Introduction
Both extracellular and intracellular signals can cause dramatic changes of cell shape that are controlled by the rapid assembly and disassembly of actin filaments. Microbial pathogens have developed a variety of sophisticated mechanisms for usurping normal cellular processes that involve actin cytoskeletal dynamics. By producing virulence factors mimicking or modifying host molecules, pathogens mediate events as diverse as bacterial invasion, antiphagocytosis and intracellular parasitism. Investigations of pathogen motility have contributed enormously to our understanding of actin-based signalling processes. The Boehringer Ingelheim Fonds (http://www.bifonds.de), an independent foundation for basic research in medicine, has brought together diverse disciplines by organising the 84th International Titisee Conference on ‘The actin cytoskeleton: from signalling to bacterial pathogenesis’. About 70 researchers from Europe, Asia and the USA met in Titisee, Germany, October 24–28, 2001, to discuss the latest progress on the reorganisation of the actin cytoskeleton in normal and bacterially infected cells. In this report, we attempt to provide an overview of the central focus of the meeting, rather than giving a comprehensive account. Thus, many interesting contributions are not mentioned.
Regulation of actin-based motility
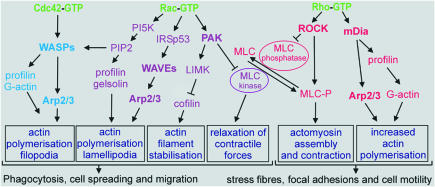
The work of many laboratories during the last decade has unravelled signalling networks that control the assembly and disassembly of filamentous actin (F-actin), providing the basis for the regulation of cell adhesion, spreading, polarisation and migration, as well as for cytokinesis and phagocytosis (Figure 1). The keynote lecturer, A. Hall (London, UK), described the pioneering experiments that showed that the reorganisation of the actin cytoskeleton is regulated by proteins of the Rho family, including Cdc42, Rac and Rho. Cdc42 activation induces the formation of tight bundles of parallel filaments that form the core of filopodia, and activation of Rac results in the formation of a network of diagonally orientated actin filaments that give rise to the thin sheets of lamellipodia, whereas Rho activation stimulates the organisation of actomyosin bundles into stress fibres (Figure 1). Coordination of the distinct roles of these GTP-binding proteins is crucial for regulating cell migration, as demonstrated by wound closure in a fibroblast monolayer: Cdc42 regulates the cell polarity, Rac the protrusion of lamellipodia at the leading edge and Rho the turnover of the highly organised structures termed focal adhesions (Nobes and Hall, 1999). Hall also discussed the control of phagocytosis by two distinct mechanisms: type I uses the immunoglobulin receptor (FcγR) and is mediated by Cdc42 and Rac, and type II uses the complement receptor (CR3) and is mediated by Rho.
Fig. 1. Regulation of actin-based motility by the Rho family GTPases. Activated Cdc42, Rac and Rho bind to and specifically activate their downstream effectors, which are either kinases (such as ROCK, PAK and PI5K) or scaffolding proteins (such as mDia, WASP and IRSp53). These effector proteins activate diverse signalling pathways with distinct effects on the actin cytoskeleton and cellular morphology. An important aspect for the cell motility is the equilibrium between the myosin light chain (MLC) and the phosphorylated MLC, which is tightly regulated.
Membrane anchoring of these small GTPases is a prerequisite for their function as molecular switches, cycling between the GDP-bound inactive and the GTP-bound active states. This cycle is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Rho GTPases also cycle between the cytosol and membranes, and this process is mediated by guanine nucleotide-dissociation inhibitors (GDIs). T. Hakoshima (Nara, Japan) described the crystal structure of RhoGDI in complex with the FERM domain of radixin, a member of the ERM (ezrin/radixin/moesin) protein family whose members link the actin cytoskeleton to the plasma membrane. The binding of the FERM domain to the GDI molecule provides a mechanism for the release of the Rho GTPase from the Rho–RhoGDI complex by the ERM proteins, an important step in the regulatory cycle of Rho.
