Abstract
DNA gyrase is the target of two plasmid-encoded toxins CcdB and microcin B17, which ensure plasmid maintenance. These proteins stabilize gyrase–DNA covalent complexes leading to double-strand breaks in the genome. In contrast, the physiological role of chromosomally encoded inhibitor of DNA gyrase (GyrI) in Escherichia coli is unclear and its mechanism of inhibition has not been established. We demonstrate that the mode of inhibition of GyrI is distinct from all other gyrase inhibitors. It inhibits DNA gyrase prior to, or at the step of, binding of DNA by the enzyme. GyrI reduces intrinsic as well as toxin-stabilized gyrase–DNA covalent complexes. Furthermore, GyrI reduces microcin B17-mediated double-strand breaks in vivo, imparting protection to the cells against the toxin, substantiating the in vitro results. Thus, GyrI is an antidote to DNA gyrase-specific proteinaceous poisons encoded by plasmid addiction systems.
INTRODUCTION
DNA gyrase is distinct from all other topoisomerases in its ability to introduce negative supercoils into DNA in the presence of ATP (Gellert et al., 1976; Wang, 1998; Champoux, 2001). The enzyme functions by making a transient break in the DNA and resealing it after passing another duplex through it. The active enzyme is a heterotetramer of GyrA and GyrB with A2B2 subunit composition. The N-terminal two-thirds of GyrA harbours the cleavage-religation activity while the C-terminal one-third wraps DNA around itself (Reece and Maxwell, 1991). On the other hand, the ATPase activity of the enzyme resides in the N-terminal half of GyrB (Ali et al., 1993), while the C-terminal half is involved in binding to DNA and GyrA (Brown et al., 1979; Tingey and Maxwell, 1996; Chatterji et al., 2000). Being an essential enzyme and present only in prokaryotes, DNA gyrase is an ideal target for antibacterials. A number of diverse chemical entities, both natural and synthetic in nature, have been found to inhibit the enzyme (Lewis et al., 1996; Maxwell, 1999). A group of synthetic inhibitors, fluoroquinolones, are used clinically as antibacterials.
Two proteinaceous inhibitors of DNA gyrase are known: CcdB and microcin B17 (Liu, 1994; Couturier et al., 1998). Genes coding for both of these proteins are present on plasmids specific to Gram-negative eubacteria. These inhibitors stabilize gyrase–DNA covalent complexes, which are converted to double-strand breaks leading to cell death (Vizan et al., 1991; Bernard et al., 1993). Another protein has been found to be associated with Escherichia coli DNA gyrase during purification of the enzyme (Nakanishi et al., 1998). Notably, the fractions containing DNA gyrase along with this protein were unable to supercoil DNA and therefore it was named the gyrase inhibitory protein, GyrI. Surprisingly, the gene coding for GyrI was found to be present on the E. coli genome itself. Moreover, in an independent study, the same protein (then called SbmC) was picked up in a screen for host factors imparting resistance to microcin B17 (Baquero et al., 1995).
Presence of a chromosomally encoded inhibitor of DNA gyrase raises many questions: (i) what is the mode of inhibition of DNA gyrase by GyrI?; (ii) why has an inhibitor of an essential enzyme been maintained through evolution?; and (iii) how does an inhibitor of DNA gyrase render protection against another? The mechanism underlying this apparent dual function of GyrI is puzzling. Moreover, the precise role of the protein in vivo has been elusive. In this study, we demonstrate that the mechanism of inhibition of DNA gyrase by GyrI is unique. We also observe that GyrI acts as an antidote for CcdB and microcin B17, and propose an interplay between the proteinaceous toxins and GyrI in vivo.
