not affect mRNA levels. The introduction of TCR-β sequences also caused a polar effect, such that nonsense codons at 61, 43 and 34 nt upstream of the last TPI intron caused ∼15-, ∼10- and ∼7-fold downregulation, respectively. This contrasts with normal TPI transcripts, which are not subject to a polar effect, as nonsense codons at most positions cause uniform (3- to 5-fold) mRNA downregulation (Cheng et al., 1994; Zhang et al., 1998).
Fig. 5. The TCR Vβ8.1Dβ2Jβ2.3 exon and adjacent intron sequences are necessary and sufficient for boundary-independent NMD regulation. (A) The TCR-β VDJ segment triggers boundary-independent NMD. Shown are TCR/TPI chimeric transcripts detected with the Vβ probe described in Figure 1. (B) A control segment of similar size (Pem exon 4 and adjacent intron sequences) does not confer boundary-independent NMD regulation. Shown are Pem/TPI (left panel) and TCR/TPI (right panel) chimeric transcripts detected with the TPI probe shown. (C) Deletion of the TCR-β VDJ region prevents boundary-independent NMD regulation. Shown are TCR-β transcripts detected with the Cβ probe shown. RPA was performed and quantitated as in Figure 1. The TPI mRNA band protected by the TPI probe (position shown) is ∼400 nt. The TCR-β mRNA band protected by the TCR-β Cβ probe (position shown) is ∼60 nt, which is the expected size based on the positions of the splice sites in TCR-β mRNA. Similar results were obtained in at least two independent transfection experiments.
As a control for this experiment, we made a chimeric TPI gene that contained an exon and adjacent intron sequences from a different gene (Figure 5B). We chose exon 4 from the Pem homeobox gene, as this exon is almost identical in size (357 nt) to the VDJβ exon (354 nt). We found that a nonsense codon 61 nt upstream of the 3′-terminal intron in this Pem–TPI chimeric gene triggered mRNA downregulation, whereas a nonsense codon 34 nt upstream failed to elicit NMD, indicating that, unlike the TCR–TPI chimeric gene, the Pem–TPI chimeric gene obeyed the –55 boundary rule.
As another test of whether the VDJ region is necessary to establish independence from the –55 boundary rule, we made a TCR gene that lacked the VDJ exon and adjacent intron sequences (Figure 5C). This VDJ-deleted TCR-β gene obeyed the –55 boundary rule, as a nonsense codon 93 nt upstream of the 3′-terminal intron triggered NMD, whereas a nonsense codon 36 nt upstream did not elicit NMD. We conclude that TCR-β sequences in or near the VDJ exon are sufficient to confer immunity from the –55 boundary rule.
DISCUSSION
In this study we have identified the first vertebrate gene that exhibits a polar effect in response to nonsense codons and does not observe the –55 boundary rule for NMD. The failure to obey the –55 boundary rule (Figures 1 and 2) suggests that the EJ complex positioned at the nt –20 to –24 position upstream of the exon–exon junction in most transcripts is unlikely to be the only signal that triggers TCR NMD. What then is the second signal for TCR NMD? First, it may be the EJ complex deposited at a unique downstream position (closer to the exon–exon junction) in TCR exons. If this were the case, then TCR sequences must be capable of conferring this unique positioning, as the introduction of the TCR VDJ region permitted TPI transcripts to be downregulated by NMD in response to proximal nonsense codons (Figure 5). Secondly, it could be an entirely different protein or protein complex that delivers the second signal for TCR NMD. Some transcripts appear to have one or more proteins bound at the ‘0’ position rather than the nt –24 position after RNA splicing (Le Hir et al., 2000a). It remains to be determined whether this ‘0’ position entity is the EJ complex or a novel protein. Thirdly, the second signal for TCR NMD could be a protein on unspliced or partially spliced mRNA rather than mature mRNA. Given the recent evidence that a proportion of cellular translation is coupled with transcription in the nucleus (Iborra et al., 2001; Wilkinson and Shyu, 2001), it is possible that pre-mRNA is scanned for nonsense codons in the nucleus and that the second signal for NMD is one or more components of the splicesome assembled at exon–intron junctions on pre-mRNAs.
