Abstract
The transcription factor E2F, which is a key element in the control of cell proliferation, is repressed by Rb and other pocket proteins in growth-arrested differentiating cells, as well as in proliferating cells when they progress through early G1. It is not known whether similar mechanisms are operative in the two situations. A body of data suggests that E2F repression by pocket proteins involves class I histone deacetylases (HDACs). It has been hypothesized that these enzymes are recruited to E2F target promoters where they deacetylate histones. Here we have tested this hypothesis directly by using formaldehyde cross-linked chromatin immunoprecipitation (XChIP) assays to evaluate HDAC association in living cells. Our data show that a histone deacetylase, HDAC-1, is stably bound to an E2F target promoter during early G1 in proliferating cells and released at the G1–S transition. In addition, our results reveal an inverse correlation between HDAC-1 recruitment and histone H4 acetylation on specific lysines.
INTRODUCTION
Rb, the product of the retinoblastoma gene, is a transcriptional co-repressor that is a key element in the control of cell proliferation and differentiation. Rb is involved in terminal differentiation in a number of tissues, and, in particular, is instrumental in the growth arrest process that is an absolute requirement for muscle cell differentiation. In this case, cell cycle exit is irreversible. Rb is also involved, in cycling cells, in cell progression through the restriction point toward S phase. In this case, however, Rb’s suppressive effect is reversible, and Rb is activated and inactivated in a cell cycle-dependent manner. Rb controls the E2F family of transcription factors (Nevins et al., 1991; Nevins, 1992), which in turn plays an essential role in the G1–S transition (Brehm et al., 1999).
E2F regulates several families of genes whose products are required for cell cycle progression, such as B-myb and cyclin A (Lam and Watson, 1993; Geng et al., 1996), or for DNA synthesis (DeGregori et al., 1995; Yan et al., 1998), such as DHFR (dihydrofolate reductase) (Fry et al., 1997). These genes are expressed in a cycle-dependent manner in proliferating cells, and are irreversibly repressed in differentiating cells. In cycling cells, these genes are silent during early G1, and are rapidly activated at the restriction point, which precedes the G1–S transition. In G0 or G1 cells, E2F sites in the promoters of these genes are generally occupied (Zwicker et al., 1996) by multimolecular complexes that include E2F proteins and Rb or other members of the pocket protein family (Takahashi et al., 2000; Wells et al., 2000). A body of experimental data indicates that the Rb–E2F repressive complex functions in association with a histone deacetylase (HDAC) (Brehm et al., 1998; Ferreira et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998; Stiegler et al., 1998; Lai et al., 1999). HDACs essentially repress transcription (Hassig et al., 1998), probably through deacetylation of histone tails that protrude from nucleosomes (Wolffe, 1996), resulting in local modification of chromatin structure (Wolffe and Guschin, 2000); they also deacetylate non-histone proteins (Kouzarides, 2000).
HDACs can be classified into two structural groups. Whereas some class II HDACs are involved in regulating cell differentiation, in particular in muscle (Miska et al., 1999; McKinsey et al., 2000), some class I HDACs (HDAC 1–3) participate in the control of cell cycle progression, as mentioned above, by cooperating with the co-repressor Rb (Brehm et al., 1998; Ferreira et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998). Rb physically associates with HDAC-1 and other class I HDACs; and in vitro as well as in live cells, a tri-molecular complex including E2F, Rb and HDAC-1 can be detected. In addition, class I HDACs and Rb cooperate in functional assays involving E2F driven reporter constructs and transient transfections. These data gave rise to a model for the mode of action of the Rb–E2F complex that postulates the recruitment of HDACs on E2F target promoters, and raises several questions: is the Rb–E2F–HDAC complex active in growth-arrested differentiating cells, in proliferating cells between early G1 and the restriction point, or in both types of cells? Is HDAC recruited to target promoters in a relatively stable manner or is the association transient? In order to test the validity of this model and answer some of these questions, we have used immunoprecipitation of crosslinked chromatin (XChIP) (Orlando et al., 1997), a technique that assays for physical associations of proteins with specific DNA sequences in live cells. The model we have used is the DHFR promoter, a well characterized E2F target gene that is silent in early G1 and switched on at the G1–S transition. Our results show a physical association between HDAC-1 and the DHFR promoter in G0 and G1 cells—in these cells, the gene is silent and histone H4 is poorly acetylated. HDAC-1 association with the DHFR promoter decreased in cells progressing through the restriction point into S phase, concomitantly with an increase in histone H4 acetylation on this promoter. Use of lysine-specific antibodies suggested that this G1–S transition-associated increase corresponded to acetylation on two specific lysines, lysines 5 and 12. Dissociation of HDAC-1 from the promoter was concomitant with an increase in DHFR mRNA steady state levels. These data demonstrate a recruitment of HDAC-1 to an E2F target promoter stable enough to be visualized by ChIP; they also show that the E2F–Rb–HDAC repressive complex is active in proliferating cells and that its recruitment to promoters is regulated in a cell cycle-dependent manner.