RESULTS
Mode of inhibition of DNA gyrase by GyrI
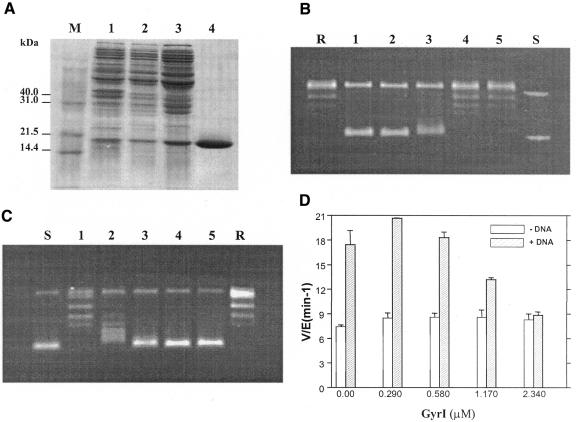
The gyrI ORF was amplified from E. coli genomic DNA, cloned and over-expressed. The protein was purified to apparent homogeneity as described in Methods (Figure 1A). The purified protein, of expected size, was able to inhibit supercoiling activity of DNA gyrase (Figure 1B). Interestingly, GyrI also inhibited the relaxation activity of DNA gyrase at similar concentrations (Figure 1C). The difference in the molar ratios of GyrI to gyrase required to inhibit supercoiling versus the relaxation reactions might be due to the differential affinity of the enzyme for the two forms of substrate DNA, namely relaxed and supercoiled closed-circular DNA (Higgins and Cozzarelli, 1982). Moreover, mechanistically the two reactions are also distinct (Williams and Maxwell, 1999). Inhibition of the ATP-independent relaxation reaction suggests that the primary mode of action of GyrI is not by interference with the ATPase activity. In agreement with such a mechanism, the intrinsic ATPase activity of GyrB was unaltered in the presence of GyrI (Figure 1D). Thus, GyrI appears to inhibit a step common to both supercoiling as well as the relaxation reactions.
Fig. 1. (A) Purification profile of GyrI. Lane 1, total cell extract after induction; lane 2, sonicate; lane 3, ammonium sulfate pellet; lane 4, pure GyrI after Superdex S200. M, molecular weight marker. The effect of GyrI on supercoiling (B) and relaxation (C) activity. Reactions were carried out with 17.5 nM (supercoiling) and 175 nM (relaxation) DNA gyrase in the presence of 0, 0.17, 0.35, 0.69 and 1.38 µM GyrI (lanes 1–5). R and S represent relaxed and supercoiled pUC18 DNA, respectively. (D) Effect of GyrI on ATPase activity. Reactions were performed with 1.44 µM GyrB in the presence of 0, 0.29, 0.58, 1.17 and 2.34 µM GyrI. ATP (2 mM) was present in all reactions. In addition to GyrB, the +DNA reactions contained 1.4 µM GyrA and 150 µg/ml sonicated salmon sperm DNA. The average of three independent experiments is denoted graphically. All reactions (in B–D) were performed at 37°C for 30 min.
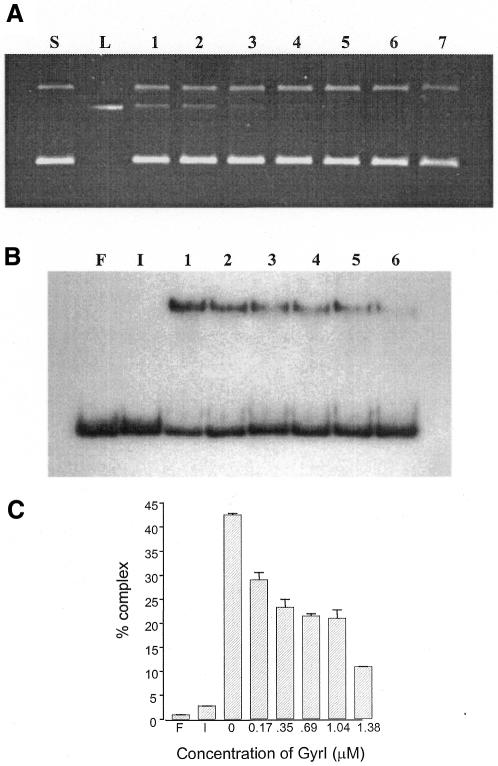
The protein–DNA covalent adduct is a transient intermediate in every topoisomerization reaction cycle. The low steady-state levels of this intermediate are visualized in the cleavage reaction by the addition of SDS followed by proteinase K digestion. Poisons of DNA gyrase like CcdB and microcin B17, which alter the cleavage-religation equilibrium, show an accumulation of the linear product in such reactions. As opposed to these toxins, GyrI did not cause any increase in the cleavage product. Instead, there was a reduction in the gyrase–DNA covalent complexes in the presence of the inhibitor (Figure 2A). To assess whether GyrI interfered with the cleavage activity of the enzyme per se or with a preceding step, its effect on the binding of substrate DNA by DNA gyrase was assessed. Electrophoretic mobility shift assays (EMSAs) were performed with a labelled fragment containing the strong gyrase site from pBR322 as the substrate, in the absence and presence of GyrI (Figure 2B). A concentration-dependent decrease in DNA binding by gyrase was observed in the presence of GyrI (Figure 2B and C). Irrespective of the order of addition of gyrase, DNA