Interactions of the activated GTP-bound Rho proteins with some of their downstream effector proteins are responsible for regulating actin-based cell motility (Figure 1). Most of these effectors are either Ser/Thr-kinases or scaffold proteins. M. Naumann (Berlin, Germany) showed that Rac1/Cdc42, and consequently Pak1, are activated during infection of gastric epithelial cells with Helicobacter pylori (Figure 1). In contrast, no activation of Rac1/Cdc42 was observed when H. pylori mutant strains (virB7 and PAI) were used. These strains lack a functional pilus-derived secretion apparatus (termed type IV), which is thought to contribute to the transfer of bacterial proteins into epithelial cells. Helicobacter pylori induces Rac1 activation for up to 60 min, which correlates with the timing of Tiam1 (RacGEF) expression. This activation may play a role in the development of chronic gastritis.
Protrusion of lamellipodia and filopodia from the cell surface requires actin polymerisation and is induced downstream of Rac/Cdc42 by a family of proteins called WASP (Wiskott–Aldrich Syndrome protein; Takenawa and Miki, 2001). This family includes WASP (specific to hematopoietic cells), N-WASP (ubiquitous) and WAVE proteins (WASP verprolin homologous proteins). Whereas WASP and N-WASP are directly activated by Cdc42, WAVE activation is mediated by the Rac effector IRSp53, which was described by T. Takenawa (Tokyo, Japan; Figure 1). He introduced the WISH and WIP proteins as regulators of N-WASP. In vitro, WISH enhances N-WASP-induced activation of the Arp2/3 complex (see below), an important regulator of the polymerisation of globular actin (G-actin) to F-actin. In contrast, WIP retards N-WASP/Cdc42-activated actin polymerisation mediated by the Arp2/3 complex.
Another major regulator of actin polymerisation is the protein Ena/VASP (enabled vasodilator-stimulated phosphoprotein; Reinhard et al., 2001) discovered by U. Walter (Würzburg, Germany). He described its properties as a molecular adaptor linking signal transduction pathways to the cytoskeleton and as an enhancer of the actin-based motility of Listeria monocytogenes. In endothelial cells, phosphorylation of VASP by cAMP- and cGMP-dependent protein kinases results in its displacement from focal adhesions. P. Chavrier (Paris, France) showed that VASP binds directly to the proline-rich domain (PRD) of WASP and enhances actin-based propulsion of WASP-coated beads in a fashion reminiscent of Listeria movement (Castellano et al., 2001). He demonstrated how the initial stages of phagocytosis (filopodia and podosome formation) are sequentially activated by Cdc42. WASP and VASP accumulation in actin-rich phagocytic cups is essential for stimulating the required actin assembly and membrane protrusion.
Integrin-mediated cell matrix adhesion regulates actin cytoskeleton organisation through distinct steps, from the formation of filopodia and lamellipodia in the early phases of focal contact establishment to the organisation of focal adhesions and stress fibres (Geiger and Bershadsky, 2001). The Rho-induced reorganisation of cytoskeletal proteins during this process is regulated by cooperation between ROCK (Rho-associated kinase) and the mDia proteins (Figure 1). A. Bershadsky (Rehovot, Israel) described the relationship between Rho-induced focal contacts and cell contractility using a micromanipulation technique in which focal complexes are induced by applying tension from the outside. He demonstrated that mDia1 mediates force-induced contact formation, even if the entire ROCK-activated pathway, including myosin II activation, is eliminated. A. Alberts (Grand Rapids, MI) showed that constitutively active mDia1 and mDia2 variants lacking their Rho-binding domains cooperate with activated ROCK to form stress fibres. At their C-terminal ends, mDia proteins contain a conserved DAD (Dia-autoregulatory domain) that interacts with the N-terminal Rho-binding domain. DAD thus acts as an autoinhibitory domain whose activity is regulated by active Rho binding, presumably much as WASP is regulated by Cdc42 (Alberts, 2001). As is the case for expression of activated mDia1 and mDia2, ectopic expression of DAD causes actin fibre formation. DAD binds to both actin and the Arp2/3 complex and induces actin polymerisation in vitro. Thus, the mDia protein, and in particular the DAD region, directly regulates the actin cytoskeleton (Figure 1).