The other unique aspect of TCR mRNA regulation reported in this paper is the polar effect of nonsense codons (Figures 1 and 2). Although this polar effect has not been reported before for any other vertebrate mRNA, it does occur in Saccharomyces cerevisiae (Losson and Lacroute, 1979; Peltz et al., 1994; Culbertson, 1999), but probably by a different mechanism, as the vast majority of S. cerevisiae genes lack introns and thus must have another downstream signal. This second signal appears to be a downstream sequence element found in most yeast transcripts (Peltz et al., 1993). Caenorhabditis elegans also appears to exert polar regulation by a mechanism that differs from that which acts on TCR transcripts, as C. elegans NMD can occur without a downstream intron (Pulak and Anderson, 1993).
How is TCR polar regulation mediated? We suggest a modified version of a surveillance model originally proposed by Peltz et al. (1994). In their model, recognition of a stop codon converts a ribosome into a post-termination surveillance complex that scans 3′ of the stop codon and then triggers NMD if it meets a second signal downstream. In the case of vertebrates, this second signal has been suggested to be a marker deposited at exon–exon junctions (such as the EJ complex). In our modification of this model, we posit that the post-termination complex becomes more ‘active’ at triggering NMD the further it travels before reaching the second signal. This modification is based on our finding that the level of NMD correlated with the distance between a nonsense codon and the 3′-terminal intron, regardless of whether this distance was altered by site-specific mutagenesis (Figures 1 and 2), deletion (Figure 3) or introduction of multiple introns and exons downstream of a nonsense codon (Figure 4). Many mechanisms could progressively activate the post-termination complex, including recruitment of positive factors, loss of negative factors, or post-translation modifications (e.g. phosphorylation events) of existing factors. We further hypothesize that this ‘progressive activation’ property of the post-termination complex can only be acquired if the translation apparatus first encounters an upstream signal, which, in the case of TCR, appears to be in the VDJ exon or flanking intron sequences (Figure 5).
Our progressive activation model is not the only explanation for the polar regulation we observed. Another possibility is that post-termination surveillance complex formation has the ability to destabilize or modify the NMD second signal and that this effect is distance dependent. Alternatively, there may be multiple NMD second signals within TCR exons that act additively to trigger NMD. Other possibilities include increased susceptibility to endonuclease attack as the distance between the nonsense codon and the second signal increases (Peltz et al., 1994; Jacobson and Peltz, 2000) or decreased ability to properly remodel mRNA (which protects mRNA from decay) as the distance between the nonsense codon and 3′ factors increases (Hilleren and Parker, 1999). To distinguish between these models, it will be critical to identify the molecules responsible for generating the NMD second signal in TCR transcripts. In addition, the nature of the upstream signal in the VDJ region must be identified and the mechanism by which it provides immunity from the –55 boundary rule must be elucidated. Recently, we found that the same region that confers the polar boundary-independent regulation also confers strong mRNA downregulation in response to nonsense codons (Gudikote and Wilkinson, 2002). Finally, it will be critical to determine whether the polar and boundary-independent characteristics of the TCR NMD response are mechanistically linked.