and GyrI, a similar reduction in binding of DNA by the enzyme was seen (data not shown). It should be noted that GyrI itself did not interact with DNA (Figure 2B, lane I). Moreover, there was no additional complex seen in the presence of GyrI, ruling out the possibility of a ternary complex between DNA, gyrase and GyrI. The interaction between GyrI and Gyr subunits was assessed by surface plasmon resonance. The dissociation constants obtained from such an experiment revealed that GyrI was able to interact with both GyrA and GyrB, albeit with different affinities. GyrI has 6-fold higher affinity for GyrA as compared to GyrB (Table I). We have considered the possibility that GyrI might interfere with GyrA–GyrB interaction and thus inhibit the formation of active holoenzyme. However, gel filtration experiments appear to rule out this possibility. Taken together, these results imply that GyrI-mediated inhibition is prior to, or at the step of, DNA binding by the enzyme. In agreement with the above conclusion, although GyrI does not affect the intrinsic ATPase of GyrB, it completely abolishes the DNA-dependent stimulation of ATPase activity (Figure 1D).
Fig. 2. (A) Effect of GyrI on DNA cleavage. Reactions were performed with 175 nM DNA gyrase at 37°C for 30 min in the presence of 0, 0.087, 0.17, 0.35, 0.69, 1.04 and 1.38 µM GyrI (lanes 1–7, respectively). The covalent gyrase–DNA complex was trapped with SDS (0.2%) and digested with proteinase K (0.8 mg/ml). S and L represent supercoiled and linearized pBR322, respectively. (B) Effect of GyrI on binding of DNA by gyrase. EMSAs were carried out with 19.7 nM of DNA and 200 nM each of GyrA and GyrB in the presence of 0, 0.17, 0.35, 0.69, 1.04 and 1.38 µM GyrI (lanes 1–6, respectively). F, free DNA; I, reaction containing 1.38 µM of GyrI in the absence of DNA gyrase. The reactions were performed at 4°C for 1 h and resolved on a 5% polyacrylamide gel. (C) The average of three independent experiments is shown.
Table I. Interaction between GyrI and Gyr subunits.
| Protein | KD (M) |
|---|---|
| GyrA | 4.52 × 10–7 |
| GyrB | 29.40 × 10–7 |
Effect of GyrI on microcin B17 and CcdB stabilized gyrase–DNA covalent complexes in vitro
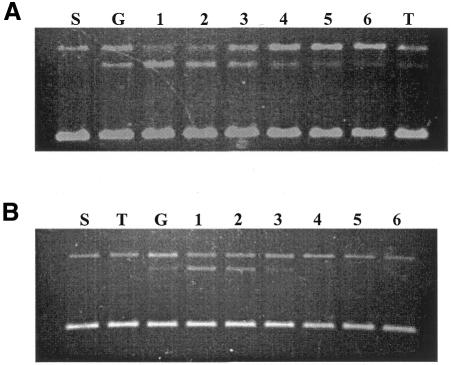
Since GyrI was able to reduce the steady-state levels of gyrase–DNA covalent complex in the absence of any inhibitor, we tested its effect on CcdB and microcin B17 stabilized complexes. Cleavage reactions were carried out wherein CcdB or microcin B17 was added along with GyrI. CcdB and microcin B17 both alter the cleavage-religation equilibrium leading to the accumulation of a gyrase–DNA covalent complex. However, in the presence of GyrI, there was a net decrease in the steady-state levels of cleavage product (Figure 3). Such a reduction of gyrase–DNA covalent complex indicates that GyrI is able to nullify the effect of CcdB and microcin B17 in vitro. Order of addition experiments were carried out where CcdB or microcin B17 was incubated with gyrase and DNA prior to the addition of GyrI. Such reactions revealed that GyrI was able to revert even a preformed CcdB stabilized gyrase–DNA complex. However, under similar experimental conditions it was unable to revert a microcin-stabilized complex probably due to the extremely slow kinetics of action of microcin B17 (Heddle et al., 2001).
