Abstract
Ehrlichia risticii, the causative agent of Potomac horse fever, has recently been isolated from many vaccinated horses with typical clinical signs of the disease. The heterogeneity of the E. risticii isolates obtained from the vaccinated horses necessitates the identification of the molecular basis of strain variations to elucidate the vaccine failure and to aid in the development of an efficient vaccine against this disease. As an attempt, two major cross-reacting surface antigen genes of 50- and 85-kDa antigens, present separately in strains 25-D (isolated in 1984) and 90-12 (isolated in 1990 from a vaccinated horse), respectively, were cloned and sequenced. A comparative sequence analysis revealed differences and similarities between these two antigens with strain-specific sizes (SSA). The 2.5- and 1.6-kb genes coding for the 85- and 50-kDa proteins, respectively, contained many different tandem repeats. The identical repeat motifs were more frequent in the middle of both genes, but the numbers and positions of the repeats were altogether different in the genes. Many of these direct repeats of both genes had exact sequence homology and coded for the same amino acids. The homology of the 5′- and 3′-flanking regions of the two genes was greater than that of the regions in the central part of the genes. A comparative analysis of the deduced amino acid sequences of these two antigen genes indicated eight common domains, which were designated identical domains. Although the sequence homologies of these identical domains were the same, the positions of the domains in their respective strains were completely different. This finding might be one of the bases of antigenic variation between the strains. In addition, there were a few unique regions in both antigen genes where no sequence homology existed. These specific regions were designated unique domains. The 50-kDa protein had two such unique domains, and the 85-kDa protein had six such unique domains. The presence of such unique domains contributed to the large size variation of these SSA. The cross-reactivity of recombinant proteins confirmed the presence of conserved epitopes between these two antigens. The SSA have been determined to be apparent protective antigens of E. risticii.
The obligate intracellular bacterium Ehrlichia risticii is the etiologic agent of Potomac horse fever (PHF). The disease is characterized by fever, leukopenia, depression, anorexia, and profuse diarrhea and affects horses of all ages, with a fatality rate as high as 20%. The mode of transmission of this disease remains unknown.
Although PHF has been well recognized and studied for a decade, the immune response against E. risticii infection is still not completely understood. Specifically, the role of individual E. risticii antigens in eliciting the immune response during infection is not well defined. In addition, the implication of strain variations in PHF vaccine failure in horses necessitates a better understanding of the molecular heterogeneity of major antigens. Studies in our laboratory indicated marked phenotypic and genotypic diversity between two strains of E. risticii (24). Heterogeneity is seen mainly in the surface antigens, as demonstrated by serological analysis (8, 24).
To describe in more detail the antigenic heterogeneity of E. risticii strains, we began a molecular analysis of two strains, with the aims of isolating the genes of major protein immunogens and of determining the strain-specific differences in the surface antigens. The two E. risticii strains used were 25-D and 90-12 (24). Strain 25-D was isolated in 1984 during the original outbreaks, and strain 90-12 was isolated in 1990 from a vaccinated horse suffering from severe clinical PHF. A preliminary serological analysis of the component antigens of strains 25-D and 90-12 indicated that antibodies to the 85-kDa antigen of strain 90-12 cross-reacted with a 50-kDa surface antigen of strain 25-D and that the molecular masses of these antigens were specific for their corresponding strains (24). That is, strain 90-12 does not possess the 50-kDa antigen and strain 25-D does not contain the 85-kDa antigen, indicating that although they cross-react in Western blots, these two antigens are strain specific for their distinct molecular masses. We previously reported the ability of these antigens to stimulate a protective immune response in a mouse model of E. risticii infection (25).
This paper describes the molecular cloning and analysis of the 50- and 85-kDa genes, which further confirmed that the immunological variations between the strains (in Western blots) is due to the diversity of genes for antigens with strain-specific sizes (SSA) and demonstrated the unique characteristics of the genes which contribute to the molecular heterogeneity of E. risticii strains.
MATERIALS AND METHODS
E. risticii strains and cultivation.
E. risticii 25-D and 90-12 were obtained from our laboratory stock cultures. Both strains were cultivated in human histiocyte cell line U937 (American Type Culture Collection) in the presence of RPMI 1640 medium (Flow Laboratories, Inc., McLean, Va.) supplemented with 4 mM l-glutamine and 15% fetal calf serum. Infected cells were incubated at 37°C in 5% CO2. The propagation of E. risticii in cell culture material was monitored by acridine orange staining (4).
Purification of E. risticii and DNA extraction.
