Abstract
The arginine methyltransferases CARM1 and PRMT1 associate with the p160 family of nuclear hormone receptor coactivators. This association enhances transcriptional activation by nuclear receptors. We describe a method for identifying arginine N-methyltransferase substrates using arrayed high-density protein membranes to perform solid-phase supported enzyme reactions in the presence of the methyl donor S-adenosyl-l-methionine. Using this screen, we identified distinct substrates for CARM1 and PRMT1. All PRMT1 substrates harbor the expected GGRGG methylation motif, whereas the peptide sequence comparisons of the CARM1 substrates revealed no such motif. The predominant CARM1 substrate identified in this screen was PABP1. We mapped the methylated region of this RNA binding molecule in vitro and demonstrate that PABP1 is indeed methylated in vivo. Prior to these findings, the only known substrate for CARM1 was histone H3. We broaden the number of CARM1 targets and suggest a role for CARM1 in regulating transcription/translation.
INTRODUCTION
Protein arginine N-methyltransferases (PRMTs) have been implicated in a variety of processes, including cell proliferation, signal transduction and protein trafficking (McBride and Silver, 2001). The methylation of arginine residues is catalyzed by two classes of PRMT enzymes: the Type I enzyme catalyzes the formation of asymmetric ω-NG,NG-dimethylarginine (aDMA) residues, and the Type II enzyme catalyzes the formation of symmetric ω-NG,N′G-dimethylarginine (sDMA) residues (Gary and Clarke, 1998). The recent molecular cloning of the yeast Type I arginine methyltransferase enzyme RMT1 (Gary et al., 1996; Henry and Silver, 1996) initiated the identification of at least six homologs in mammals: PRMT1–6 (McBride and Silver, 2001; Frankel et al., 2002). PRMT5 is the only Type II enzyme identified to date (Branscombe et al., 2001).
Work in yeast unveiled the first identified biological role of arginine methylation: shuttling of the hnRNP protein Npl3p between the nucleus and cytoplasm requires Rmt1 activity (Shen et al., 1998). More recently, it was shown that protein–protein interactions are regulated by arginine methylation, both inhibiting and permitting interactions in different signaling scenarios. The asymmetric dimethylation of arginine residues flanking proline-rich motifs can inhibit the binding of SH3 domains in vitro (Bedford et al., 2000). A diametric effect is seen with interactions between the survival of motor neurons protein (SMN) and the snRNP proteins SmD1 and SmD3, where binding is promoted by symmetrical arginine dimethylation of the snRNP proteins (Friesen et al., 2001).
Apart from controlling protein–protein interactions, arginine methylation has also been implicated in the regulation of transcription. The coactivator-associated arginine methyltransferase (CARM1) binds the p160 family of nuclear hormone receptor coactivators. This interaction enhances transcriptional activation by nuclear receptors, possibly as a result of the specific methylation of histone H3 by CARM1 (Chen et al., 1999). PRMT1, like CARM1, can also bind p160, and these two PRMTs have been shown to act in a synergistic manner to enhance reporter gene activation by nuclear receptors (Koh et al., 2000). The arginine methylation of histone H4 by PRMT1 facilitates the subsequent acetylation of H4 tails by p300 and thus plays a central part in transcription regulation (Wang et al., 2001).
Other proteins, apart from the histones, contain aDMA residues, including Sam68, hnRNP A1 and A2 and nucleolin (Gary and Clarke, 1998; McBride and Silver, 2001). By compiling data on the sequence surrounding actual methylated residues, a consensus sequence (RGG) can be generated for the Type I PRMTs (Rawal et al., 1995). However, CARM1 is unable to methylate PRMT1 substrates, indicating that CARM1 substrates harbor a distinct motif. The only identified substrate for CARM1 is histone H3 (Chen et al., 1999), and it seems plausible that additional physiologically relevant substrates remain undiscovered. Here, we used high-density protein filter arrays to screen for proteins that are methylated by PRMT1 and CARM1.