Mechanisms behind actin-based cell motility
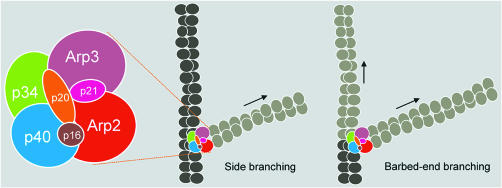
The most generally expressed view is that the Arp2/3 complex is the major factor that drives actin nucleation and actin filament branching and initiates filopodial/lamellipodial membrane protrusion (Higgs and Pollard, 2001; Pantaloni et al., 2001). The Arp2/3 complex consists of two actin-related proteins (Arp2 and Arp3) and five other subunits (p40, p34, p21, p20 and p16) (Figure 2). The activation of the Arp2/3 complex is regulated by the C-terminal VCA (verprolin homology, cofilin homology, acidic) domains of WASP proteins in concert with their activators Rac/Cdc42 and phosphatidylinositol-4,5-biphosphate (PIP2; Figure 1). Other proteins play key roles in the dynamic properties of actin filaments by coupling them to the cell surface (Ena/VASP family), capping their ends (capping proteins), severing them (gelsolin, ADF/cofilin), promoting dissociation from the pointed ends (ADF/cofilin), regulating association at the barbed end (profilin) and crosslinking them (α-actinin, fascin, filamin). T. Stossel (Boston, MA) described filamin A as a large actin-binding protein that localises at the leading edge of migrating cells, at junctions between orthogonally intersecting actin filaments. Filamin A induces actin filament crosslinking, which is required for specific and organised movement at the leading edge of migrating cells. As a scaffold for the spatial organisation of diverse signalling pathways, filamin A binds about 30 different signalling proteins, amongst them small GTP-binding proteins such as Rac, Rho, Cdc42 and RalA. From the phenotype of filamin A-deficient cells, Stossel concluded that filamin A is necessary for stabilising orthogonal actin networks and that Arp2/3 complex-mediated branching of actin alone is not sufficient for establishing an orthogonal actin network and maintaining mechanical stability at the leading edge.
Fig. 2. Current models of Arp2/3 complex organisation and actin filament branching. Subunit organisation of the Ar2/3 complex as suggested by Robinson et al. (2001) and Gournier et al. (2001).
M.D. Welch (Berkeley, CA) demonstrated that a fully active human Arp2/3 complex can be reconstituted when its seven subunits are expressed as recombinant proteins in insect cells. Based on the reconstitution experiments, he proposed two alternative models of Arp2/3 complex subunit organisation and interaction with actin filaments. The main difference between these two models is the orientation of Arp2 and Arp3 within the complex at the binding site for the slow-growing (pointed) end of actin filaments (see Gournier et al., 2001).
A highlight of the meeting was T. Pollard’s (New Haven, CT) presentation of the crystal structure of the 225 kDa Arp2/3 complex, prepared from bovine brain. In the crystallised, inactive complex, both Arps are in ATP-free, open conformations. Activation of the complex is proposed to require a conformational change that brings the two Arps close together to resemble the subunits in an actin filament at the pointed end (Figure 2; Robinson et al., 2001).
No agreement has been reached concerning the mechanism by which the Arp2/3 complex causes the filaments to branch in solution (Figure 2; Condeelis, 2001). The first model, according to Pollard, proposes that branches form along the side of mother filaments. His time-lapse movies of actin filaments growing in the presence of the Arp2/3 complex showed that daughter filaments arise randomly along the sides of pre-existing actin filaments rather than at barbed ends. The second model described by M.-F. Carlier (Gif-sur-Yvette, France) proposes that the activated Arp2/3 complex binds to and branches free barbed ends, resulting in the formation of two new filaments (barbed-end branching model). She also reported that the Arp2/3 complex exists in different structural/functional states: the inactive Arp2/3, the activated VCA–Arp2/3 complex and the ternary G-actin–VCA–Arp2/3 complex. The latter nucleates the branching of actin filaments. Carlier showed that ATP affinity to Arp2/3 is enhanced by more than two orders of magnitude upon activation of the Arp2/3 complex by the VCA domains of WASP, implying a large structural change. She also presented a motility assay using solid particles together with ATP as an energy source, the Arp2/3 complex and actin filaments in the presence of ADF, profilin and capping proteins. The assay allows testing of the functions of putative regulators of processes like phagocytosis, discovery of new activators of the Arp2/3 complex and screening for inhibitors of motility.