E. risticii organisms were purified by centrifugation over a linear Renografin gradient according to procedures described previously (5). A procedure described earlier (6) was used for the extraction of DNA from E. risticii. Briefly, Renografin-purified organisms suspended in 50 mM Tris–25 mM EDTA–0.9% NaCl buffer were treated with lysozyme (2 mg/ml; 37°C for 10 min), 0.5% sodium dodecyl sulfate (65°C for 30 min), and proteinase K (400 μg/ml; Sigma Chemical Co., St. Louis, Mo.) (56°C for 6 h). Finally, the DNA was extracted by phenol-chloroform treatment and ethanol precipitation and then dissolved in 10 mM Tris–0.5 mM EDTA (pH 8.0) to a concentration of 1.0 μg/ml.
Cloning of E. risticii genomes.
Fragments of the genomic DNA of E. risticii (strain 25-D) were cloned in vector λgt11, and a recombinant expressing the complete 50-kDa protein antigen was identified in our laboratory (7). Additional cloning of E. risticii (strain 90-12) was performed with vector λZAP (Stratagene, La Jolla, Calif.). Briefly, genomic DNA (6 μg) of strain 90-12 was restriction digested with Sau3AI enzyme (10 U) for 1 h at 37°C. After the size distribution was determined in a 1% agarose gel, the restricted DNA fragments were inserted into vector λZAP by use of conversion adaptors as described by Stover et al. (22). Custom-made single-stranded oligonucleotides (Oligos ET Inc., Wilsonville, Oreg.) of different lengths, when mixed in equimolar quantities, formed duplex adaptors of three different lengths, so that gene fusions at all three reading frames were possible. The conversion adaptors carried an EcoRI cohesive end at one terminus for ligation to the λZAP arms and a restriction enzyme-specified cohesive end at the opposite terminus for ligation to restriction DNA fragments. The restricted DNA fragments were ligated to the conversion adaptors with 6 U of T4 DNA ligase at 15°C for 6 h. After completion of the ligation reaction, the enzyme was heat inactivated at 70°C for 10 min. The adaptor-modified insert DNA was phosphorylated with 10 U of T4 polynucleotide kinase at 37°C for 30 min, followed by the removal of excess adaptors by spin-column chromatography with a Sephacryl S-400 matrix (Promega Corp., Madison, Wis.). Finally, dephosphorylated, adaptor-modified insert DNA was ligated to EcoRI-digested and dephosphorylated vector λZAP DNA (Stratagene) with 4 Weiss U of T4 DNA ligase at 15°C for 6 h. The ligation mixture was packaged in a packaging mix (Gigapack II Gold; Stratagene) at 22°C for 2 h.
Immunoscreening of recombinants for antigen expression.
The recombinants were screened by a previously described procedure (7) with E. risticii 90-12 antisera (rabbit and mouse) absorbed with lysates of Escherichia coli XL1-Blue and λZAP phage (7). The antigen-positive recombinants were purified by single-plaque isolation and stored in 0.1 M NaCl–10 mM Tris (pH 7.9)–10 mM MgSO4.
Western immunoblotting and identification of recombinant antigens.
Western blotting was performed according to a procedure described previously (6) with mouse antisera collected at 3 weeks postinfection. The identification of recombinant antigens was performed by Western blotting with specific recombinant clone-specific antibodies prepared by eluting the absorbed rabbit and mouse anti-E. risticii 90-12 antibodies from a nitrocellulose membrane containing the recombinant λZAP phages expressing the E. risticii antigens according to a previously described procedure (6, 7, 17).
Recombinant DNA procedures.
The E. risticii antigens were expressed by several recombinant λZAP phages. In vivo excision of the pBluescript SK(−) phagemids from these λZAP recombinant phages was done according to the instructions of the manufacturer (Stratagene). All restriction enzymes, T4 DNA ligase, and calf intestinal alkaline phosphatase were obtained from New England BioLabs (Beverly, Mass.) and used according to the instructions of the manufacturer. Phagemid DNA was prepared for PCR, sequencing, and other recombinant procedures by a modification of the procedure of Birnboim and Doly (2). PCR-amplified products were cloned into vector pCRII (Invitrogen). PCR mixtures and cycles were set up according to a procedure described earlier (3).
The recombinant phagemid and plasmid DNAs were transformed into their appropriate competent cells by electroporation or heat shock according to manufacturers’ (Bio-Rad and Invitrogen) instructions.