RESULTS AND DISCUSSION
Designing a screen to identify novel arginine methyltransferases substrates
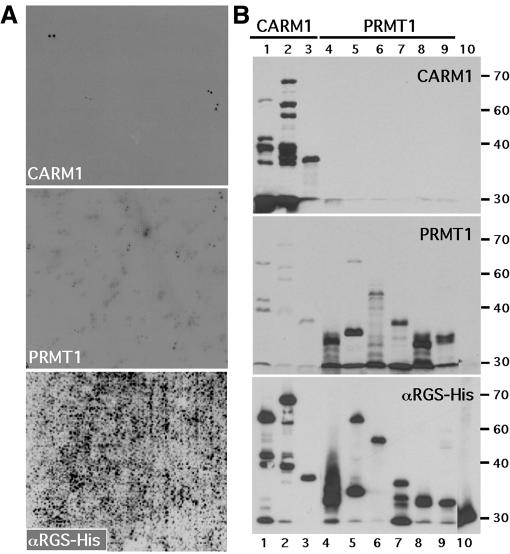
In the quest for kinase substrates, successful screens have been employed using phage expression libraries (Fukunaga and Hunter, 1997). We reasoned that a similar approach could be used for the identification of PRMT substrates. All of the arginine methyltransferase enzymes, with the exception of PRMT2, are active as GST fusion proteins. This allowed the isolation of sufficient quantities of active enzyme to perform large-scale enzyme reactions on a solid support. We initially attempted these experiments using a phage expression library induced on a PVDF membrane and then incubated with recombinant PRMT in the presence of 3H-labeled S-adenosyl-l-methionine ([3H]AdoMet), the universal methyl group donor. Using this approach we had very little success, probably due to the fact that the levels of protein induced, and the tritium signal we were attempting to detect, were relatively low. We next undertook a similar experiment but this time used an arrayed protein library (Bussow et al., 1998). The high-density protein expression filters used in these experiments represent a novel product for high-throughput proteomics and were generated by the German Human Genome Project (DHGP). This human brain protein library contains 37 200 clones of His-tagged fusion protein arrayed and spotted in a duplicate pattern on two PVDF membranes, providing extensive coverage of the human proteome. We used active recombinant arginine methyltransferases to perform large-scale enzyme reactions on the membranes. This approach has successfully identified substrates for two arginine methyltransferases, PRMT1 and CARM1 (Figure 1A). It was immediately obvious, when comparing the methylation patterns, that these two enzymes exhibit mutually exclusive substrate specificity. Identified substrates with the strongest signal were obtained as bacterial stocks from the DHGP in Germany and re-tested to confirm that they harbored the expected His-tagged fusion proteins and that the appropriate enzyme specifically methylated the recombinant protein substrates (Figure 1B).
Fig. 1. Methylation screen for PRMT1 and CARM1 substrates. (A) High-density protein filters were methylated in situ with the indicated recombinant PRMT and [3H]AdoMet as described in Methods. The lower panel is a western analysis of the array using an αHis-tag antibody, which will bind all the induced recombinant proteins. The field represented contains ∼10 000 different His-tagged fusion proteins spotted in duplicate. (B) PRMT substrates are specifically methylated in the secondary screen. Recombinant proteins identified in the primary substrate screen were induced and 10 µg of total bacterial lysate was methylated by the indicated recombinant PRMT. Clones identified in the CARM1 screen (lanes 1–3) and in the PRMT1 screen (lanes 4–9) were compared; lane 10 harbors a negative control. The lower panel was probed with αHis-tag antibody. Molecular mass markers (kDa) are shown on the right. Clones were identified by sequence analysis (Table I).
The identity of the PRMT substrates
To determine the identity of the methylated fusion proteins, we isolated the expression vectors and sequenced the plasmid DNA. Five different proteins were identified as PRMT1 substrates (Table I). Two of these proteins (hnRNPU and ILF3) have been described previously as harboring methylated arginine residues (Liu and Dreyfuss, 1995; Tang et al., 2000), thus providing proof that this in vitro screen can indeed identify proteins that are arginine methylated in vivo. All five substrates are RNA binding proteins, and they also contain RGG or RGRG repeats that represent the methylation consensus sequence for known PRMT1 substrates.
Table I. The identity of substrates that were methylated in vitro specifically by CARM1 and PRMT1.
| Substrates | No. of clonesa | Laneb | GenBank accession No. |
|---|---|---|---|
| CARM1 | |||
| Poly(A)-binding protein 1 | 6 | 1 and 2 | P11940 |
| TARPP | 1 | 3 | AAK48713 |
| PRMT1 | |||
| hnRNPU | 2 | 4 | NM_004501 |
| CIRP | 4 | 5 | NM_001280 |
| ILF3 | 2 | 6 and 7 | AF167570 |
| FUS/TLS | 1 | 8 | Q28009 |
| RNA helicase A | 1 | 9 | NM_001357 |
aSome of the protein clones were identified multiple times.
bThe lane number refers to the corresponding sample position in Figure 1B.