U. Aebi (Basel, Switzerland) presented actin’s remarkable propensity for ‘supramolecular patterning’. Not only can actin be induced in vitro to polymerise into various types of filaments, sheets, tubes and ribbons, but it also forms, in the absence of any actin-binding proteins, filament bundles and branches. Based on kinetic observations, Aebi argued that filament branching, for example, is induced by actin itself, so that the role of the Arp2/3 complex in this dynamic process is to stabilise rather than to induce filament branching.
Rocketing pathogens
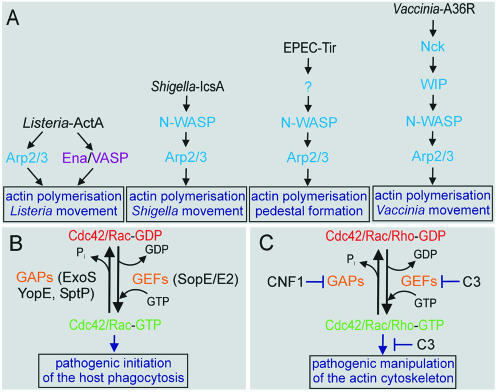
Several pathogenic bacteria have evolved strategies to ‘hijack’ the Arp2/3 complex to their own advantage. Intracellular pathogens such as Listeria and Shigella induce their own uptake by non-professional phagocytes and spread from cell to cell by organising actin into ‘comet tails’ (Figure 3A; Cossart and Bierne, 2001). P. Cossart (Paris, France) described two bacterial proteins that allow L. monocytogenes to invade eukaryotic cells. Indeed, phagocytosis of the bacteria is induced by the contact between these so-called internalins (InlA and InlB) and host cell receptors. For human cells, it has been shown that E-cadherin allows entry of bacteria expressing InlA. In contrast, murine E-cadherin does not interact with InlA. This specificity is due to a single amino acid difference at position 16 in E-cadherin. Transgenic mice producing human E-cadherin have allowed the analysis of InlA function in a mouse model, and the protein has been shown to promote crossing of the intestinal barrier. InlB promotes the entry of bacteria, as well as of InlB-coated beads, into various cell types such as hepatocytes, fibroblasts and endothelial cells. Further signalling involves Rac1 and Cdc42, LIM kinase, cofilin and the Arp2/3 complex as key players, suggesting a tightly controlled process of nucleation and branching of actin filaments, followed by disassembly during the process of phagocytosis.
Fig. 3. Manipulation of the actin cytoskeleton by pathogens. (A) Arp2/3-based motility of pathogens. In contrast to Shigella and Listeria, the intracellular motility of Vaccinia and EPEC is dependent on the tyrosine phosphorylation of the pathogen-derived proteins A36R and Tir, respectively, and in addition involves the host cell Nck adaptor protein. (B) Structural mimicry in bacterial virulence. (C) Modifying bacterial protein toxins.
J. Wehland (Braunschweig, Germany) reported on actin filament nucleation and organisation using the Listeria system, in which the bacterial ActA protein is the key player in bacterial movement within infected cells. By mimicking the WASP protein, the surface-exposed ActA directly interacts with G-actin, the Arp2/3 complex and members of the Ena/VASP protein family (Figure 3A). Motility of Listeria in Ena/VASP-deficient cells is substantially retarded but is restored by the introduction of Ena/VASP cDNAs, making possible detailed functional studies using deleted and mutated constructs. Wehland also described new insights into N-WASP functions using cells derived from conditional knockout mice. Whereas loss of N-WASP function led to embryonic lethality, fibroblasts deficient in N-WASP surprisingly supported the formation of numerous filopodia (Nobes and Hall , 1999). However, comet-tail formation and motility of intracellular Shigella flexneri was completely abolished, as was actin pedestal formation induced by attaching enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC; Figure 3A).