All preparative and nonpreparative agarose (Bio-Rad) gel electrophoresis procedures were performed with Tris-acetate buffer, and the DNA was visualized with ethidium bromide (Sigma). Cloned inserts were separated from recombinant plasmid or phagemid DNAs by appropriate restriction enzyme digestion. The insert bands of recombinants, as ascertained by electrophoresis, were excised and processed for purification of DNA with GenecleanII (Bio 101, Inc., La Jolla, Calif.) silica matrix.
DNA sequence determination and analysis.
Denatured double-stranded DNA sequencing was accomplished by a modification of the dideoxy chain termination method of Sanger et al. (15, 21) with the Sequenase version 2.0 kit (Amersham).
The nucleotide and deduced amino acid sequences were analyzed with IBI Pustell (IBI Limited, Cambridge, England) and PepPlot software. PepPlot was written by Michael Gribskov and John Deverenux of the Genetics Computer Group and is available through the National Institutes of Health, Bethesda, Md.
Expression of cloned E. risticii antigens.
All of the recombinant antigens were expressed in E. coli JM109 cells and purified under denaturing conditions according to the manufacturer’s (Invitrogen) pRSET-ABC expression system protocol.
Nucleotide sequence accession numbers.
The sequences of the 50- and 85-kDa antigen genes were deposited in the GenBank data library under accession numbers AF059672 and AF059673, respectively.
RESULTS
Cloning and sequencing of the 50- and 85-kDa-antigen genes.
A single recombinant clone, designated pB50-6, expressing the 50-kDa antigen was identified from the λgt11 library of strain 25-D. Two different recombinant clones, designated pB85-11 and pB85-17, expressing the 85-kDa antigen were isolated from the λZAP library of strain 90-12. Initial Western blot analysis of these clones revealed that the 85-kDa antigen was expressed in a fusion with the Lac-Zα peptide (data not shown). Further, clone-specific antibodies of either recombinant antigen reacted with the 50- and 85-kDa proteins of strains 25-D and 90-12, respectively (data not shown). Also, in Southern blot analysis, probes made from the inserts of the recombinant clones of either antigen hybridized with genomic DNA of both strains (24). Based on this antigenic and genomic relatedness and because of the different molecular sizes of the antigens, the 50- and 85-kDa proteins were designated SSA homologs for strains 25-D and 90-12, respectively.
The 3.9-kb insert of clone pB50-6 was digested with various restriction enzymes, and the resulting smaller fragments were subcloned in pBluescript SK(+). By sequencing of these fragments, the complete nucleotide sequences of the 50-kDa-antigen gene and its 5′- and 3′-flanking regions were obtained.
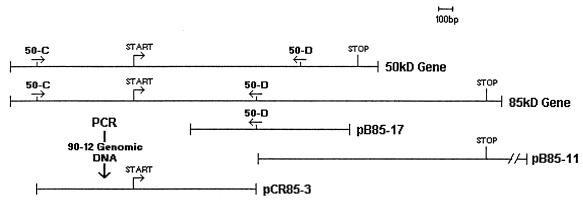
Clones pB85-11 and pB85-17 contained inserts of 4.5 and 1.1 kb, respectively. These two clones contained overlapping regions, and together they contained 845 bp of the 85-kDa-antigen gene sequence (Fig. 1). Further analysis of clone pB85-17 revealed partial sequence homology with the 50-kDa-antigen gene. Thus, the remaining 165 bp of the 5′ region of the gene was separately cloned by PCR from strain 90-12 genomic DNA with primers 50-C (5′ GAA TGT TCA GCT TTC CGG 3′) and 50-D (5′ AGC TGT ATC GTT CGT GAG 3′). The 1.5-kb amplified fragment was cloned in vector pCRII, designated pCR85-3, and sequenced (Fig. 1).
FIG. 1.
Composite profile of three overlapping clones of the 85-kDa-antigen gene and their positions with respect to the gene. The two overlapping clones, pB85-17 and pB85-11, were identified from the genomic library of strain 90-12. The insert, pCR85-3, was amplified directly from the genomic DNA of strain 90-12 by use of two specific primers, 50-C and 50-D. The sequence information of primer 50-C was obtained from the upstream region of the 50-kDa-antigen gene sequence, whereas that of primer 50-D was obtained from the sequence of clone pB85-17.
Nucleotide sequence analysis.
Analysis of the nucleotide sequence of the 50-kDa-antigen gene revealed a 1,617-bp-long open reading frame (ORF) and 896-bp upstream and 146-bp downstream noncoding regions (Fig. 2). The complete ORF was capable of coding for a 59.83-kDa protein consisting of 539 amino acids. The upstream region contained nearly perfect prokaryotic promoter sequences (13, 26). A purine-rich region present upstream of the start codon was identified as a potential ribosome binding site (Fig. 2).
