Abstract
The organization of eukaryotic chromatin has a major impact on all nuclear processes involving DNA substrates. Gene expression is affected by the positioning of individual nucleosomes relative to regulatory sequence elements, by the folding of the nucleosomal fiber into higher-order structures and by the compartmentalization of functional domains within the nucleus. Because site-specific acetylation of nucleosomal histones influences all three aspects of chromatin organization, it is central to the switch between permissive and repressive chromatin structure. The targeting of enzymes that modulate the histone acetylation status of chromatin, in synergy with the effects mediated by other chromatin remodeling factors, is central to gene regulation.
Introduction
Since the discovery of the basic principles of chromatin organization, which involves the wrapping of DNA around histone octamers to form nucleosomes and the folding of the nucleosomal fiber into higher-order structures (Luger and Richmond, 1998; Hayes and Hansen, 2001; Woodcock and Dimitrov, 2001), the question of how such extensive packaging can be compatible with reactions that involve ‘reading’ the DNA has stimulated extensive research. It soon became apparent that many aspects of chromatin structure could be explained by interactions between nucleosomal histones and DNA, neighboring nucleosomes and non-histone proteins. Most of these interactions involve the N-terminal ‘tails’ of the core histones, which reach out from the rather compact nucleosomal core particle (Fletcher and Hansen, 1996).
The N-termini of the extensively studied histones H4 and H3 are among the most highly conserved sequences in eukaryotes. Although rather short (19 and 26 amino acids, respectively), their documented and suspected interactions suggest central roles for these domains in chromatin structure and function. Post-translational modifications of conserved tail amino acids, notably phosphorylation, methylation and acetylation, modulate the interaction potential of the tail domains, and hence influence the folding and functional state of the chromatin fiber (Grunstein, 1997; Howe et al., 1999; Berger, 2001; Jenuwein and Allis, 2001).
With the identification of transcription activators and co-activators as dedicated histone acetyltransferases (HATs), it became possible to document the relationships between histone acetylation and gene activation in many cases (Sterner and Berger, 2000; Chen et al., 2001; Roth et al., 2001). Without exception, multi-protein assemblies determine the functions, substrate specificities and targeting of integral HAT subunits (Wolffe and Hayes, 1999; Nakatani, 2001; Ogryzko, 2001). The acetylation of histones, and hence all effects on structure, can be reversed by dedicated histone deacetylases (HDACs), and many repression phenomena involve histone deacetylation (Khochbin et al., 2001). Thus, the interplay between HDACs and HATs results in dynamic transitions in chromatin structure and, hence, in switches between activity states. The functional importance of the HATs and HDACs is highlighted by the fundamental regulatory roles that they have in developmental processes, and by the fact that their deregulation has been linked to the progression of cancers (e.g. leukemia, colorectal and breast cancer) and diverse human disorders, like the Rubinstein–Tabi and fragile X syndromes (Timmermann et al., 2001).
Broad, domain-wide histone acetylation
Acetylation of histones H3 and H4 counteracts the tendency of nucleosomal fibers to fold into highly compact structures in vitro (Garcia-Ramirez et al., 1995; Tse et al., 1998) and acetylated chromatin is more accessible to interacting proteins in vivo, as illustrated by its increased sensitivity to DNase I (Hebbes et al., 1994; Krajewski and Becker, 1998). The importance of histone acetylation as an epigenetic marker of chromosomal domains has recently been corroborated by advanced chromatin immunoprecipitation (X-CHIP) studies using histone isotype-specific antisera (Suka et al., 2001). An important conclusion from such studies in yeast was that the ground state of chromatin, be it transcribed or not, is characterized by intermediate levels of H3 and H4 acetylation, a state brought about by a mix of untargeted HAT and HDAC activities (Vogelauer et al., 2000). In this context, site-specific acetylation or deacetylation leads to locally restricted activation or repression of transcription, respectively. However, this overall level of flexibility appears to be a specific feature of the highly active yeast genome. In differentiated, higher eukaryotic cells, most of the genome consists of hypoacetylated, inactive chromatin, which may be considered the ‘ground state’. Activation of house-keeping and cell-type-specific genes involves the acetylation of histones across broad chromatin domains. A well-characterized example is provided by the β-globin loci, which reveal broad acetylation throughout domains with defined boundaries as a function of transcriptional competence (Litt et al., 2001; Schübeler et al., 2001). Another prominent example of a broad acetylation effect is provided by the dosage compensated male X chromosome in Drosophila (Lucchesi, 1998), where the acetylation of histone H4 at lysine 16 (H4K16) by MOF (Males absent on the First) correlates with increased transcription of many genes throughout most of the male X chromosome (Akhtar and Becker, 2000; Smith et al., 2001).