CARM1 methylates fewer recombinant proteins on the high-density protein expression filters than PRMT1, thus displaying greater substrate specificity than PRMT1 (Figure 1A). We identified two new substrates for CARM1 (Table I). The poly(A)-binding protein 1 (PABP1) (Blobel, 1973) was the predominant substrate and was isolated six times in the screen. The second identified substrate, TARPP, was recently described as an abundant protein in immature thymocytes and may be involved in early T-cell development (Kisielow et al., 2001). Neither of the CARM1 substrates harbor RGG motifs. In order to identify the methylation motif recognized by CARM1, we concentrated on PABP1, the most prevalent substrate identified in the screen (Table I).
Mapping of the methylated region in PABP1
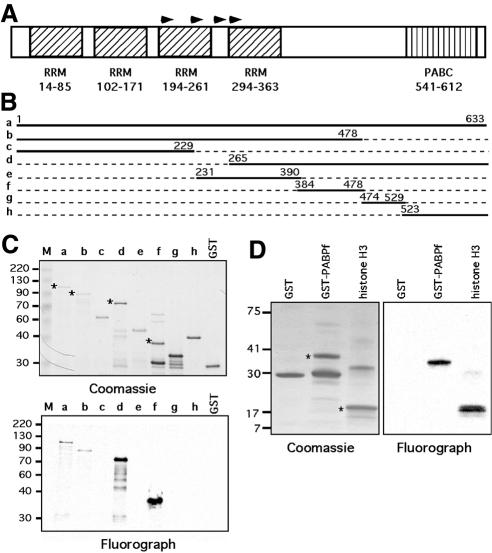
To determine the region of PABP1 methylation by CARM1, we generated a full-length GST–PABP1 fusion construct and a set of seven deletion mutants (Figure 2B). For in vitro methylation, GST fusion proteins containing the sequences depicted in Figure 2B were incubated with recombinant CARM1 in the presence of [3H]AdoMet. The smallest methylated region was mapped to a 100-amino-acid stretch of PABP1 (Figure 2C, construct f). PABP1 has four RRM domains that mediate RNA and protein interactions (Figure 2A). In addition to binding the poly(A) tail of mRNA, PABP1 mediates mRNA circularization through binding of the eIF4F translation initiation complex, which in turn associates with the mRNA 5′ cap structure (Imataka et al., 1998). This circularization enhances translation by allowing recycling of ribosomes (Varani, 2001). PABP1 also harbors a C-terminal domain (PABC) that is involved in protein–protein interactions (Deo et al., 2001) (Figure 2A). The methylated region, which has no function attributed to it, lies between the RRM domain and the PABC domain. Furthermore, this region appears to be methylated as efficiently as is histone H3 by CARM1 (Figure 2D).
Fig. 2. Deletion analysis of PABP1 identifies the methylated region. (A) Diagram of the domain structure of PABP1. Arrowheads represent the start sites of the clones identified in the primary screen. (B) The regions of PABP1 that were fused to GST for deletion analysis. (C) Purified GST fusion proteins were incubated with recombinant CARM1 in the presence of [3H]AdoMet, separated by SDS–PAGE, and the methylated proteins visualized by fluorography (lower panel). A duplicate substrate gel was Coomassie stained to demonstrate loading (upper panel). (D) Histone H3 and GST–PABPf are both efficiently methylated by CARM1. The asterisks on the Coomassie gels indicate the protein bands that are methylated on the fluorograph.