FIG. 2.
Nucleotide sequence of the 50-kDa-antigen gene and flanking regions and deduced amino acid sequence. Putative −10, −35, and ribosome binding site regions are underlined, and the putative start of transcription is denoted with +1. Regions of dyad symmetry and adjacent thymine-rich regions are underlined.
The ORF of the 85-kDa-antigen gene was 2,547 bp long and capable of coding for a protein of 94.33 kDa consisting 849 amino acids. The 5′- and 3′-flanking regions contained regulatory elements similar to those described for the 50-kDa-antigen gene. The most important finding of the nucleotide analyses was the presence of several direct repeats in both genes. These repeats were more frequent in the middle of the genes, and many of these repeats were identical in both genes. All of the identical repeats coded for the same amino acids, but the positions and frequencies of these repeats were different in both genes. Also, the lengths and sequences of the direct repeats varied between both genes. The composite profiles of the repeats and their abundances in the 50- and 85-kDa-antigen gene sequences are presented in Fig. 3. Because of the abundance of 11-nucleotide-long repeats in both genes, these repeats were further analyzed for their sequence composition (Table 1).
FIG. 3.
Bar diagrams of direct repeats of 50- and 85-kDa-antigen gene sequences. The repeats are categorized according to their base-pair size, denoted by the numbers (55 to 10) placed under the bars. Each bar was then subdivided according to the sequence homologies of the repeats, as denoted by the block(s) in each bar.
TABLE 1.
Different types of 11-nucleotide-long repeats in 50- and 85-kDa-antigen genesa
| Type of repeat in 50-kDa-antigen gene | Repeat sequence | No. of repeats | Type of repeat in 85-kDa-antigen gene | Repeat sequence | No. of repeats | |
|---|---|---|---|---|---|---|
| I | AAAGAAATACT | 6 | I | AAAGAAATACT | 7 | |
| II | GAAATACTCAC | 5 | II | GAAATACTCAC | 5 | |
| III | AAATTTAAAGA | 5 | III | AAATTTAAAGA | 8 | |
| IV | CTAAAAGAGAT | 4 | IV | CTAAAAGAGAT | 7 | |
| V | AAAGACATACT | 2 | V | AAAGACATACT | 7 | |
| VI | TTTAAAGAGCT | 2 | VI | ATACTTACAGA | 9 | |
| VII | ATTTTTTATAA | 2 | VII | ACAGCTAAAGA | 6 | |
| VIII | AACTTTAAACG | 2 | VIII | TTTAAAGAACT | 6 | |
| IX | AAGTTTAAAGA | 2 | IX | GAAATACTTAC | 5 | |
| X | AGTTTAAAAAA | 2 | X | AGCACTGGTAA | 5 | |
| XI | TACTCACTAAT | 2 | XI | GATAAATTTAA | 5 | |
| XII | ATAAGTTTAAA | 2 | XII | CTTATAGAAAG | 5 | |
| XIII | TACTTACAGAT | 7 | ||||
| XIV | ACCGGTAACTT | 4 | ||||
| XV | ATGCAACAAAA | 3 | ||||
| XVI | GCTAAAGAAGT | 3 | ||||
| XVII | CTTACAGATAA | 3 | ||||
| XVIII | GCAATAACTGG | 2 | ||||
| XIX | ATGGTAAGGAC | 2 | ||||
| XX | ACTTATAGAAG | 2 |
There were 12 different types of repeat in the 50-kDa-antigen gene and 20 different types of repeat in the 85-kDa-antigen gene. The first five repeat sequences were identical in both genes.
Analyses of deduced amino acid sequences of SSA homologs.
Analyses of the deduced amino acid sequences of the 50- and 85-kDa antigens indicated considerable homology between these two SSA homologs. The identical repeats of their two genes code for the same amino acids, indirectly indicating the conserved regions of these two antigens. Substitutions or additions of one or several contiguous amino acid residues were identified throughout the molecules, but the significant homology in the amino acid sequences of the 50- and 85-kDa antigens was very pronounced in certain regions. These specific areas were designated identical domains (ID). The most interesting feature of these ID was their unique distribution in the amino acid sequences of individual antigens. The domains were positioned one after another (ID I to ID VIII) in the 50-kDa antigen, whereas the positioning of the same domains was totally different in the 85-kDa antigen (Fig. 4). In these ID, the similarities in the amino acid sequences between the two antigens varied from more than 94% to less than 79%.