RESULTS AND DISCUSSION
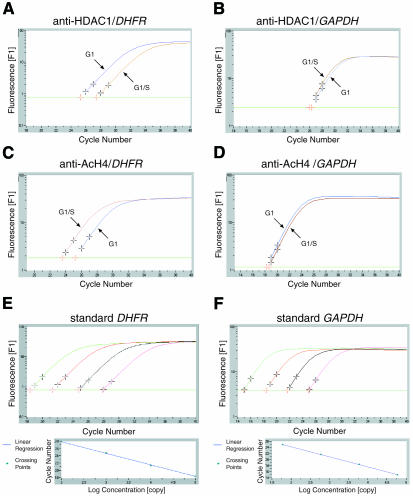
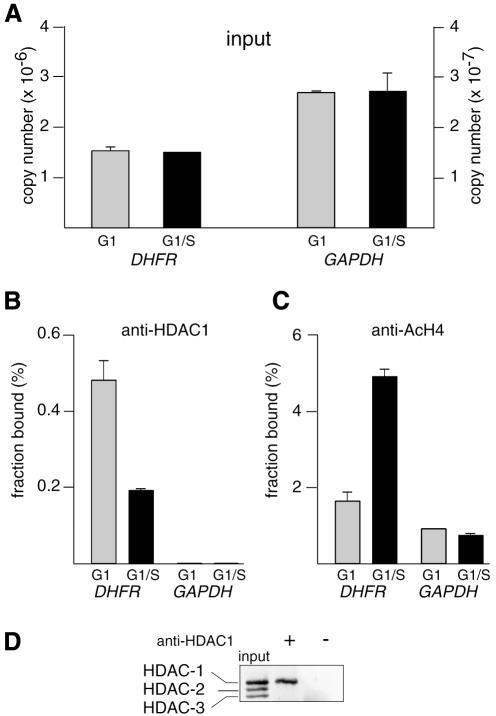
The association between HDAC-1 and the promoter of the DHFR gene, an E2F target promoter, was monitored by XChIP with chromatin prepared from NIH 3T3 fibroblast cells at different phases of the cell cycle (determined by FACS analysis, see Supplementary data, available at EMBO reports Online). Histone H4 acetylation was followed in parallel on the same promoter. Results of a typical experiment are shown in Figures 1 and 2. Target sequences (either the DHFR promoter between –49 and +131 bp with reference to the transcription start site and encompassing the E2F binding site, or else a GAPDH sequence that was used here as a constitutively expressed, internal control) were detected by quantitative PCR, using a LightCycler (Roche Diagnostics). In this assay, the amount of promoter DNA in the samples was estimated by real time monitoring of the accumulation of the amplified sequence, using SYBR green dye fluorescence. The amplified product did indeed correspond to the promoter sequence as demonstrated by its melting curve and by gel analysis of the final product (see Supplementary data). Numbers of copies were estimated by reference to a standard curve, obtained from PCR run in parallel using known concentrations of a plasmid harboring the sequence to be amplified (Figure 1E and F). Standardization of the chromatin inputs for immunoprecipitation was assessed in each experiment (Figure 2A; Methods). A fraction of DHFR sequence was found in association with the anti-HDAC-1 immunoprecipitate in chromatin extracted from G1 cells (Figures 1A and 2B). In these precipitates, the HDAC-1 protein was readily detectable (Figure 2D). As a control, chromatin was immunoprecipitated in the absence of specific antibodies, and in these samples, neither the HDAC-1 protein (Figure 2D) nor significant amounts of DHFR promoter (see legend to Figure 2) were detected. HDAC-1 associated with the DHFR promoter decreased as cells progressed through the G1–S transition (Figures 1A and 2B). Identical results were obtained using a different anti-HDAC antibody (see Supplementary data). Levels of GAPDH, which is constitutively expressed, were also evaluated as a negative control: as expected the sequence was found to be barely detectable in the immunoprecipitates (Figures 1B and 2B).