Mapping of the methylated sites in PABP1 and mutational analysis
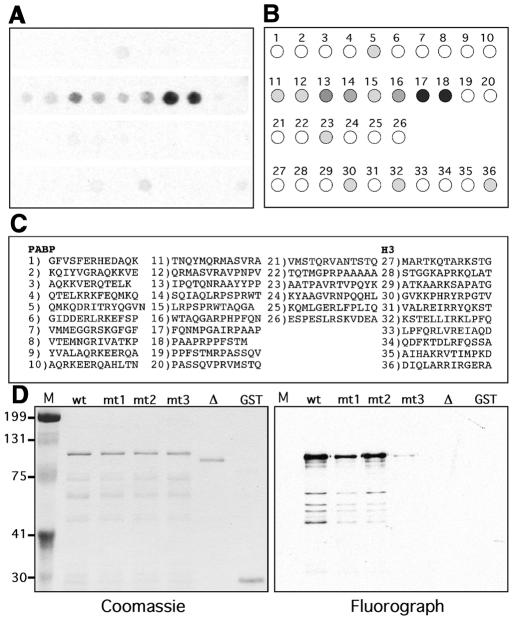
To elucidate further the motif that is methylated by CARM1, we synthesized 13mer peptides that contained an arginine residue flanked by six amino acids, using the ‘SPOTs’ technique of multipeptide synthesis on derivatized cellulose (Linn et al., 1997). Every arginine within the PABP1 fragment spanning the region d (Figure 2B) was synthesized in this arrayed fashion. As a control, all arginine residues within histone H3 were also synthesized as peptides. We then used active recombinant CARM1 to perform a large-scale in situ enzyme reaction on peptide filters (Figure 3A and B). A patch of eight consecutive peptides (11–18) is methylated efficiently by CARM1, with the strongest substrates being 17 and 18. These two peptides are contiguous and share some of the same sequence generating a RPAAPR motif. Portions of this motif are found in the other two CARM1 substrates, histone H3 (APR) and TARPP (RPLA). Close analysis of the PABP1-methylated peptides reveals that all of the peptides do contain at least one proline residue, which may create a structural kink for substrate presentation. Remarkably, this methylated patch of eight consecutive peptides lies within the PABP1f GST fusion protein that represents the smallest region methylated in the GST mapping experiments (Figure 2C).
Fig. 3. Motif mapping and mutational analysis of PABP1. (A–C) CARM1 methylation of arginine containing peptides from PABP1 (GenBank accession No. NP_002559) and histone H3 (GenBank accession No. CAB11424). Cellulose filters containing the synthesized peptides were methylated with recombinant CARM1 and [3H]AdoMet. (A) Fluorograph of the membrane exposed overnight. (B) Graphic depiction of the results, showing the orientation of the derivatized spots on which peptides were synthesized and the relative intensity of the signal. (C) Sequences of the peptides corresponding to numbered spots. (D) Mutational analysis of PABP1. Purified GST fusion proteins, GST–PABP1 (wt), R455 to A (mt1), R460 to A (mt2), R455 460 to A double mutant (mt3) and the ‘patch’ [384–478] deletion mutant (Δ), were methylated with CARM1 in the presence of [3H]AdoMet. A duplicate substrate gel was Coomassie stained to demonstrate equal loading.
Next, we examined which arginine residues in PABP1 were essential for methylation by CARM1. The two arginine residues (spot 17–R455 and spot 18–R460) that were most strongly methylated in Figure 3A were replaced with an alanine residue, either individually or in combination. In addition, a deletion mutant (GST–PABPΔ) was generated that was missing the methylated patch (amino acids 384–478, harboring consecutive peptides 11–18 in Figure 3C). This site-specified mutagenesis was performed on full-length PABP1, within the context of GST–PABP1. The in vitro methylation assay using recombinant CARM1 demonstrates that the R455 460A double mutation strongly influenced the methylation of PABP1 and that the deletion of the methylation ‘patch’ totally abolished the in vitro methylation of PABP1 (Figure 3D).
Full-length PABP1 is methylated in vivo
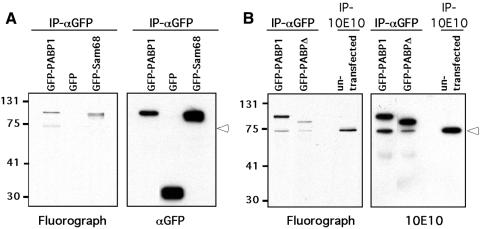
To determine whether PABP1 is methylated in cells, we used the in vivo methylation assay described by Liu and Dreyfuss (1995). HeLa cells were transiently transfected with GFP, GFP–PABP1 and GFP–Sam68, and the methylated proteins were labeled as described in Methods. Sam68 serves as a positive control for in vivo methylation (Bedford et al., 2000). GFP is the negative control to confirm that translation was indeed inhibited. Both the GFP fusion of PABP1 and Sam68 are methylated in HeLa cells, whereas GFP alone remains unmodified (Figure 4A), demonstrating that cellular PABP1 (fused to GFP) is methylated. Next, we show that GFP–PABPΔ displayed much-reduced methylation in vivo and that endogenous PABP1 was a methylated protein (Figure 4B). The fact that PABPΔ is lightly methylated in vivo but not in vitro suggests that there are additional methylation sites for either CARM1 (requiring structural integrity or auxiliary molecules) or another arginine methyltransferase. Interestingly, αGFP immunoprecipitation of GFP–PABP1 co-immunoprecipitated endogenous PABP1, either through direct (GFP–PABP/PABP) or indirect [GFP–PABP/poly(A)-tail/PABP] association.