While broad acetylation of histone H3/H4 leads to partial decondensation of chromosomal domains, this opening is not tightly correlated with active transcription per se, but rather marks regions of transcriptional competence. It is known that a domain that has been rendered ‘permissive’ by broad acetylation will never be found close to repressive heterochromatic structures in nuclei (Schübeler et al., 2000). Nevertheless, the causal relationship between residence in euchromatin and histone acetylation has not yet been established.
Local, targeted histone acetylation
Transcriptional activation within a permissive domain frequently correlates with additional, targeted acetylation of histones at promoter nucleosomes (Brown et al., 2000; Forsberg and Bresnick, 2001), although notable exceptions exist (Deckert and Struhl, 2001). With the availability of more diagnostic reagents and more sophisticated analyses, the old idea (Turner, 1993) that what matters are patterns of acetylation at specific lysines within the histone N-termini, rather than simple charge neutralization by non-specific modification, receives convincing confirmation (Vogelauer et al., 2000; Deckert and Struhl, 2001). However, the exact requirements are not at all transparent. Whereas in some instances targeted acetylation of histone H3 is found in conjunction with broader acetylation of histone H4 (Schübeler et al., 2000; Vignali et al., 2000), these observations cannot be generalized (Litt et al., 2001).
Given the importance of HATs as co-activators of transcription and the frequent association of repression with HDACs, transcriptional regulation must involve targeting of these enzymes to specific sites. Localized histone acetylation is observed in promoter and enhancer elements, but can also be found enriched at boundary or insulator elements of chromosome domains and other DNase I hypersensitive sites in nuclei (Litt et al., 2001), supporting the idea that histone acetylation facilitates protein–DNA interactions within chromatin in general. Many cases in which activating transcription factors recruit HAT-containing co-activators to specific promoters have now been documented (Brown et al., 2000). Yeast HAT complexes can associate with the transactivation domains of activators like VP16, Gcn4, Gal4 and Hap4 (Utley et al., 1998), although this may require an adapter protein. For example, the yeast SAGA and NuA4 complexes, which contain the HATs Gcn5 and Esa1, respectively, are recruited via the shared subunit Tra1 (Brown et al., 2001). The human homolog of Tra1, TRRAP, is also implicated in recruiting HAT complexes, in this case to transcription complexes containing c-myc (McMahon et al., 2000; Bouchard et al., 2001; Frank et al., 2001) and E2F (Lang et al., 2001). Although some basic principles are emerging, our present understanding of HAT involvement in gene activation remains dominated by inherent complexities. Activator–co-activator selectivity is inferred from the fact that different activators induce different acetylation patterns in vivo (Deckert and Struhl, 2001). For example, the recruitment of SAGA by VP16 leads to local H3 acetylation near the promoter, while targeting of NuA4 by the same protein results in broad acetylation of H4 over a domain of >3 kb (Vignali et al., 2000). The targeting of diverse HAT complexes via sequence-specific DNA-binding proteins leads to stimulation of transcription from chromatin templates in vitro (Ikeda et al., 1999; Kundu et al., 2000) and in yeast (Bhaumik and Green, 2001; Larschan and Winston, 2001). Although the HAT subunits of co-activator complexes are currently the center of interest, large HAT complexes like SAGA contain other functions as well. This is illustrated by the observation that SAGA has been found to be an essential co-activator for Gal4-activated transcription in vivo, but this activation function relied mainly on the SAGA components Spt3 and Spt20 and less on the HAT subunit Gcn5 (Bhaumik and Green, 2001; Larschan and Winston, 2001).