Fig. 3. Effect of GyrI on CcdB (A) and microcin B17 (B) stabilized gyrase–DNA covalent complex. Cleavage reactions were carried out with 175 nM DNA gyrase, 2 µM CcdB or 1.2 U/µl microcin B17 in the presence of 0, 0.17, 0.35, 0.69, 1.04 and 1.38 µM GyrI (lanes 1–6, respectively). S, G and T represent supercoiled pBR322 DNA, reaction performed with DNA gyrase alone and reaction performed with CcdB/microcin B17 alone, respectively. All reactions were carried out at 37°C for 1 h.
Action of GyrI in vivo
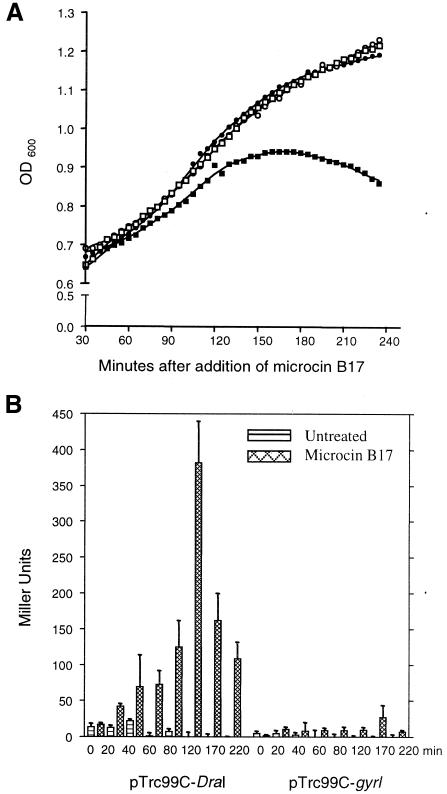
To test the ability of GyrI to counteract microcin B17 inside the bacterial cell, we analysed the sensitivity of cells expressing GyrI to the toxin. Microcin B17 was spotted on a lawn of DH5α cells harbouring either vector or plasmid producing GyrI and the zone of inhibition was monitored. Uninduced expression levels of GyrI imparted an 8-fold protection to the cells against the microcin B17, and this was further enhanced when its expression was induced with IPTG (Table II). Furthermore, the growth of AP1-200-9 transformed with pTrc99C-DraI or pTrc99C-gyrI was monitored in the presence of microcin B17. Cells lacking the GyrI over-expressing plasmid appeared to die 2 h after the addition of the toxin (Figure 4A). On the other hand, cells expressing GyrI continue to grow in the presence of microcin B17, suggesting that GyrI was also able to negate the effect of the toxin in vivo. Thus, GyrI is a defence mechanism of the cell against proteinaceous poisons targeting DNA gyrase.
Table II. Resistance imparted by GyrI against microcin B17.
| Plasmids | MIC (–IPTG) | MIC (+IPTG) |
|---|---|---|
| PTrc99C-DraI | 1 U | 1 U |
| PTrc99C-gyrI | 8 U | >16 U |
MIC, minimal inhibitory concentration of microcin B17.
Fig. 4. (A) Effect of GyrI on cell survival. Growth curve of AP1-200-9 transformed with either pTrc99C-DraI (squares) or pTrc99C-gyrI (circles) in the absence (open symbols) or presence (filled symbols) of microcin B17. (B) Effect of GyrI on microcin B17-induced double-strand breaks in vivo. β-galactosidase activity of AP1-200-9 cells transformed with either pTrc99C-DraI or pTrc99C-gyrI after different durations (as indicated) of treatment with microcin B17.
Action of microcin B17 on its target, DNA gyrase, ultimately leads to generation of double-strand breaks in the genome and cell death. To show that the resistance imparted by GyrI was due to decrease in the double-strand breaks, an AP1-200-9 strain harbouring the lacZ gene under the SOS inducible dinD promoter was used (Piekarowicz et al., 1991). Treatment of the cells with microcin B17 leads to double-strand breaks. The consequent SOS response in these cells was visualized by increase in β-galactosidase activity (Figure 4B). The β-galactosidase activity peaked after 2 h of treatment; however, prolonged exposure led to lower levels of induction probably due to cell death. In contrast, in cells producing GyrI, there was no significant induction of β-galactosidase, suggesting a suppression of double-strand breaks. Thus, these results demonstrate that the resistance imparted by GyrI is by inhibiting the formation of lethal double-strand breaks in the cell.