Fig. 1. Cell cycle-dependent recruitment of HDAC-1 and histone H4 deacetylation on DHFR promoter: detection of DHFR promoter in anti-HDAC-1 and anti-acetylated histone H4 immunoprecipitates. Chromatin from synchronized NIH 3T3 cells, either in G1 or at the G1–S transition, as indicated, was analyzed by the XChIP procedure, using anti-HDAC-1 (A and B) or anti-acetylated histone H4 (anti-AcH4) (C and D), followed by PCR analysis of eluted DNA using a LightCycler. The curves show the accumulation of PCR products plotted against the number of cycles (A and C, DHFR; B and D, GAPDH). For the sake of clarity, the results are shown for only one dilution of the immunoprecipitates, but three dilutions were analyzed for each sample. Crosses indicate data points used by the software in calculating copy numbers. (E and F) Curves obtained with reference plasmid DNA for DHFR (E) or GAPDH (F), using 1, 10, 100 and 1000 fg of plasmid.
Fig. 2. Cell cycle-dependent recruitment of HDAC-1 and histone H4 deacetylation on DHFR promoter: compilation of results from Figure 1. Chromatin from synchronized NIH 3T3 cells, either in G1 phase (gray bars) or at the G1–S transition (black bars), was analyzed by the XChIP procedure, followed by quantitative PCR of eluted DNA. Equal amounts of chromatin (A) were subjected to immunoprecipitation using anti-HDAC-1 (B) or anti-AcH4 (C) antibodies. DHFR or GAPDH sequences (as indicated) were detected by quantitative PCR. Copy numbers were estimated by reference to a plasmid containing the promoter sequence and used as a standard; (A) number of copies in the inputs; (B) and (C) fraction of the total number of copies detected as antibody-bound material; a control sample run in parallel in the absence of antibodies revealed little association of DHFR (0.01% of the input) or GAPDH (0.002% of the input). Shown are the results of a typical experiment, with range bars indicating standard deviations of triplicates. This experiment has been reproduced four times with similar results. (D) Chromatin, immunoprecipitated by anti-HDAC-1 (+) or control (–) antibodies or before immunoprecipitation (input), was incubated for 15 min at 95°C and analyzed by western blotting, using an antibody that recognizes HDAC-1, HDAC-2 and HDAC-3 (H80920; Transduction Laboratories).