Whereas tethering of HATs to defined sites via activators explains local hyperacetylation, it is less obvious how the acetylation of large domains is achieved. Potential mechanisms include the recruitment of HATs to distinct ‘entry sites’ from which they ‘spread’ throughout a domain (Kelley and Kuroda, 2000), possibly by attachment to a tracking protein such as RNA polymerase II (Wittschieben et al., 1999). Alternatively, the residence of a particular chromosomal domain within an acetylation-competent nuclear compartment may ensure relatively uniform modification (Schübeler et al., 2000). If acetylation itself were to generate high-affinity binding sites for HATs, propagation schemes could be envisaged (Gu et al., 2000; Forsberg and Bresnick, 2001).
How does histone acetylation work?
Currently, it is widely assumed that particular histone acetylation patterns lead to altered folding of the nucleosomal fiber that renders chromosomal domains more accessible. As a consequence, the transcription machinery may be able to access promoters and hence initiate transcription more frequently. In addition, the unfolding of chromosomal domains also facilitates the process of transcription elongation itself. Nucleosomes are obstacles to the elongating RNA polymerase, which may need to transfer the histone octamers it encounters to acceptor DNA in the wake of elongation (Studitsky et al., 1997; Orphanides and Reinberg, 2000). Transcription is found to stutter less on hyperacetylated nucleosomal templates than on non-acetylated ones (Protacio et al., 2000). Therefore, some HATs may be responsible for facilitating the passage of the elongating polymerase, either as part of dedicated elongation factor complexes such as FACT (John et al., 2000), or as an integral activity of the elongating polymerase machinery itself (Cho et al., 1998; Wittschieben et al., 1999).
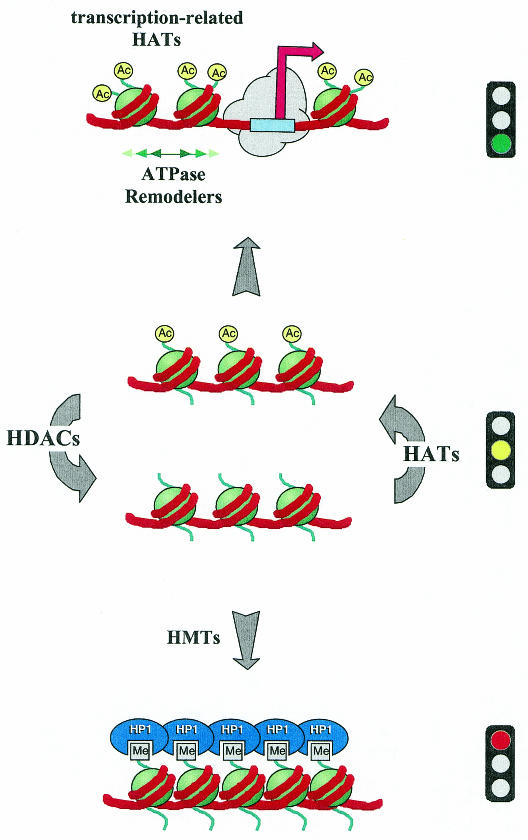
If the loosening of higher-order chromatin structure by domain-wide histone acetylation is necessary but not sufficient for transcription, what is the role of further, targeted acetylation of individual nucleosomes at regulatory elements? Even in the absence of a folded nucleosomal fiber, single nucleosomes are still considerable obstacles to the interactions between many transcription factors and their binding partners (Struhl, 1998). It may be that hyperacetylated nucleosomes, which appear to be somewhat less restrictive towards interacting factors in some cases (Vettese-Dadey et al., 1996; Anderson et al., 2001; Sewack et al., 2001), function by increasing the flexibility of the DNA associated with the ends of nucleosomes (Krajewski and Becker, 1998). Overall, the effect of acetylation per se on nucleosome structure appears rather modest (Wang et al., 2000). However, specific acetylation patterns displayed by the histone tails may also function to recruit further modulators of chromatin structure. The dramatic changes in promoter structure that accompany transcriptional activation are, therefore, presumably not the direct result of acetylation, but due to the synergistic actions of several factors. These include other covalent modifications such as phosphorylation, as well as the rearrangement of histones/nucleosomes relative to the DNA by nucleosome remodeling factors (see Figure 1).