DISCUSSION
DNA gyrase is the only enzyme that introduces negative supercoils into DNA. In addition to supercoiling, DNA gyrase catalyses catenation/decatenation as well as knotting/unknotting reactions in vitro (Reece and Maxwell, 1991). Furthermore, in the absence of ATP the enzyme catalyses relaxation of negatively supercoiled DNA. Each of these topoisomerization reactions involves a complex series of steps. The enzyme binds to DNA and introduces a break with concomitant formation of a protein–DNA covalent complex. This is followed by passage of another duplex through this molecular gate and finally resealing of the break. Although the actual covalent protein–DNA link is formed with GyrA, recent work shows that both subunits contribute to the cleavage-religation catalysis (Berger et al., 1998; Liu and Wang, 1999). It is not surprising because GyrB not only harbours the ATPase domain but is also involved in interacting with DNA including the transfer segment (Tingey and Maxwell, 1996; Chatterji et al., 2000).
Mechanistically, all inhibitors of DNA gyrase can be classified into two groups (Lewis et al., 1996; Maxwell, 1999). The first class, exemplified by coumarins and cyclothialidines, prevent the binding of ATP to the enzyme and, as a result, inhibit supercoiling. The second class, e.g. quinolones, CcdB and microcin B17, act by stabilizing enzyme–DNA covalent intermediates. These protein–DNA adducts act as roadblocks for DNA tracking machines: RNA and DNA polymerases. Collisions of polymerases with these adducts lead to the generation of double-strand breaks in DNA that are lethal to the cells (Drlica and Zhao, 1997). In contrast, GyrI inhibits DNA gyrase by a mechanism distinct from the above modes of inhibition. As opposed to ATPase inhibitors, the ATP hydrolysis by GyrB is unchanged in the presence of GyrI. Moreover, unlike CcdB and microcin B17, there was a decrease in cleavage complex in the presence of GyrI. This was paralleled by a concomitant decrease in the binding of the enzyme to DNA. Since GyrI does not bind to DNA, it might act by sequestering DNA gyrase resulting in a reduction in the effective concentration of the enzyme. Alternatively, it could act by directly inhibiting DNA binding. Both mechanisms would cause reactions to be favoured in the reverse direction leading to a decrease in the non-covalent as well as covalent complexes. In contrast, Baquero et al. (1995) had earlier postulated that GyrI (SbmC) imparts resistance to the cells against microcin B17 by binding to the toxin and sequestering it. However, it is unlikely that GyrI operates by such a mechanism, since it was able to reduce both CcdB and microcin B17 stabilized gyrase–DNA complexes. Thus, GyrI acts on the target of these toxins and not on the toxins per se. Such a mechanism envisages that GyrI would protect cells against other gyrase inhibitors that also act by stabilizing gyrase–DNA covalent complexes. Indeed, we observed that GyrI imparts resistance to ciprofloxacin and, hence, increases cell survival (M. Chatterji and V. Nagaraja, unpublished data).
At first glance, GyrI appears to be a burden for the cell as it inhibits supercoiling activity. This study reveals that despite its inhibitory activity, GyrI is useful to E. coli as it reduces double-strand breaks mediated by gyrase inhibitors. While decrease in gyrase activity may be tolerated to a certain degree, even a few double-strand breaks can prove lethal to the cells (Miguel and Tyrrell, 1986). In addition, the reduction in gyrase activity might be compensated by various mechanisms available for homeostatic regulation of supercoiling, e.g. altered expression of other topoisomerases (Pruss et al., 1982; Tse-Dinh and Beran, 1988). Therefore, we propose that chromosomally placed GyrI has been maintained as an antidote to toxins that target DNA gyrase.
Both proteinaceous inhibitors of DNA gyrase are plasmid-borne. Along with the toxins, the plasmids also code for their antidote namely CcdA (for CcdB) and MccG (for microcin B17). In the case of an F-plasmid encoded CcdA–CcdB system, CcdA protein is more unstable than CcdB and hence host cells are dependent on the presence of the plasmid for continued production of the antidote for their survival (Couturier et al., 1998). Upon cell division, daughter cells devoid of the plasmid are killed by the residual toxin. On the other hand, the microcin-producing plasmid employs a variant mechanism wherein the toxin is secreted out of the cell and targets neighbouring cells that lack the plasmid. The target cells could either be daughter cells that have lost the plasmid during cell division or other unrelated bacteria. At the same time, the cells harbouring the plasmid are protected against the toxin by MccG. As in other plasmid-addiction systems, the antidote is part of the same operon that codes for and post-translationally modifies microcin B17 (Liu, 1994). Thus, both systems select for cells harbouring the plasmid. Since GyrI confers resistance against the toxins even in the absence of their antidote, it represents a protective mechanism adopted by the host cells against these selfish plasmids.