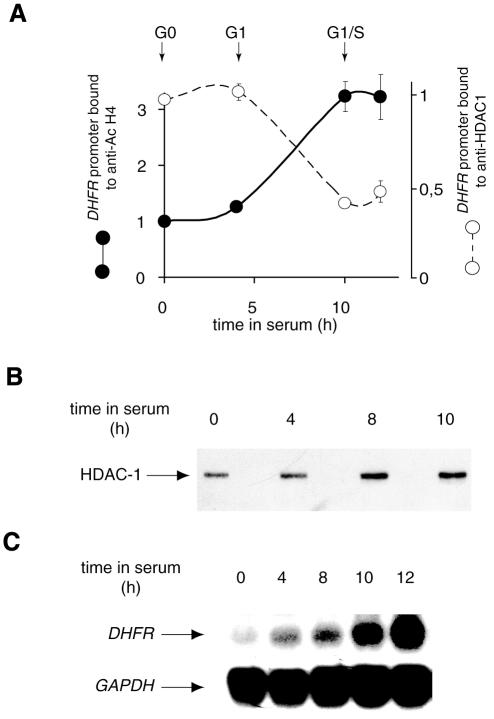
A reciprocal result was observed when the same XChIP approach was used to assay acetylated histone H4 on the DHFR promoter (Figures 1C and 2C): acetylation was minimal in G1 cells and increased at the G1–S transition. In contrast, histone H4 acetylation on the GAPDH gene did not vary (Figures 1D and 2C). Note that the detection of acetylated histone was more sensitive than the detection of associated HDAC-1, a larger fraction of the input being retained with anti-acetylated H4 antibodies than with anti-HDAC-1. This is most likely related to the fact that, in contrast to the histones, HDAC-1 does not bind directly to DNA. Its detection is thus highly dependent on the efficiency of the cross-linking procedure. A time course analysis of histone H4 acetylation and HDAC-1 association (Figure 3A) indicated that HDAC-1 is associated with the DHFR promoter in G0 cells and during early G1, and released at the G1–S transition. The observed decrease in HDAC-1 association with the DHFR promoter did not reflect a general decrease in the HDAC-1 protein level in cell extracts (the amount of HDAC-1 protein actually increased near the G1–S transition; Figure 3B). HDAC-1 release from the DHFR promoter (observed beyond 10 h post-serum addition) was concomitant with the increase in the steady state level of DHFR mRNA (Figure 3C). This release thus correlates with increased histone acetylation and activity of the promoter. This suggests that HDAC-1 is responsible for histone deacetylation and repression of the promoter during early G1. This hypothesis is supported by the observed effect of a histone deacetylase inhibitor, trichostatin A (TSA), on the promoter; in quiescent cells, TSA treatment resulted in the acetylation of histone H4 to levels similar to those seen at the G1–S transition in serum-treated cells (see Supplementary data). Under these conditions, however, the gene was not induced, indicating that histone H4 acetylation on the promoter is not sufficient to trigger transcription. It must be emphasized that TSA prevents cell progression into S phase. Our results thus strongly suggest that an additional cell cycle-regulated event, distinct from histone H4 acetylation, is required for DHFR gene activation.
Fig. 3. Inverse correlation between HDAC-1 recruitment and histone H4 acetylation on DHFR promoter. (A) Chromatin was extracted from cells at different phases of the cell cycle, and immunoprecipitated with anti-HDAC-1 or anti-AcH4 as indicated. The results are expressed as fold variation with reference to the resting cells (mean values from three determinations in three independent experiments, with range bars indicating standard deviations). (B) HDAC-1 protein in immunoprecipitates was monitored in parallel experiments by western blotting with an anti-HDAC antibody. (C) Total RNA was extracted from cells at indicated time points and analyzed by northern blotting using a DHFR or a GAPDH probe, as indicated.
In order to determine which of the acetylatable lysines of histone H4 were in fact acetylated at the G1–S transition, chromatin was immunoprecipitated from G1 or G1–S cells, using antibodies directed against the acetylated forms of specific H4 lysines. Results (Figure 4) indicated that acetylation was preferentially increased on lysines 5 and 12. This result confirms the modification of histone H4 acetylation at the G1–S transition, and suggests that lysines 5 and 12 are targets for HDAC-1 on the DHFR promoter. Since these same lysines are known to be targets for cytoplasmic HAT-B, which acetylates de novo synthesized histones (Sobel et al., 1995), it could be postulated that their acetylation is related to early replication of the DHFR promoter. However, our data are consistent with a previous report in yeast, in which inactivation of the rpd3 gene, of which HDAC-1 is an ortholog, resulted in increased acetylation of histone H4 on lysines 5 and 12 (Rundlett et al., 1998). In that study, acetylation of lysines 5 and 12 on histone H4 was clearly involved in transcription at the target promoters. Furthermore, it is important to note that some transcriptional co-activators which display a histone acetyltransferase activity are able to acetylate these lysines, at least in vitro; for example, CBP/p300 acetylates all lysines of histone H4 with a preference for lysine 5 (Schiltz et al., 1999).