Fig. 1. The histone acetylation switch. Targeted HAT and HDAC activities negotiate the acetylation status of chromatin. Acetylation establishes a structure that permits ATP-dependent chromatin remodeling factors to open promoters. Deacetylation, frequently followed by histone methylation, may form a solid base for highly repressive structures, such as heterochromatin. Acetylated histone tails are shown as yellow circles. Methylations are indicated as gray rectangles. HAT, histone acetyltransferase; HDAC, histone deacetylase; HMT, histone methyltransferase; HP1, heterochromatin protein 1.
Histone modification: signal integration at promoters
Signal transduction cascades are known to employ covalent protein modification, most notably phosphorylation, to modulate gene expression in response to extracellular stimuli. Not surprisingly, these phosphorylation cascades also impact on chromatin organization. Following the ‘classical’ route of signal transduction, histone acetyltransferase activity may be activated or repressed by phosphorylation or acetylation (Cheung et al., 2000a; Kouzarides, 2000). More indirectly, phosphorylation of nucleosomes at the histone N-termini has profound effects on chromatin organization at specific sites. In yeast, many genes are co-regulated by pathways involving HATs and kinases (Lo et al., 2001). A striking example of this kind of synergism is the growth-factor induction of immediate early genes in higher eukaryotes, which involves co-ordinated phosphorylation of H3S10 and acetylation at H3K14 on the same tail (Clayton et al., 2000). Co-ordination can be explained, at least in part, by the sensitivity of Gcn5, the HAT involved in this particular activation event, to phosphorylation of the H3 N-terminus. In vitro, Gcn5 acetylates a phosphorylated H3S10 peptide much more avidly than an unmodified one (Cheung et al., 2000b; Lo et al., 2001). The factor(s) that respond to phospho-acetylation remain unknown, but the fact that it was possible to raise an antiserum that selectively recognizes the double-modified tail (Clayton et al., 2000) indicates that these may exist.
During the past 2 years, several histone methyltransferases (HMT) that function as epigenetic repressors have been characterized (Rice and Allis, 2001; Zhang and Reinberg, 2001). A synergism, similar to that between histone acetylation and phosphorylation, is also suggested between histone acetylation and methylation at specific sites by the physical interaction of CBP with HMT activity (Vandel and Trouche, 2001). Interestingly, histone deacetylation and the subsequent methylation also appear to be co-ordinated activities, since both have been found to occur in one large complex (Czermin et al., 2001).
HAT complexes themselves may be tethered to distinct regulatory sites via modified histones, i.e. by interaction with the nucleosome itself. Combinations of acetylation, phosphorylation and methylation may be the code for recognition and binding by chromatin regulators, such as HATs (Strahl and Allis, 2000; Turner, 2000; Imhof and Becker, 2001). Indeed, many chromatin-modifying enzymes share domains that can selectively interact with modified nucleosomes. For example, the bromodomain, found in both Gcn5 and Swi2 (Winston and Allis, 1999), has an affinity for the H4 tail, and this is much enhanced by acetylation at defined lysines (Jacobson et al., 2000; Owen et al., 2000). While the recognition of specific histone isoforms by dedicated protein domains emerges as one fundamental targeting principle, the initial establishment of the modification patterns presumably relies on the above-mentioned targeting principles.
Switching between activation and repression pathways
Histone acetylation emerges as a central switch that allows interconversion between permissive and repressive chromatin structures and domains (Figure 1, center panel). These principles are not only at the heart of transcriptional regulation but are also likely to govern other processes involving chromatin substrates, including replication, site-specific recombination and DNA repair (Wolffe and Hayes, 1999; Roth et al., 2001). As indicated above, domain-wide and site-specific histone acetylation are necessary for transcription, but are not sufficient to establish full chromatin accessibility. However, the switch to a permissive chromatin structure is conducive to synergistic actions between nucleosome remodeling complexes containing ATPases of the SWI2/SNF2 family. These enzymes increase the accessibility of nucleosomal DNA by weakening histone–DNA interactions leading to, for example, histone octamer relocation (Figure 1, upper panel). Detailed analyses of this kind of interplay are presented in recent reviews (Fry and Peterson, 2001; Becker and Hörz, 2002).
The switch to repressive chromatin involves histone deacetylation, which promotes the condensation of the nucleosomal fiber and invites repressive factors. One way of switching the state of chromatin more permanently is to prevent acetylation by methylation of the corresponding lysine residues (Figure 1, lower panel). Methylation at specific tail lysines may attract heterochromatin protein 1 (HP1) to lock chromatin in a repressive, inaccessible configuration.