As a corollary to the model, since GyrI interferes with an essential activity of the cell, its production is expected to be under strict control. In agreement with this hypothesis, overproduction of GyrI is not tolerated by the cells and also leads to alteration in the cellular morphology (Nakanishi et al., 1998; M. Chatterji and V. Nagaraja, unpublished data). Therefore, it is not surprising that the expression of GyrI is induced only under stress conditions such as stationary phase or SOS response (Baquero et al., 1995). In summary, cells express GyrI only to combat the dire situation of breaks in the DNA even at the cost of partial inhibition of an essential function.
By virtue of its ability to reduce gyrase-mediated double-strand breaks, GyrI has the potential to play a more general role. Although GyrI seems to have evolved as an antidote to toxins, any signal that is amplified by gyrase action leading to DNA damage, e.g. quinolones and intrinsic gyrase-mediated lesions, can be combated by production of this protein by the cells. In addition, in a recent study, it was observed that gyrI when present in a multicopy plasmid imparted resistance against mitomycin C (Wei et al., 2001). Mitomycin C is a DNA damaging agent that intercalates into DNA and alkylates it. Bacterial cells turn on the SOS regulon in response to the toxic effects of this compound. Resistance to mitomycin C by GyrI might be due to reduced intercalation as a result of reduced negative supercoiling of DNA. Molecular details of a wider role of GyrI as an arsenal of cellular rescue machinery need further investigation.
METHODS
Bacterial strains and plasmids. Escherichia coli DH10B was used for all cloning experiments. Over-expression plasmids pPH3, pAG111 (Hallett et al., 1990) and pJW312-SalI (Lynn and Wang, 1989) were employed for E. coli GyrA, GyrB and topoisomerase I purification, respectively. Microcin B17 was purified from ZK650 cells (Yorgey et al., 1993). AP1-200-9 was used for the detection of SOS response in the cells (Piekarowicz et al., 1991).
Cloning of gyrI. gyrI was PCR amplified using E. coli genomic DNA as template and primers PF (5′-ATCAATGGATCCGTCATGAACTACG) and PR (5′-CCTGAGATCTATTAGTGATGTTTTGG) containing RcaI and BglII sites, respectively. The reaction was carried out using Pfu polymerase. RcaI- and BglII-digested PCR product was ligated to NcoI–BamHI cut pTrc99C-DraI. pTrc99C-DraI is a derivative of pTrc99C, which contains a DraI site downstream of the Ptrc promoter.
Enzyme and substrate preparation. GyrA and GyrB were purified as described previously (Maxwell and Howells, 1999). Specific activity of purified DNA gyrase was calculated with 1 U defined as the amount of enzyme required to completely supercoil 500 ng of relaxed pUC18 DNA at 37°C in 30 min. Supercoiled pUC18 and pBR322 were prepared by standard DNA purification protocols (Sambrook et al., 1989). Escherichia coli topoisomerase I and relaxed pUC18 were prepared as described by Lynn and Wang (1989).
Purification of proteinaceous inhibitors of DNA gyrase. Microcin B17 was purified from ZK650 cells as described previously (Chatterji et al., 2001). The unit strength of microcin B17 was determined as described previously (Davagnino et al., 1986), except that DH5α was used as the tester/sensitive strain. Purified CcdB was a kind gift from R. Varadarajan. For purification of GyrI, DH5α cells harbouring pTrc99C-gyrI were grown until an OD600 of 0.6 was reached, and induced with 0.05 mM IPTG for 4 h. The cells were harvested, resuspended in 30 mM Tris–HCl pH 7.5 containing 30 mM NaCl and sonicated. After centrifugation at 10 000 g for 90 min, the supernatant (S100) was subjected to an ammonium sulfate fractionation (55% saturation). The pellet was dissolved in 10 mM Tris–HCl pH 7.5, 1 mM EDTA and 1 mM DTT, and resolved on a high-resolution Superdex S-200 column (150 ml). More than 99% homogenous preparation of protein was obtained as detected by silver staining.