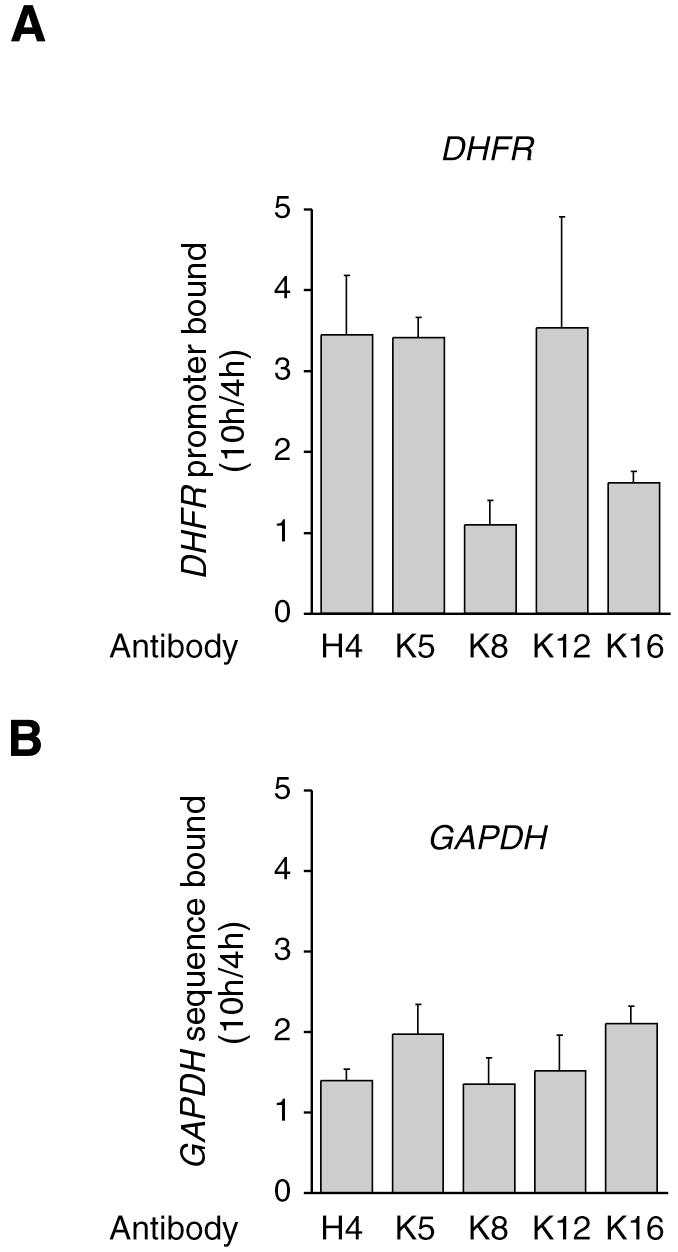
Fig. 4. Lysines 5 and 12 are deacetylated on repressed DHFR promoter. Chromatin was extracted from G1 (4 h) or G1–S (10 h) cells, and immunoprecipitated with anti-AcH4 or anti-acetylated H4 K5, K8, K12 or K16 antibodies, as indicated. The results are expressed as fold variation with reference to the G1 cells (mean values from three determinations in three independent experiments, with range bars indicating standard deviations). (A) DHFR; (B) GAPDH.
Our data show a stable and cell cycle-dependent recruitment of a histone deacetylase to an E2F target promoter, and thus provide the first direct experimental evidence for the presumed mode of action of these enzymes on these promoters. In addition, an inverse correlation between HDAC-1 recruitment and H4 histone acetylation was observed (Figure 3A). These results indicate a balance between two states for the DHFR promoter during the cell cycle. When the gene is silent, HDAC-1 is physically associated with the promoter and histone H4 is deacetylated. Upon activation of the gene, HDAC-1 is released and histone H4 is acetylated. In that regard, the DHFR promoter seems to be regulated in a canonical manner. It should be noted, however, that this may not be a general scheme, and on other promoters, such a correlation between gene activity and histone acetylation might not be observed. In any case, our results suggest that HDAC-1 dissociation from the DHFR promoter is a key event of the G1–S transition and that the same process could regulate the expression of other genes at this stage of the cell cycle. Determining the mechanisms involved and the composition of the recruited complexes will be central to understanding cell proliferation control.