FIG. 4.
Schematic diagram of the diversity and similarity of the 50- and 85-kDa antigens. The numbers at the top show ID. The homology in the amino acid sequences for the corresponding ID regions of the tested antigens varied from more than 94% to less than 79%. The unmarked areas indicate no homology between the two antigens.
ID I is the largest identical domain, consisting of 129 amino acids. In ID I the amino acid sequences of the 50- and 85-kDa antigens are very similar, and the estimated homology is 89.15% (87.08% in nucleotide sequence), with 14 amino acid conversions. The position of this particular domain is the same in the primary structures of both antigens. ID II consists of 51 amino acids. In a comparison of SSA homologs, this particular domain is found further downstream in the 85-kDa antigen. The estimated homology is 88.24% (89.54% in nucleotide sequence), with six amino acid conversions, between the 50- and 85-kDa antigens. ID III consists of 42 amino acids. The estimated homology in amino acid sequence is 92.85% (92.06% in nucleotide sequence), with three amino acid conversions. This particular domain is also found further downstream in the 85-kDa antigen than in the 50-kDa antigen. ID IV consists of 21 amino acids. The estimated homology in amino acid sequence is 90.48% (85.71% in nucleotide sequence), with two amino acid conversions. With respect to the 50-kDa antigen, this particular domain is found further upstream in the 85-kDa antigen. ID V consists of 39 amino acids. Among all the domains, this domain has the lowest homology, 79.49% (80.34% in nucleotide sequence), in SSA homologs. This domain is found further upstream in the 85-kDa antigen than in the 50-kDa antigen. ID VI has the highest homology, 94.55% (93.82% in nucleotide sequence), between the two antigens. Similarly, ID VII and ID VIII possess high homology. ID VII has 92.11% homology (85.08% in nucleotide sequence) and ID VIII has 94.12% homology (96.73% in nucleotide sequence) in their respective areas of the SSA homologs.
By comparison of the positions of all the ID in SSA homologs (Fig. 4), it is clear that six domains out of eight are changed with respect to their positions in these antigens. The domains are further apart from each other in the 85-kDa antigen than in the 50-kDa antigen, and the gaps are filled with unique sequences. This observation indicates the presence of six major unique domains in the 85-kDa antigen and two unique domains in the 50-kDa antigen.
Hydropathy analysis showed that the SSA of both strains have alternative hydrophilic and hydrophobic motifs, which are characteristic of transmembrane proteins (data not shown). The largest hydrophobic stretch belonged to ID I and formed the hydrophobic core region of the predicted signal peptide.
Characteristics of recombinant antigens.
The complete ORFs of the 50- and 85-kDa antigens were constructed by PCR and cloned in the pRSET-C expression vector (Invitrogen). The complete 50- and 85-kDa-antigen genes were amplified separately from the genomic DNAs of the original and variant strains of E. risticii by use of two modified primers, which created BamHI and EcoRI sites at the extreme 5′ and 3′ ends of the genes, respectively. After double digestion with BamHI and EcoRI, amplified genes were cloned separately in the multiple cloning region of the pRSET-C expression vector. Sequence analyses of these recombinant inserts confirmed the correct amplification of the SSA genes directly from their respective strains. The identities of the expressed proteins were established as the 50- and 85-kDa antigens by the reactivities of E. risticii (strains 25-D and 90-12) polyclonal antisera and the 85-kDa clone-specific antibody (Fig. 5). Both the 50- and the 85-kDa antigens migrated anomalously during electrophoresis and appeared to be 9.0 kDa smaller than the predicted sizes.
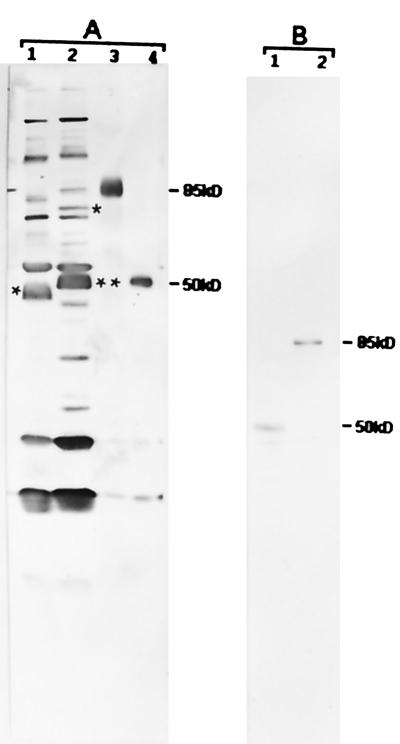
FIG. 5.