At present, the field of chromatin research appears to be as dynamic as the subject of its studies. Without doubt, advances in understanding the targeting and effects of histone acetylation will contribute significantly to unraveling the principles of gene regulation.

Anton Eberharter & Peter B. Becker

Acknowledgments
Acknowledgements
We would like to apologize to our colleagues that, due to the inverse relationship between original publications on the subject and the available print space, we were unable to refer to many original publications. Special thanks to R. Aasland for his comments and his assistance in designing the figure and several thoughtful referees.
References
- Akhtar A. and Becker, P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 367–375. [DOI] [PubMed] [Google Scholar]
- Anderson J.D., Lowary, P.T. and Widom, J. (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 307, 977–985. [DOI] [PubMed] [Google Scholar]
- Becker P.B. and Hörz, W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., in press. [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene, 20, 3007–3013. [DOI] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green, M.R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev., 15, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Dittrich, O., Kiermaier, A., Dohmann, K., Menkel, A., Eilers, M. and Luscher, B. (2001) Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev., 15, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Lechner, T., Howe, L. and Workman, J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S. and Workman, J.L. (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science, 292, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Chen H., Tini, M. and Evans, R.M. (2001) HATs on and beyond chromatin. Curr. Opin. Cell Biol., 13, 218–224. [DOI] [PubMed] [Google Scholar]
- Cheung P., Allis, C.D. and Sassone-Corsi, P. (2000a) Signaling to chromatin through histone modifications. Cell, 103, 263–271. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner, K.G., Cheung, W.L., Sassone-Corsi, P., Denu, J.M. and Allis, C.D. (2000b) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- Cho H., Orphanides, G., Sun, X., Yang, X.J., Ogryzko, V., Lees, E., Nakatani, Y. and Reinberg, D. (1998) A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol., 18, 5355–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A.L., Rose, S., Barratt, M.J. and Mahadevan, L.C. (2000) Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J., 19, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Schotta, G., Hulsmann, B.B., Brehm, A., Becker, P.B., Reuter, G. and Imhof, A. (2001) Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO rep., 2, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J. and Struhl, K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol., 21, 2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher T.M. and Hansen, J.C. (1996) The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr., 6, 149–188. [DOI] [PubMed] [Google Scholar]
- Forsberg E.C. and Bresnick, E.H. (2001) Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. BioEssays, 23, 820–830. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder, M., Fernandez, P., Taubert, S. and Amati, B. (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev., 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C.J. and Peterson, C.L. (2001) Chromatin remodeling enzymes: who’s on first? Curr. Biol., 11, 185–197. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Rocchini, C. and Ausio, J. (1995) Modulation of chromatin folding by histone acetylation. J. Biol. Chem., 270, 17923–17928. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Gu W., Wei, X., Pannuti, A. and Lucchesi, J.C. (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J., 19, 5202–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J.J. and Hansen, J.C. (2001) Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev., 11, 124–129. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton, A.L., Thorne, A.W. and Crane-Robinson, C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L., Brown, C.E., Lechner, T. and Workman, J.L. (1999) Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr., 9, 231–243. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Steger, D.J., Eberharter, A. and Workman, J.L. (1999) Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol., 19, 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A. and Becker, P.B. (2001) Modifications of the histone N-terminal domains. Evidence for an “epigenetic code”? Mol. Biotechnol., 17, 1–13. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner, A.G., King, D.S. and Tjian, R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis, C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- John S., Howe, L., Tafrov, S.T., Grant, P.A., Sternglanz, R. and Workman, J.L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes Dev., 14, 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L. and Kuroda, M.I. (2000) The role of chromosomal RNAs in marking the X for dosage compensation. Curr. Opin. Genet. Dev., 10, 555–561. [DOI] [PubMed] [Google Scholar]
- Khochbin S., Verdel, A., Lemercier, C. and Seigneurin-Berny, D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev., 11, 162–166. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski W.A. and Becker, P.B. (1998) Reconstitution of hyperacetylated, DNase I-sensitive chromatin characterized by high conformational flexibility of nucleosomal DNA. Proc. Natl Acad. Sci. USA, 95, 1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu T.K., Palhan, V.B., Wang, Z., An, W., Cole, P.A. and Roeder, R.G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell, 6, 551–561. [DOI] [PubMed] [Google Scholar]