METHODS
Plasmids. Construct C (β-290) contains a wild-type TCR-β gene with a full-length open reading frame (pAc/IF; Carter et al., 1995). Constructs A, B, E, F and G (β-697, -696, -716, -694 and -695, respectively) are derivatives of C that contain single nucleotide mutations in the Cβ2.3 exon. Construct D (β-317) contains a nonsense codon in the Cβ2.3 exon (pAc/FS3; Carter et al., 1996). Construct O (β-337) is a TCR-β minigene that contains a nonsense codon in the VDJ exon (construct C; Li et al., 1997). Construct H (β-556) is identical to O except that it lacks the nonsense codon. Constructs I–N (β-612, -788, -693, -700, -790 and -789, respectively) and constructs P–U (β-706, -705, -708, -707, -691 and -692, respectively) are derivatives of H that contain single nucleotide mutations in the VDJ exon. Constructs V and W (β-717 and -718, respectively) were generated by an inverse PCR deletion method (Wang and Wilkinson, 2001) with the inverse primers MDA-791 (5′-TCGGTAAGTTGGGAGCTAGTAATGAAGGGGAGGGAG-3′) and MDA-793 (5′-CCAGTCCCGACTGCTGGCACAGAAATATACAG-3′) using H and O as templates, respectively. Constructs X and Y (β-755 and -754, respectively) were generated by inverse PCR with the inverse primers MDA-791 (5′-TCGGTAAGTTGGGAGCTAGTAATGAAGGGGAGGGAG-3′) and MDA-792 (5′-CATCAGGGATATCTCCTTTCTCCGTGCTGTCAGC-3′) using H and O as templates, respectively. Constructs A′–I′ (β-583, -584, -376, -368, -595, -585, -586, -593 and -577, respectively) are derivatives of C containing single nucleotide mutations in the VDJ exon. Construct J′ (β-652) was prepared by inserting the TCR-β gene VDJ segment (containing 23 nt of the 3′ end of IVS-Lβ, the entire Vβ8.1Dβ2Jβ2.3 exon and 674 nt of the 5′ end of IVS-JCβ) between the TPI gene BglII site and nt 131 of TPI IVS-4, thereby replacing TPI exons 2, 3 and 4. Construct K′ (β-726) is identical to J′ except it has a TGA nonsense codon 61 nt upstream of the last intron. Constructs L′–O′ (β-794, -793, -796 and -795, respectively) are derivatives of J′ containing single nucleotide mutations in TPI exon 6. Construct P′ (G-396) is identical to J′ except that a mouse Pem gene fragment was inserted instead of a TCR-β fragment. This Pem fragment contained 23 nt of the 3′ end of IVS-3, 357 nt of exon 4, and 512 nt of the 5′ end of IVS-4 (Maiti et al., 1996; note that a single T 12 nt from the 3′ end of exon 4 was deleted to shift frame to that of the downstream TPI exon). Construct Q′ (G-395) is identical to P′ except it has a TGA nonsense codon 61 nt upstream of the last intron. Construct R′ and S′ (G-403 and -404, respectively) are derivatives of P′ containing single nucleotide mutations in TPI exon 6. Construct T′ (β-780) is identical to C except that a VDJ segment (same size as the inserted VDJ segment in construct J′) was removed from the construct. Constructs U′–X′ (β-887, -888, -883 and -884, respectively) are derivatives of T′ that contain single nucleotide mutations in the Cβ2.3 exon. All nucleotide substitutions were generated by the site-specific mutagenesis protocol described in Wang and Wilkinson (2000).
Transfection, RNA isolation and RPA. DNA constructs were transiently transfected into HeLa cells using Lipofectamine according to the manufacturer’s instructions (Gibco-BRL, Gatersburg, MD). Total RNA was isolated and RPA was performed as described previously using specific riboprobes (Lindsey and Wilkinson, 1996; Li et al., 1997). The TCR-β Vβ and neo riboprobes were prepared as described by Mühlemann et al. (2001). The TCR-β Cβ riboprobe is a 162 nt fragment generated by PCR that contained 102 nt of the 3′ end of IVS-JC and 60 nt of the 5′ end of Cβ2.1. The TPI riboprobe is a 390 nt fragment generated by PCR that contained 250 nt of the 3′ end of TPI exon 7 and 140 nt of the downstream SV40 exon. mRNA levels were determined using a direct radioactivity scanner (Instant Imager, Packard Instruments, Downers Grove, IL). We determined that our RPA assay was quantitative by performing titration experiments: increasing the amount of input RNA linearly increased the level of the protected TCR-β bands, whereas increasing the amount of riboprobe had no effect on the protected bands, indicating that excess probe was present in the annealing reaction (data not shown).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Gilbert Cote (U.T.M.D.A.C.C.), Maureen Goode (U.T.M.D.A.C.C.) and Ann-Bin Shyu (University of Texas) for their valuable comments on the manuscript. We thank Drs Phil Anderson (University of Wisconsin), Rachel Aronoff (Max-Planck-Institute for Medical Research), Michael Culbertson (University of Wisconsin), Allan Jacobson (University of Massachusetts Medical School) and Roy Parker (University of Arizona) for helpful discussions. This research was supported by National Institutes of Health grant GM 58595 and National Science Foundation grant MCB-9808936.