Enzyme assays. Supercoiling and cleavage assays were performed as described previously (Chatterji et al., 2001). Relaxation reactions were carried out in supercoiling buffer except spermidine and ATP were omitted and supercoiled pUC18 was used as substrate. The reactions were performed at 37°C for 30 min. ATPase and EMSAs were carried out as described previously (Chatterji et al., 2000). Surface plasmon resonance experiments were performed on a BIAcore 2000 system (BIAcore). GyrI was immobilized on the CM5 sensor chip via amine coupling in acetate buffer (pH 3.0). The surface was blocked with ethanolamine hydrochloride. The interaction was assessed in 10 mM HEPES–NaOH pH 7.4 containing 150 mM NaCl, 1 mM EDTA and 1 mM DTT. Different concentrations of GyrA and GyrB were passed over the immobilized GyrI and the subsequent changes in resonance units were recorded. Proteins used for the experiment were dialysed against running buffer prior to the experiment. Bovine serum albumin was used as the negative control.
In vivo assays. The effect of GyrI on the in vivo toxicity of microcin B17 was tested by spotting various concentrations of the microcin B17 on a lawn of DH5α cells containing either pTrc99C-DraI or pTrc99C-gyrI in the presence and absence of IPTG. The presence of a zone of inhibition and its size is indicative of the sensitivity of the cells to microcin B17. Furthermore, exponentially growing cultures transformed with GyrI containing plasmid or plasmid alone were treated with microcin B17 (2.7 U/µl) and their OD600 were recorded at different time intervals. As negative control, cells were treated with equivalent amounts of ethanol, and used as a solvent for microcin B17. AP1-200-9, harbouring the lacZ gene under the SOS inducible dinD promoter, was used to monitor double-strand breaks induced by gyrase in the presence of microcin B17. Cells harbouring either pTrc99C-DraI or pTrc99C-gyrI were grown to an OD600 of 0.6 and treated with 0.76 U/µl of microcin B17 for varied time and β-galactoside activity of the cells were measured by a standard procedure (Miller, 1992).
Acknowledgments
ACKNOWLEDGEMENTS
We thank J.C. Wang and A. Maxwell for over-expressing constructs of E. coli topoisomerase I and DNA gyrase, respectively, R. Kolter for ZK4 and ZK650 strains, A. Piekarowicz and D.C. Stein for AP1-200-9, and R. Varadarajan for CcdB. S. Unniraman is acknowledged for discussion and critical reading of the manuscript, and J. Jacob for technical assistance. This work is supported by research grants to V.N. from the Department of Science and Technology, Government of India.
REFERENCES
- Ali J.A., Jackson, A.P., Howells, A.J. and Maxwell, A. (1993) The 43-kDa N-terminal fragment of the gyrase B protein hydrolyses ATP and binds to coumarin drugs. Biochemistry, 32, 2717–2724. [DOI] [PubMed] [Google Scholar]
- Baquero M.R., Bouzon, M., Varea, J. and Moreno, F. (1995) sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol. Microbiol., 18, 301–311. [DOI] [PubMed] [Google Scholar]
- Berger J.M., Fass, D., Wang, J.C. and Harrison, S.C. (1998) Structural similarities between topoisomerases that cleave one or both strands. Proc. Natl Acad. Sci. USA, 95, 7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Kezdy, K.E., Van Melderen, L., Steyaert, J., Wyns, L., Pato, M.L., Higgins, P.N. and Couturier, M. (1993) The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol., 234, 534–541. [DOI] [PubMed] [Google Scholar]
- Brown P.O., Peebles, C.L. and Cozzarelli, N.R. (1979) A topoisomerase from Escherichia coli related to DNA gyrase. Proc. Natl Acad. Sci. USA, 76, 6110–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J.J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem., 70, 369–413. [DOI] [PubMed] [Google Scholar]
- Chatterji M., Unniraman, S., Maxwell, A. and Nagaraja, V. (2000) The additional 165 amino acids in the B protein of Escherichia coli DNA gyrase have an important role in DNA binding. J. Biol. Chem., 275, 22888–22894. [DOI] [PubMed] [Google Scholar]
- Chatterji M., Unniraman, S., Mahadevan, S. and Nagaraja, V. (2001) Effect of different classes of inhibitors on DNA gyrase from Mycobacterium smegmatis. J. Antimicrob. Chemother., 48, 479–485. [DOI] [PubMed] [Google Scholar]
- Couturier M., Bahassi, el-M. and Van Melderen, L. (1998) Bacterial death by DNA gyrase poisoning. Trends Microbiol., 6, 269–275. [DOI] [PubMed] [Google Scholar]