Western blot analysis of recombinant-expressed proteins of the 50- and 85-kDa-antigen genes. Lanes 1 contain the antigens from E. risticii 25-D, and lanes 2 contain the antigens from E. risticii 90-12. Lanes 3 and 4 contain E. coli-expressed recombinant 85- and 50-kDa fusion proteins, respectively. (A) Blot reacted with sera from strain 90-12-infected mice. The asterisk in lane 1 represents the location of the 50-kDa antigen. This antigen is not distinguishable as a separate band, as it overlaps with the 51-kDa antigen band. The single asterisk in lane 2 represents the location of the 85-kDa antigen band, and double asterisks represent the location of the 51-kDa antigen band. The changes in the sizes of the recombinant 85- and 50-kDa proteins (lanes 3 and 4, respectively) are in close agreement with the expected fusion product of the pRSET-ABC expression system, which adds a 3.5-kDa protein mass to its fusion products. (B) Blot reacted with recombinant 85-kDa clone-specific antibody from strain 90-12. The recombinant 85-kDa clone-specific antibody recognized only the 50-kDa antigen in strain 25-D (lane 1) and only the 85-kDa antigen in strain 90-12 (lane 2). Numbers at right are molecular masses in kilodaltons.
DISCUSSION
The existence of antigenic variation among field isolates of E. risticii has been well demonstrated (8, 9). This antigenic heterogeneity, along with the poor efficacies of present vaccines, has been implicated in PHF vaccine failure (9). An effective vaccine against E. risticii must elicit a protective immune response against all the strains present in the field. This situation requires identifying and characterizing the antigens of E. risticii that are involved in stimulating protective immunity as well as in contributing to the antigenic variation. Previously, we reported the major pathogenic, immunogenic, and molecular differences between E. risticii 25-D and 90-12 (24). One important antigenic difference was the presence of an 85-kDa antigen in strain 90-12 and its cross-reactive homolog, a 50-kDa antigen, in strain 25-D. We also demonstrated that these two antigens could stimulate protective immune responses in a murine model of PHF (25). The present study revealed the molecular basis of the size and antigenic variations of the two antigens. The presence of variable numbers of tandem repeat sequences accounts for the large size variation between the two antigens. This type of size difference in immunological cross-reactive antigens has been observed among isolates of other rickettsial and ehrlichial organisms (10, 12, 19, 20, 23, 28). However, the E. risticii SSA appear to be unique in the number and complexity of the repeats units.
The lengths of the repeats in the 50- and 85-kDa-antigen genes are comparatively shorter than those of the repeats in the gene for the 120-kDa immunodominant protein of E. chaffeensis (28) and the rompA gene of Rickettsia rickettsii, R. conorii, and R. akari (10), but the number of repeats is higher in E. risticii than in any other Rickettsia or Ehrlichia species. The most abundant repeats in the 50- and 85-kDa-antigen gene sequences are 11 nucleotides long, whereas in the 120-kDa-protein and rompA genes the average length of the repeats is between 200 and 250 nucleotides. Repeat structures such as those in SSA homologs are thought to develop by unequal homologous recombination (11, 14, 27), slip strand mispairing during replication (16, 18), or both.
In the SSA genes, the homology of the 5′- and 3′-flanking regions is greater than that of the regions encoding the two variant proteins. The degree of homology of the flanking regions suggests that transcriptional regulation of the SSA genes is similar among the strains.
Southern blot hybridization of restriction endonuclease (EcoRI and HindIII) digests of the genomic DNAs from these two strains with a cloned probe for the 85-kDa antigen showed only one hybridizable restriction fragment (24). This result suggests that the SSA homolog genes reside in a similar genomic context in each antigenic variant and that only one SSA allele is represented in each strain of E. risticii. Further research also indicates that the sizes of the SSA appear to be very stable during passage through animal or in vitro cell cultures. In our studies, the sizes of these antigens remained unchanged for 100 cell culture or three horse and mouse passages (unpublished data). This result indicates that the observed size variations are not caused by the laboratory manipulation of E. risticii. Recently, we demonstrated size variations of SSA from several isolates of E. risticii obtained from typical cases of PHF in the field (9). To understand the complete evolution of SSA genes, it is necessary to know at what stage of the E. risticii life cycle the events leading to size and sequence variations of SSA occur; one possible location could be in an as-yet-unidentified transmission vector.