- Lang S.E., McMahon, S.B., Cole, M.D. and Hearing, P. (2001) E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem., 276, 32627–32634. [DOI] [PubMed] [Google Scholar]
- Larschan E. and Winston, F. (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev., 15, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M.D., Simpson, M., Recillas-Targa, F., Prioleau, M.N. and Felsenfeld, G. (2001) Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J., 20, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.S., Duggan, L., Tolga, N.C.E., Belotserkovskya, R., Lane, W.S., Shiekhattar, R. and Berger, S.L. (2001) Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science, 293, 1142–1146. [DOI] [PubMed] [Google Scholar]
- Lucchesi J.C. (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev., 8, 179–184. [DOI] [PubMed] [Google Scholar]
- Luger K. and Richmond, T.J. (1998) The histone tails of the nucleosome. Curr. Opin. Genet. Dev., 8, 140–146. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Wood, M.A. and Cole, M.D. (2000) The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol., 20, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y. (2001) Histone acetylases—versatile players. Genes Cells, 6, 79–86. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V. (2001) Mammalian histone acetyltransferases and their complexes. Cell. Mol. Life Sci., 58, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G. and Reinberg, D. (2000) RNA polymerase II elongation through chromatin. Nature, 407, 471–475. [DOI] [PubMed] [Google Scholar]
- Owen D.J., Ornaghi, P., Yang, J.C., Lowe, N., Evans, P.R., Ballario, P., Neuhaus, D., Filetici, P. and Travers, A.A. (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J., 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protacio R.U., Li, G., Lowary, P.T. and Widom, J. (2000) Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol. Cell. Biol., 20, 8866–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J.C. and Allis, C.D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol., 13, 263–273. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu, J.M. and Allis, C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Schübeler D., Francastel, C., Cimbora, D.M., Reik, A., Martin, D.I. and Groudine, M. (2000) Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev., 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Schübeler D., Groudine, M. and Bender, M.A. (2001) The murine β-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl Acad. Sci. USA, 98, 11432–11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewack G.F., Ellis, T.W. and Hansen, U. (2001) Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol., 21, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Allis, C.D. and Lucchesi, J.C. (2001) Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem., 276, 31483–31486. [DOI] [PubMed] [Google Scholar]
- Sterner D.E. and Berger, S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Studitsky V.M., Kassavetis, G.A., Geiduschek, E.P. and Felsenfeld, G. (1997) Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science, 278, 1960–1963. [DOI] [PubMed] [Google Scholar]
- Suka N., Suka, Y., Carmen, A.A., Wu, J. and Grunstein, M. (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell, 8, 473–479. [DOI] [PubMed] [Google Scholar]
- Timmermann S., Lehrmann, H., Polesskaya, A. and Harel-Bellan, A. (2001) Histone acetylation and disease. Cell. Mol. Life Sci., 58, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C., Sera, T., Wolffe, A.P. and Hansen, J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. (1993) Decoding the nucleosome. Cell, 75, 5–8. [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda, K., Grant, P.A., Cote, J., Steger, D.J., Eberharter, A., John, S. and Workman, J.L. (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498–502. [DOI] [PubMed] [Google Scholar]
- Vandel L. and Trouche, D. (2001) Physical association between the histone acetyl transferase CBP and a histone methyl transferase. EMBO rep., 2, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettese-Dadey M., Grant, P.A., Hebbes, T.R., Crane- Robinson, C., Allis, C.D. and Workman, J.L. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J., 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Steger, D.J., Neely, K.E. and Workman, J.L. (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J., 19, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu, J., Suka, N. and Grunstein, M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Wang X., Moore, S.C., Laszckzak, M. and Ausio, J. (2000) Acetylation increases the α-helical content of the histone tails of the nucleosome. J. Biol. Chem., 275, 35013–35020. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis, C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Wittschieben B.O. et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Hayes, J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C.L. and Dimitrov, S. (2001) Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev., 11, 130–135. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg, D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360. [DOI] [PubMed] [Google Scholar]