METHODS
Cell culture and synchronization.
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (BRL, Life Technologies) supplemented with 10% bovine calf serum. Cells were synchronized by serum deprivation for 48 h (G0 cells) and treated with serum for 4 h (G1 cells), 10 h (G1–S) or 12 h, unless otherwise indicated.
Formaldehyde cross-linking and chromatin immunoprecipitation.
Cells were treated with formaldehyde at a final concentration of 1% for 8 min at 37°C. Cross-linking was stopped by addition of glycine to a final concentration of 0.125 M. Cross-linked cells were harvested, washed in phosphate-buffered saline supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 5 mM sodium butyrate (NaB). Subsequent procedures were performed on ice, with buffers supplemented with 1 mM PMSF, 5 mM NaB and a protease inhibitor mix (Roche Diagnostics). Cells were lysed in lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40). After homogenization with a Dounce homogenizer, nuclei were pelleted and lysed by incubation in nuclear lysis buffer (50 mM Tris–HCl pH 8.1, 10 mM EDTA, 1% SDS). Chromatin was sonicated with eight 10 s pulses (50 W, amplitude 80%, Bioblock Vibra Cell 72434). DNA contents in nuclear extracts were standardized by non-denaturing gel electrophoresis and standardization was verified by quantitative PCR on the LightCycler. After centrifugation, the supernatant was diluted 10-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1, 167 mM NaCl). Diluted extracts were pre-cleared with protein A/protein G agarose beads (Sigma) and incubated with anti-HDAC-1 antibodies (raised against the synthetic peptide EEKPEAKGVKEEVKLA in Bryan Turner’s laboratory; this antibody is specific for HDAC-1 and does not cross react with the other HDACs; see Figure 2D), anti-acetylated H4 antibodies (Upstate biotechnology), anti-acetylated H4 K5, K8, K12 and K16 (raised in Bryan Turner’s laboratory) or irrelevant antibodies, and immunoprecipitated with protein A/protein G agarose beads. Following extensive washing (details available upon request), bound DNA fragments were eluted by overnight incubation at 65°C followed by treatment with proteinase K. Samples were analyzed by quantitative PCR (LightCycler, Roche Diagnostics) using SYBR green dye (Figure 1). The primers used were: GCCTAAGCTGCGCAAGTGGT and GTCTCCGTTCTTGCCAATCC for the DHFR sequence; and CCAATGTGTCCGTCGTGGATCT and GTTGAAGTCGCAGGAGACAACC for GAPDH. Numbers of copies of the specific sequences were calculated by reference to a log-linear standard curve constructed from the number of cycles necessary to detect product accumulation after amplification of a plasmid harboring the sequence (Figure 1E and F). Two to three dilutions of each sample were analyzed and results are expressed as the fraction of the total number of input copies that were detected in each immunoprecipitate.
Immunoprecipitation and western blotting.
For determination of the amount of HDAC-1 protein in NIH 3T3 cells during the cell cycle, equivalent numbers of cells (50 × 106) were lysed with lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM MgCl2, 0.4% NP-40, supplemented with a protease inhibitor mix) for 15 min at 4°C. After centrifugation, whole cell extracts were diluted 1-fold with dilution buffer (50 mM Tris pH 8.0, 0.4% NP-40). Diluted extracts were precleared with protein A/protein G agarose beads, incubated with an anti-HDAC-1 antibody (06-720 from Upstate Biotechnology) or mouse IgGs as a control, and immunoprecipitated with protein A/protein G agarose beads. Proteins were analyzed by western blotting using an antibody that recognizes HDAC-1, HDAC-2 and HDAC-3 (H80920 from Transduction Laboratories) and standard procedures.
Northern blotting.