A 125I surface labeling experiment determined that the 50-kDa protein of strain 25-D is an apparent surface antigen (6). The immunological cross-reactivity and sequence homology proved that the 85-kDa protein of strain 90-12 is the homolog of the 50-kDa protein; thus, this particular protein also is present on the surface of strain 90-12. Comparative analyses of deduced amino acid sequences of SSA homologs from the two different strains of E. risticii revealed the presence of eight major ID in both strains, but the positions of the respective domains in comparison to each other were completely different in each strain. This finding might be a source of antigenic variation because some of the exposed epitopes in one of the proteins could have become buried in the membrane or otherwise become inaccessible in the other protein and vice versa. It is also possible that the exposed epitopes of these two proteins are strain specific and that their common determinants are normally buried in the membrane. Moreover, the presence of considerable amounts of new sequences in the 85-kDa-antigen gene increase the chance of the appearance of different epitopes on the surface of strain 90-12. Finally, the presence of point mutations in each ID should also be considered a possible source of antigenic variation, but this variation may not be caused solely by the accumulation of point mutations. Possibly the combination of point mutations and recombinational processes is responsible for the antigenic variation.
Analyses of the amino acid compositions and estimation of the isoelectric points (6.45 for the 50-kDa antigen and 6.33 for the 85-kDa antigen) of these two proteins indicated that they were almost identical in these aspects. Although one of the interesting features is the increase in acidic and basic residues (equal amounts) in the 85-kDa antigen amino acid composition, this fact does not increase the total charge on the surface of the molecule but produces more hydrophilic regions which can be exposed as outer membrane domains. Thus, the possibility of generating more epitopes on the surface is much higher for strain 90-12 than for strain 25-D. The migration patterns of the two expressed proteins in sodium dodecyl sulfate gels matched those of their respective native proteins, yet the sequential sizes of both of the proteins differed by 9.0 kDa from the observed sizes in the gels. This type of anonymous migration is a common feature of proteins which have tandem repeats in their gene sequences (1).
The possibility of using the nucleotide sequences of the 50- and 85-kDa-antigen genes as a starting point for a recombinant or subunit vaccine is attractive. Previously it was observed that in cross-protection studies, mice immunized with strain 90-12 were completely protected from a challenge infection with strain 25-D, but mice immunized with strain 25-D were partially protected against challenge with strain 90-12 (24). A similar but more pronounced result was obtained when the purified recombinant 50- and 85-kDa antigens were used for immunization (25). Mice immunized with the 50-kDa antigen were protected against homologous strain 25-D but not against heterologous strain 90-12, whereas mice immunized with the 85-kDa antigen were protected against both strains. This difference in immunoprotection correlates with the molecular difference between the antigens of the respective strains. The cumulative results of all of these experiments suggest that variation in the surface antigens of E. risticii, such as the 50- and 85-kDa antigens, may contribute to vaccine failure. However, before use as a recombinant vaccine, it is necessary to confirm that the above proteins or their homologs in different strains are indeed highly protective immunogens.
Additionally, with the nucleotide sequences of the SSA genes, a PCR assay can be developed to quickly identify strain differences among field isolates of E. risticii. However, characterization of the highly unusual repetitive structure of the 50- and 85-kDa antigens of E. risticii strains should be pursued for a better understanding of the role of these proteins in the Ehrlichia-host interaction.