Fig. 4. PABP1 is methylated in vivo. (A) The GFP fusion protein of PABP1 is methylated in vivo. HeLa cells were transiently transfected with GFP, GFP–PABP1 and GFP–Sam68 plasmid DNA. Methylated proteins were labeled and analyzed as described in Methods. Immunoprecipitations (IP) were performed with anti-GFP antibodies. The 3H-labeled proteins were visualized by fluorography (left panel), and the same membrane was immunoblotted with αGFP antibodies (right panel). (B) Endogenous PABP1 but not the ‘patch’ deletion is efficiently methylated in vivo. HeLa cells were transiently transfected with GFP–PABP1 and GFP–PABPΔ plasmid DNA. Methylated proteins were labeled and analyzed as described in Methods. IP were performed with either anti-GFP or anti-PABP1 (10E10) antibodies. The 3H-labeled proteins were visualized by fluorography (left panel), and the same membrane was immunoblotted with the 10E10 antibody (right panel). Open arrowheads mark the position of the endogenous PABP1.
In this study, we have designed a novel screening method for discovering arginine methylated substrates. The identification of PABP1 as a CARM1-specific substrate expands the repertoire of targets for this enzyme. RNA binding proteins and, in particular, the hnRNPs are highly post-translationally modified through arginine methylation. The hnRNPs are likely methylated by PRMT1 at RGG motifs. Unlike the hnRNPs, histone H3 and PABP1 contains no RGG motifs. The mapping experiments described here identified the region in PABP1 that is methylated by CARM1. This 100-amino-acid patch contains a propensity of proline and arginine residues. Moreover, the two arginine residues that are most strongly methylated in vitro (RPAAPR) (Figure 3) lie within a similar motif to that described for the Arg17 (APR) site in histone 3 that is efficiently methylated in vivo (Schurter et al., 2001). In actual fact, the PABP1 motif is a mirror image duplication of the histone H3 motif.
Arginine methylation may regulate the shuttling abilities of PABP1 or affect its competence to interact with binding partners. One alluring possibility is that the methylation of PABP1 could tag those transcripts that are derived from nuclear receptor transactivated loci.
METHODS
Antibodies and plasmids. All PABP1–GST fusion proteins were subcloned in pGEX6P-1 (Amersham Pharmacia Biotech). The anti-His antibody and the anti-GFP antibody were purchased from Qiagen and Clontech, respectively. The 10E10 monoclonal antibody was a gift from Dr G. Dreyfuss. Site-directed mutagenesis was performed on full-length PABP1 in pGEX6P-1 by PCR using mutant primer sets. For GFP fusion constructs, pGEX6P-1 fusion constructs were cut and directly subcloned into pEGFP-C1 (Clontech). Histone H3 (calf thymus) was purchased from Boehringer Mannheim. The source of GST–PRMTs has been described previously (Frankel et al., 2002).
High-density protein filters. The high-density protein expression filters were obtained from the Resource Center of the DHGP (www.rzpd.de).
Peptide-on-paper synthesis. This synthesis was performed by Jerini (www.jerini.de).
In situ methylation assay. High-density protein filters were pre-blocked with BSA and then incubated overnight with 100 µg of recombinant PRMT and 250 µCi of S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet; 79 Ci/mmol from a 12.6 µM stock solution in dilute HCl–ethanol 9:1, pH 2.0–2.5; Amersham Pharmacia Biotech) in 10 ml of PBS. Filters were then washed three times in TBST (25 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween 20), sprayed with Enhance (NEN) and exposed to film overnight. Cellulose filters containing the SPOTs-synthesized peptides were methylated in the same fashion.
In vitro methylation assay. In vitro methylation reactions were performed in a final volume of 30 µl PBS. The reaction contained 0.5–1 µg of substrate and 1 µg of recombinant PRMT. All methylation reactions were carried out in the presence of 0.42 µM [3H]AdoMet.