REFERENCES
- Carter M.S., Doskow, J., Morris, P., Li, S., Nhim, R.P., Sandstedt, S. and Wilkinson, M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- Carter M.S., Li, S. and Wilkinson, M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Belgrader, P., Zhou, X. and Maquat, L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Gudikote J.P. and Wilkinson, M.F. (2002) T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J., 21, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik, A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker, R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra F.J., Jackson, D.A. and Cook, P.R. (2001) Coupled transcription and translation within nuclei of mammalian cells. Science, 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Jacobson A. and Peltz, S.W. (2000) Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae. In Sonenberg, N., Hershey, J.W.B. and Mathews, M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 827–847.
- Kim V.N., Yong, J., Kataoka, N., Abel, L., Diem, M.D. and Dreyfuss, G. (2001a) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Kataoka, N. and Dreyfuss, G. (2001b) Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science, 293, 1832–1836. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde, E., Maquat, L.E. and Moore, M.J. (2000a) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Moore, M.J. and Maquat, L.E. (2000b) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E. and Moore, M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson, M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Li S., Leonard, D. and Wilkinson, M.F. (1997) T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med., 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey J.S. and Wilkinson, M.F. (1996) Pem: a testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Dev. Biol., 179, 471–484. [DOI] [PubMed] [Google Scholar]
- Losson R. and Lacroute, F. (1979) Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl Acad. Sci. USA, 76, 5134–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D. and Steitz, J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 22, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu, M.D. and Steitz, J.A. (2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science, 293, 1836–1839. [DOI] [PubMed] [Google Scholar]
- Maiti S., Doskow, J., Sutton, K., Nhim, R.P., Lawlor, D.A., Levan, K., Lindsey, J.S. and Wilkinson, M.F. (1996) The Pem homeobox gene: rapid evolution of the homeodomain, X chromosomal localization and expression in reproductive tissue. Genomics, 34, 304–316. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O., Mock-Casagrande, C.S., Wang, J., Li, S., Custódio, N., Carmo-Fonseca, M., Wilkinson, M.F. and Moore, M.J. (2001) Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell, 8, 33–43. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat, L.E. (1998). A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., Brown, A.H. and Jacobson, A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., He, F., Welch, E. and Jacobson, A. (1994) Nonsense-mediated mRNA decay in yeast. Prog. Nucleic Acid Res. Mol. Biol., 47, 271–298. [DOI] [PubMed] [Google Scholar]
- Pulak R. and Anderson, P. (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev., 7, 1885–1897. [DOI] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson, M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Sun X. and Maquat, L.E. (2000) mRNA surveillance in mammalian cells: the relationship between introns and translation termination. RNA, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M.W. and Kulozik, A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. and Wilkinson, M.F. (2000) Site-directed mutagenesis of large (13-kb) plasmids in a single-PCR procedure. Biotechniques, 29, 976–978. [DOI] [PubMed] [Google Scholar]
- Wang J. and Wilkinson, M.F. (2001) Deletion mutagenesis of large (12-kb) plasmids by a one-step PCR protocol. Biotechniques, 31, 722–724. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.F. and Shyu, A.B. (2001) Multifunctional regulatory preteins that control gene expression in both the nucleus and cytoplasm. BioEssays, 23, 775–787. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun, X., Qian, Y., LaDuca, J.P. and Maquat, L. (1998) At least one intron is required for the nonsense-mediated decay of triosephosphate ismoerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]