REFERENCES
- 1.Anderson B E, McDonald G A, Jones D C, Regnery R L. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun. 1990;58:2760–2769. doi: 10.1128/iai.58.9.2760-2769.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas B, Mukherjee D, Mattingly-Napier B L, Dutta S K. Diagnostic application of polymerase chain reaction for detection of Ehrlichia risticii in equine monocytic ehrlichiosis (Potomac horse fever) J Clin Microbiol. 1991;29:2228–2233. doi: 10.1128/jcm.29.10.2228-2233.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta S K, Myrup A C, Rice R M, Roble M G, Hammond R C. Experimental reproduction of Potomac horse fever in horses with a newly isolated Ehrlichia organism. J Clin Microbiol. 1985;22:265–269. doi: 10.1128/jcm.22.2.265-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta S K, Rice R M, Hughes T D, Savage P K, Myrup A C. Detection of serum antibodies against Ehrlichia risticii in Potomac horse fever by enzyme-linked-immunosorbent assay. Vet Immunol Immunopathol. 1987;14:85–92. doi: 10.1016/0165-2427(87)90077-8. [DOI] [PubMed] [Google Scholar]
- 6.Dutta S K, Shankarappa B, Thaker S R, Mattingly-Napier B L. DNA restriction endonuclease cleavage pattern and protein antigenic profile of Ehrlichia risticii. Vet Microbiol. 1990;25:29–38. doi: 10.1016/0378-1135(90)90090-i. [DOI] [PubMed] [Google Scholar]
- 7.Dutta S K, Shankarappa B, Mattingly-Napier B L. Molecular cloning and analysis of recombinant major antigen of Ehrlichia risticii. Infect Immun. 1991;59:1162–1169. doi: 10.1128/iai.59.3.1162-1169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta S K, Shankarappa B, Mattingly-Napier B. Antigenic analysis of Ehrlichia risticii isolates. In: Plowright W, Rossdale P D, Wade J F, editors. Equine infectious diseases. VI. Cambridge, England: R&W Publication Ltd.; 1991. pp. 61–65. [Google Scholar]
- 9.Dutta S K, Vemulapalli R, Biswas B. Association of deficiency in vaccine antibody response and heterogeneity of Ehrlichia risticii strains with vaccine failure in Potomac horse fever. J Clin Microbiol. 1998;36:506–512. doi: 10.1128/jcm.36.2.506-512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilmore R D. Comparison of the romp A gene repeat regions of Rickettsiae reveals species-specific arrangement of individual repeating units. Gene. 1993;125:97–102. doi: 10.1016/0378-1119(93)90752-o. [DOI] [PubMed] [Google Scholar]
- 11.Haas R, Meyer T F. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 12.Hanson B. Identification and partial characterization of Rickettsia tsutsugamushi major protein immunogens. Infect Immun. 1985;50:603–609. doi: 10.1128/iai.50.3.603-609.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 15.Kraft R, Tardiff J, Krauter K S, Leinwand L A. Using mini-prep plasmid DNA for sequencing double stranded templates with SequenaseTM. BioTechniques. 1988;6:544–546. [PubMed] [Google Scholar]
- 16.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 17.Lyon J A, Geller R H, Haynes J G, Chuley J G, Weber J L. Epitope map and processing scheme for the 195,000-dalton surface glycoprotein of Plasmodium falciparum merozoites deduced from cloned overlapping segments of the gene. Proc Natl Acad Sci USA. 1986;83:2989–2993. doi: 10.1073/pnas.83.9.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy G L, Connell T D, Barritt D S, Koomey M, Cannon J G. Phase variation of gonococcal protein II: regulation of the gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 19.Oaks E V, Stover C K, Rice R M. Molecular cloning and expression of Rickettsia tsutsugamushi genes for two major protein antigens in Escherichia coli. Infect Immun. 1987;55:1156–1157. doi: 10.1128/iai.55.5.1156-1162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oaks E V, Rice R M, Kelly D J, Stover C K. Antigenic and genetic relatedness of eight Rickettsia tsutsugamushi antigens. Infect Immun. 1989;57:3116–3122. doi: 10.1128/iai.57.10.3116-3122.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stover C K, Vodkin M H, Oaks E V. Use of conversion adaptors to clone antigen genes in λgt11. Anal Biochem. 1987;163:398–407. doi: 10.1016/0003-2697(87)90241-7. [DOI] [PubMed] [Google Scholar]
- 23.Stover C K, Marana D P, Carter J M, Roe B A, Mardis E, Oaks E V. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect Immun. 1990;58:2076–2084. doi: 10.1128/iai.58.7.2076-2084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemulapalli R, Biswas B, Dutta S K. Pathogenic, immunogenic, and molecular differences between two Ehrlichia risticii strains. J Clin Microbiol. 1995;33:2987–2993. doi: 10.1128/jcm.33.11.2987-2993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vemulapalli R, Biswas B, Dutta S K. Studies with recombinant proteins of Ehrlichia risticii; identification of strain specific antigen as a protective antigen. Vet Parasitol. 1998;76:189–202. doi: 10.1016/s0304-4017(97)00209-4. [DOI] [PubMed] [Google Scholar]
- 26.von Hipple P H, Ress W A, Rippe K, Wilson K S. Specificity mechanisms in the control of transcription. Biophys Chem. 1996;59:231–246. doi: 10.1016/0301-4622(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 27.Williams R O, Young J R, Majiwa P A O. Genomic environment of T. brucei VSG genes: presence of a minichromosome. Nature (London) 1982;299:417–421. doi: 10.1038/299417a0. [DOI] [PubMed] [Google Scholar]
- 28.Yu X J, Crocquet-Valdes P, Walker D H. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene. 1997;184:149–154. doi: 10.1016/s0378-1119(96)00586-0. [DOI] [PubMed] [Google Scholar]