Transient transfection and in vivo methylation assay. GFP fusion constructs were transiently transfected into HeLa cells using lipofectin (Gibco-BRL). Twenty-four hours after transfection, the cells were labeled using a previously described in vivo methylation assay (Liu and Dreyfuss, 1995). The cells were lysed in RIPA buffer, and immunoprecipitations were performed with anti-GFP antibodies or 10E10. Samples were separated on a 10% SDS–polyacrylamide gel, transferred to a PVDF membrane, sprayed with Enhance and exposed to film overnight.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs P. Leder and A. Frankel for provided critical discussion. M.T.B. is supported by the Damon Runyon Cancer Research Foundation Scholar Award DRS-28-02 and NIH Center Grant ES07784. J.L. is a postdoctoral fellow supported by the postdoctoral fellowships program from the Korea Science and Engineering Foundation (KOSEF).
REFERENCES
- Bedford M.T., Frankel, A., Yaffe, M.B., Clarke, S., Leder, P. and Richard, S. (2000) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem., 275, 16030–16036. [DOI] [PubMed] [Google Scholar]
- Blobel G. (1973) A protein of molecular weight 78 000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc. Natl Acad. Sci. USA, 70, 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscombe T.L., Frankel, A., Lee, J.H., Cook, J.R., Yang, Z., Pestka, S. and Clarke, S. (2001) Prmt5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem., 276, 32971–32976. [DOI] [PubMed] [Google Scholar]
- Bussow K., Cahill, D., Nietfeld, W., Bancroft, D., Scherzinger, E., Lehrach, H. and Walter, G. (1998) A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res., 26, 5007–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Ma, H., Hong, H., Koh, S.S., Huang, S.M., Schurter, B.T., Aswad, D.W. and Stallcup, M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- Deo R.C., Sonenberg, N. and Burley, S.K. (2001) X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc. Natl Acad. Sci. USA, 98, 4414–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A., Yadav, N., Lee, J., Branscombe, T.L., Clarke, S. and Bedford, M.T. (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem., 277, 3537–3543. [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Massenet, S., Paushkin, S., Wyce, A. and Dreyfuss, G. (2001) SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell, 7, 1111–1117. [DOI] [PubMed] [Google Scholar]
- Fukunaga R. and Hunter, T. (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J., 16, 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D. and Clarke, S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol., 61, 65–131. [DOI] [PubMed] [Google Scholar]
- Gary J.D., Lin, W.J., Yang, M.C., Herschman, H.R. and Clarke, S. (1996) The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem., 271, 12585–12594. [DOI] [PubMed] [Google Scholar]
- Henry M.F. and Silver, P.A. (1996) A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol., 16, 3668–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi, A. and Sonenberg, N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow J., Nairn, A.C. and Karjalainen, K. (2001) TARPP, a novel protein that accompanies TCR gene rearrangement and thymocyte education. Eur. J. Immunol., 31, 1141–1149. [DOI] [PubMed] [Google Scholar]
- Koh S.S., Chen, D., Lee, Y.H. and Stallcup, M.R. (2000) Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem., 276, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Linn H., Ermekova, K.S., Rentschler, S., Sparks, A.B., Kay, B.K. and Sudol, M. (1997) Using molecular repertoires to identify high-affinity peptide ligands of the WW domain of human and mouse YAP. Biol. Chem., 378, 531–537. [DOI] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss, G. (1995) In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol., 15, 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A.E. and Silver, P.A. (2001) State of the arg: protein methylation at arginine comes of age. Cell, 106, 5–8. [DOI] [PubMed] [Google Scholar]
- Rawal N., Rajpurohit, R., Lischwe, M.A., Williams, K.R., Paik, W.K. and Kim, S. (1995) Structural specificity of substrate for S-adenosylmethionine:protein arginine N-methyltransferases. Biochim. Biophys. Acta, 1248, 11–18. [DOI] [PubMed] [Google Scholar]
- Schurter B.T. et al. (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry, 40, 5747–5756. [DOI] [PubMed] [Google Scholar]
- Shen E.C., Henry, M.F., Weiss, V.H., Valentini, S.R., Silver, P.A. and Lee, M.S. (1998) Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev., 12, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Kao, P.N. and Herschman, H.R. (2000) Protein-arginine methyl-transferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem., 275, 19866–19876. [DOI] [PubMed] [Google Scholar]
- Varani G. (2001) Delivering messages from the 3′ end. Proc. Natl Acad. Sci. USA, 98, 4288–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science, 293, 853–857. [DOI] [PubMed] [Google Scholar]