- Davagnino J., Herrero, M., Furlong, D., Moreno, F. and Kolter, R. (1986) The DNA replication inhibitor microcin B17 is a forty-three-amino-acid protein containing sixty percent glycine. Proteins, 1, 230–238. [DOI] [PubMed] [Google Scholar]
- Drlica K. and Zhao, X. (1997) DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev., 61, 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi, K., O’Dea, M.H. and Nash, H.A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA, 73, 3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett P., Grimshaw, A.J., Wigley, D.B. and Maxwell, A. (1990) Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene, 93, 139–142. [DOI] [PubMed] [Google Scholar]
- Heddle J.G., Blance, S.J., Zamble, D.B., Hollfelder, F., Miller, D.A., Wentzell, L.M., Walsh, C.T. and Maxwell, A. (2001) The antibiotic microcin B17 is a DNA gyrase poison: characterisation of the mode of inhibition. J. Mol. Biol., 307, 1223–1234. [DOI] [PubMed] [Google Scholar]
- Higgins N.P. and Cozzarelli, N.R. (1982) The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res., 10, 6833–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.J., Tsai, F.T.F. and Wigley, D.B. (1996) Molecular mechanisms of drug inhibition of DNA gyrase. BioEssays, 18, 661–671. [DOI] [PubMed] [Google Scholar]
- Liu J. (1994) Microcin B17: posttranslational modifications and their biological implications. Proc. Natl Acad. Sci. USA, 91, 4618–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. and Wang, J.C. (1999) Similarity in the catalysis of DNA breakage and rejoining by IA and IIA DNA topoisomerases. Proc. Natl Acad. Sci. USA, 96, 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn R.M. and Wang, J.C. (1989) Peptide sequencing and site-directed mutagenesis identify tyrosine-319 as the active site tyrosine of Escherichia coli DNA topoisomerase I. Proteins, 6, 231–239. [DOI] [PubMed] [Google Scholar]
- Maxwell A. (1999) DNA gyrase as a drug target. Biochem. Soc. Trans., 27, 48–53. [DOI] [PubMed] [Google Scholar]
- Maxwell A. and Howells, A.J. (1999) Protocols for DNA Topoisomerases I: DNA Topology and Enzyme Purification. Humana Press, Totawa, NJ, pp. 135–144.
- Miguel A.G. and Tyrrell, R.M. (1986) Repair of near-ultraviolet (365 nm)-induced strand breaks in Escherichia coli DNA. The role of the polA and recA gene products. Biophys. J., 49, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 71–74.
- Nakanishi A., Oshida, T., Matsushita, T., Imajoh-Ohmi, S. and Ohnuki, T. (1998) Identification of DNA gyrase inhibitor (GyrI) in Escherichia coli. J. Biol. Chem., 273, 1933–1938. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan, R. and Stein, D.C. (1991) A new method for the rapid identification of genes encoding restriction and modification enzymes. Nucleic Acids Res., 19, 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G.J., Manes, S.H. and Drlica, K. (1982) Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell, 31, 35–42. [DOI] [PubMed] [Google Scholar]
- Reece R.J. and Maxwell, A. (1991) DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol., 26, 335–375. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tingey A.P. and Maxwell, A. (1996) Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res., 24, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh Y.C. and Beran, R.K. (1988) Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J. Mol. Biol., 202, 735–742. [DOI] [PubMed] [Google Scholar]
- Vizan J.L., Hernandez-Chico, C., del Castillo, I. and Moreno, F. (1991) The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J., 10, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C. (1998) Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys., 31, 107–144. [DOI] [PubMed] [Google Scholar]
- Wei Y., Vollmer, A.C. and LaRossa, R.A. (2001) In vivo titration of mitomycin C action by four Escherichia coli genomic regions on multicopy plasmids. J. Bacteriol., 183, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N.L. and Maxwell, A. (1999) Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry, 38, 13502–13511. [DOI] [PubMed] [Google Scholar]
- Yorgey P., Davagnino, J. and Kolter, R. (1993) The maturation pathway of microcin B17, a peptide inhibitor of DNA gyrase. Mol. Microbiol., 9, 897–905. [DOI] [PubMed] [Google Scholar]