Structural mimicry in bacterial virulence
Protein secretion via the so-called type III apparatus allows several bacterial pathogens such as Salmonella spp., Pseudomonas aeruginosa, Yersinia spp. and EPEC to inject bacterial proteins directly into eukaryotic cells to subvert cell functions. Some of these injected proteins exert their activity by functioning as molecular mimics of cellular proteins (Figure 3B; Stebbins and Galan, 2001). J.E. Galan (New Haven, CT) reported on the mechanisms by which the bacterial pathogen Salmonella modulates cellular functions through proteins delivered into the host cell via a type III secretion apparatus. The pathogen injects a GEF for Cdc42/Rac (SopE) to induce actin cytoskeleton rearrangements and membrane ruffling, leading to bacterial internalisation into the host cell. Remarkably, once internalised, Salmonella injects a GAP (SptP) that reverses the induced cytoskeletal changes. In addition to its GAP activity located within its N-terminus, the C-terminal domain of SptP possesses potent tyrosine phosphatase activity. Salmonella uses two additional mimics to modulate cellular responses. One is the actin-binding protein (SipA), which lowers the critical concentration of G-actin and stabilises F-actin. The second is an inositol phosphatase (SopB), which mediates actin cytoskeleton rearrangements and bacterial entry in a Cdc42-dependent manner. Structural studies based on X-ray data show that the bacterial RhoGAPs SptP from Salmonella (Galan) and ExoS from P. aeruginosa (M. Würtele, Dortmund, Germany) have no recognisable structural similarities to the eukaryotic RhoGAPs (Stebbins and Galan, 2001; Wurtele et al., 2001). However, they use the same mechanism of stabilising the transition state of the GTPase reaction, via an arginine finger.
W.-D. Hardt (München, Germany) showed that Salmonella typhimurium employs two homologous SopE proteins (SopE and E2) with slightly different specificities. SopE is an efficient GEF for both Cdc42 and Rac1, whereas SopE2 efficiently interacts only with Cdc42. Now that the SopE/Cdc42 complex structure has been solved by G. Buchwald (Dortmund, Germany), it is clear that, in addition to lacking any sequence relationship to known mammalian GEFs, SopE also has no similarity to host protein GEFs at the tertiary structure level. Nevertheless, SopE uses a similar mechanism as the mammalian RacGEF Tiam1 in accelerating the nucleotide exchange.
A large virulence plasmid representing the anti-host defence function of all pathogenic species of Yersinia was presented by J. Heesemann (München, Germany). It carries genes encoding a type III secretion apparatus, at least six effector proteins (Yersinia outer proteins; Yops), secretion regulators and the outer membrane protein YadA (Yersinia adhesin). Some of these proteins act on Rho GTPases by conventional means, as is the case for YopE, which is a RhoGAP. YopT, on the other hand, modifies RhoA by an unknown mechanism and induces the release of Rho from the membrane.
Bacterial toxins that modify host proteins
In order to invade eukaryotic cells, several bacterial protein toxins modulate the functions of Rho proteins by covalent modification, such as ADP-ribosylation or glucosylation, or by activation via deamidation and transglutamination (Aktories et al., 2000). These bacterial toxins can be used as potent and selective tools for the study of the cellular functions of their eukaryotic targets (Figure 3C). K. Aktories (Freiburg, Germany) reported on a new type C toxin produced by Staphylococcus aureus (C3stau), with 35% sequence identity to the ADP-ribosyltransferase C3 from Clostridium botulinum. C3stau modifies RhoE/Rnd3, as well as RhoA, RhoB and RhoC. RhoE/Rnd3 is a member of the Rho family, but it is deficient in GTP hydrolysis activity and, therefore, constitutively active. Furthermore, C3stau does not possess any translocation domain that is typical for other bacterial protein toxins.
Escherichia coli cytotoxic necrotising factor 1 (CNF1) converts a glutamine residue on Rho GTPases (Gln63 of RhoA) to glutamic acid. This glutamine deamidation blocks both the intrinsic and the GAP-stimulated GTP hydrolysis reaction, and as a result of the CNF1 activity the Rho is permanently activated, inducing a profound reorganisation of the host cell actin cytoskeleton. P. Boquet (Nice, France) showed that the crystal structure of the catalytic domain of CNF1 exhibits a novel protein fold, further illustrating the differences to eukaryotic transglutaminases to which it has neither sequence nor structural similarity. Furthermore, Boquet presented new results on the degradation of the toxin-activated Rho proteins by mammalian proteasomes, suggesting that bacterial toxin activity combined with the degradation of the modified Rho proteins by the host may provide a way to transiently influence the actin cytoskeleton.