Total RNA extracted from NIH 3T3 cells was purified using a kit from Promega (RNAgents) and analyzed by northern blotting using standard procedures and cDNA probes for the mouse DHFR or GAPDH gene, previously labeled using a Redi-prime II random primer labeling kit (Amersham).
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr B.M. Turner for materials and for critical reading of the manuscript. This work was supported by grants from the Association pour la Recherche sur le Cancer, from the Comité du Val de Marne de la Ligue Contre le Cancer and from the European 5th PCRDT (grant QLG1-1999-00866). R.F. was supported by the Comité du Val d’Oise de la Ligue Contre le Cancer, I.N. by the Association pour la Recherche sur le Cancer and M.M. by the CNRS.
REFERENCES
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J. and Kouzarides, T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska, E., Reid, J., Bannister, A. and Kouzarides, T. (1999) The cell cycle-regulating transcription factors E2F-RB. Br. J. Cancer, 80 Suppl 1, 38–41. [PubMed] [Google Scholar]
- DeGregori J., Kowalik, T. and Nevins, J.R. (1995) Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes [published erratum appears in Mol. Cell. Biol., 1995, 15, 5846–5847]. Mol. Cell. Biol., 15, 4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Magnaghi-Jaulin, L., Robin, P., Harel-Bellan, A. and Trouche, D. (1998) The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl Acad. Sci. USA, 95, 10493–10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry C.J., Slansky, J.E. and Farnham, P.J. (1997) Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol. Cell. Biol., 17, 1966–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Eaton, E.N., Picon, M., Roberts, J.M., Lundberg, A.S., Gifford, A., Sardet, C. and Weinberg, R.A. (1996) Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene, 12, 1173–1180. [PubMed] [Google Scholar]
- Hassig C.A., Tong, J.K., Fleischer, T.C., Owa, T., Grable, P.G., Ayer, D.E. and Schreiber, S.L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl Acad. Sci. USA, 95, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A., Lee, J.M., Yang, W.M., DeCaprio, J.A., Kaelin, W.G., Jr, Seto, E. and Branton, P.E. (1999) RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol., 19, 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E.W. and Watson, R.J. (1993) An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J., 12, 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo, A.A. and Dean, D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman, R., Naguibneva, I., Robin, P., Lorain, S., Le Villain, J.P., Troalen, F., Trouche, D. and Harel-Bellan, A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang, C.L., Lu, J. and Olson, E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Karlsson, C., Langley, E., Nielsen, S.J., Pines, J. and Kouzarides, T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Nevins J.R., Chellappan, S.P., Mudryj, M., Hiebert, S., Devoto, S., Horowitz, J., Hunter, T. and Pines, J. (1991) E2F transcription factor is a target for the RB protein and the cyclin A protein. Cold Spring Harb. Symp. Quant. Biol., 56, 157–162. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt, H. and Paro, R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen, A.A., Suka, N., Turner, B.M. and Grunstein, M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen, C.A., Vassilev, A., Cook, R.G., Allis, C.D. and Nakatani, Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem., 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- Sobel R.E., Cook, R.G., Perry, C.A., Annunziato, A.T. and Allis, C.D. (1995) Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA, 92, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., De Luca, A., Bagella, L. and Giordano, A. (1998) The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res., 58, 5049–5052. [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B. and Dynlacht, B.D. (2000) Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev., 14, 804–816. [PMC free article] [PubMed] [Google Scholar]
- Wells J., Boyd, K.E., Fry, C.J., Bartley, S.M. and Farnham, P.J. (2000) Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol., 20, 5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. (1996) Histone deacetylase: a regulator of transcription. Science, 272, 371–372. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Guschin, D. (2000) Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol., 129, 102–122. [DOI] [PubMed] [Google Scholar]
- Yan Z., DeGregori, J., Shohet, R., Leone, G., Stillman, B., Nevins, J.R. and Williams, R.S. (1998) Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker J., Liu, N., Engeland, K., Lucibello, F.C. and Muller, R. (1996) Cell cycle regulation of E2F site occupation in vivo. Science, 271, 1595–1597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.