Perspectives
The 84th International Titisee Conference mobilised scientists from many different research areas to report on their latest discoveries and developments on the mechanisms behind actin nucleation and filament branching, signal transduction pathways regulating actin-based cytoskeletal (re)organisation, pathogen–host cell adhesion and the manipulation of the actin cytoskeleton by pathogens. This multidisciplinary meeting was a perfect platform for lively discussions that continued for many hours at the bar. Obviously, many questions remained unanswered and many more mechanisms need to be clarified. However, considering the clever ways in which bacterial pathogens manipulate Rho GTPase signalling and actin cytoskeletal rearrangements, we would be well advised to keep a close eye on the infection pathways of these organisms in order to gain further insights into the relevant eukaryotic signal transduction pathways.
Mohammad Reza Ahmadian, Alfred Wittinghofer & Gudula Schmidt
Acknowledgments
Acknowledgements
We would like to thank all participants of the 84th International Titisee Conference and, last but not least, the Boehringer Ingelheim Fonds.
References
- Aktories K., Schmidt, G. and Just, I. (2000) Rho GTPases as targets of bacterial protein toxins. Biol. Chem., 381, 421–426. [DOI] [PubMed] [Google Scholar]
- Alberts A.S. (2001) Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem., 276, 2824–2830. [DOI] [PubMed] [Google Scholar]
- Castellano F., Le Clainche, C., Patin, D., Carlier, M.F. and Chavrier, P. (2001) A WASp–VASP complex regulates actin polymerization at the plasma membrane. EMBO J., 20, 5603–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J. (2001) How is actin polymerization nucleated in vivo? Trends Cell Biol., 11, 288–293. [DOI] [PubMed] [Google Scholar]
- Cossart P. and Bierne, H. (2001) The use of host cell machinery in the pathogenesis of Listeria monocytogenes. Curr. Opin. Immunol., 13, 96–103. [DOI] [PubMed] [Google Scholar]
- Geiger B. and Bershadsky, A. (2001) Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol., 13, 584–592. [DOI] [PubMed] [Google Scholar]
- Gournier H., Goley, E.D., Niederstrasser, H., Trinh, T. and Welch, M.D. (2001) Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell, 8, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Higgs H.N. and Pollard, T.D. (2001) Regulation of actin filament network formation through Arp2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem., 70, 649–676. [DOI] [PubMed] [Google Scholar]
- Lommel S., Benesch, S., Rottner, K., Franz, T., Wehland, J. and Kuhn, R. (2001) Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO rep., 2, 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes C.D. and Hall, A. (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol., 144, 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D., Le Clainche, C. and Carlier, M.F. (2001) Mechanism of actin-based motility. Science, 292, 1502–1506. [DOI] [PubMed] [Google Scholar]
- Reinhard M., Jarchau, T. and Walter, U. (2001) Actin-based motility: stop and go with Ena/VASP proteins. Trends Biochem. Sci., 26, 243–249. [DOI] [PubMed] [Google Scholar]
- Robinson R.C., Turbedsky, K., Kaiser, D.A., Marchand, J.B., Higgs, H.N., Choe, S. and Pollard, T.D. (2001) Crystal structure of Arp2/3 complex. Science, 294, 1679–1684. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E. and Galan, J.E. (2001) Structural mimicry in bacterial virulence. Nature, 412, 701–705. [DOI] [PubMed] [Google Scholar]
- Takenawa T. and Miki, H (2001) WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci., 114, 1801–1809. [DOI] [PubMed] [Google Scholar]
- Wurtele M., Wolf, E., Pederson, K.J., Buchwald, G., Ahmadian, M.R., Barbieri, J.T. and Wittinghofer, A. (2001) How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nature Struct. Biol., 8, 23–26. [DOI] [PubMed] [Google Scholar]