Highlights
-
•
Transcriptomics of postoperative tissue from 32 patients and quantitative proteomics of 19 patients were used to explore potential biomarkers in invasive pituitary adenomas.
-
•
Multiple methods were used to verify the potential association of target gene with pituitary tumor invasiveness and mitophagy, immune-related processes.
-
•
To guide the clinical diagnosis and therapy of patients with invasive pituitary adenomas.
Keywords: Pituitary adenoma; Mitophagy, Invasion; HSPD1; Diagnosis
Abstract
Background
The crucial role of mitophagy in tumor progression has been recognized. Therefore, our study aimed to investigate the potential correlation between pituitary adenoma invasiveness and the mitophagy processes.
Methods
In this study, we used transcriptomics of postoperative tissue from 32 patients and quantitative proteomics of 19 patients to screen for mitophagy-related invasion genes in pituitary adenomas. The invasive predictive value of target genes was analyzed by Lasso regression model, CytoHubba plugin and expression validation. Co-expression correlation analysis was used to identify paired proteins for target genes, and a predictive model for pituitary adenoma invasiveness was constructed by target genes and paired proteins and assessed using ROC analysis, calibration curves and DCA. GO function, pathway (GSEA or GSVA) and immune cell analysis (ssGSEA or CIBERSORT) were further utilized to explore the action mechanism of target gene. Finally, immunohistochemistry and cell function experiments were used to detect the differential expression and key roles of the target genes in pituitary adenomas.
Results
Finally, Heat shock protein family D member 1 (HSPD1) was identified as a target gene. The quality of a predictive model for pituitary adenoma invasiveness consisting of HSPD1 and its paired protein expression profiles was satisfactory. Moreover, the expression of HSPD1 was significantly lower in invasive pituitary adenomas than in non-invasive pituitary adenomas. Downregulation of HSPD1 may be significantly related to invasion process, mitochondria-related pathway and immune cell regulation in pituitary adenomas.
Conclusion
The downregulation of HSPD1 may serve as a predictive indicator for identifying invasive pituitary adenomas.
Introduction
Pituitary adenomas, though primarily benign tumors, encompass certain subtypes referred to as invasive pituitary adenomas, which are characterized by their invasive growth, rapid progression, and high recurrence rates [1], [2], [3]. Patients with invasive pituitary adenomas experience poor prognoses and are at a significant risk of recurrence even after undergoing standard surgical treatment [4]. Consequently, the World Health Organization designated them as "high-risk" pituitary tumors in 2017 [5,6]. Mitophagy, a selective form of autophagy responsible for removing damaged and aging mitochondria, plays a critical role in maintaining cellular homeostasis by ensuring the quantity and quality of mitochondrial populations [7,8]. Its involvement has been observed in tumor prevention, metabolic program rearrangement, enhancement of tumor resistance, and promotion of tumor invasion [9], [10], [11]. In various cancers, such as hepatocellular carcinoma, glioma, and non-small cell lung cancer, targeting mitophagy has demonstrated efficacy in inhibiting tumor progression, suggesting its potential as a promising strategy in suppressing malignant tumor advancement [12], [13], [14], [15]. Despite this, limited research exists on mitophagy in the context of invasive pituitary tumors, and the development of related therapeutic targets remains an area of exploration. Therefore, our study aims to investigate mitophagy-related biomarkers associated with invasiveness in pituitary adenomas to predict the risk of invasive progression and identify potential therapeutic targets for this condition.
Materials and methods
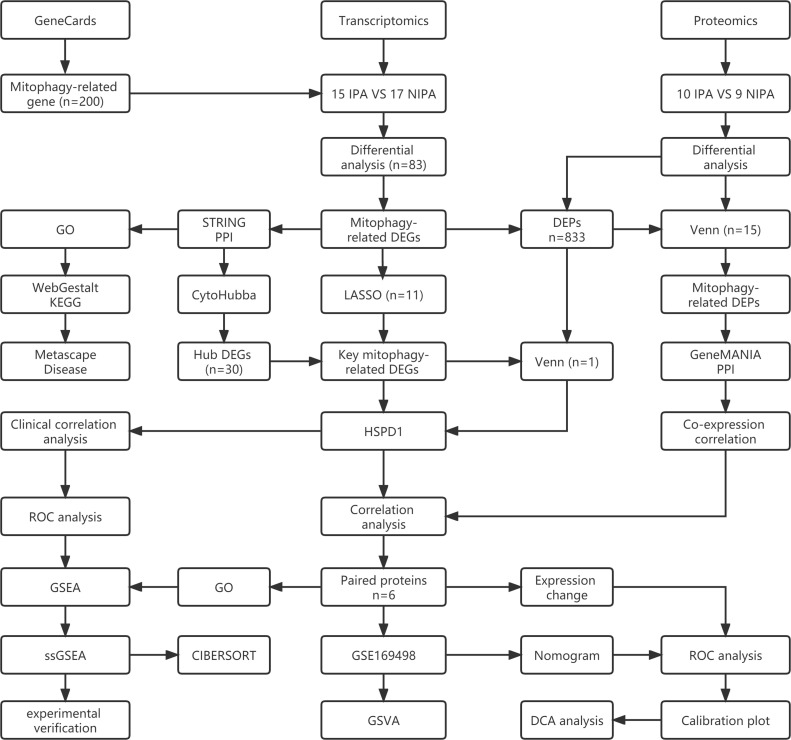
The analysis process of this study is shown in Fig. 1.
Fig. 1.
The analysis flow chart of this study.
Data source and preparation
Transcriptomic data included postoperative tissue sample data from 15 patients with invasive pituitary adenomas and 17 patients with non-invasive pituitary adenomas. First, the samples were subjected to quality inspection, and the mRNA was isolated after passing quality inspection, and mRNA interruption and cDNA synthesis were performed. The double-stranded cDNA ends were then repaired, and an A base was added at the 3 'end to prepare the adaptor ligation reaction system to connect the adaptors to the cDNA. Then the products were amplified by PCR, and the corresponding detection methods were selected for quality inspection of the library. After the PCR product is denatured into single strand, the cyclization reaction system is prepared, the reaction procedure is set up to obtain single strand circular product, the uncyclized linear DNA molecules are digested, and finally the PCR product is sequenced on the machine (MGISEQ 2000).
Proteomic data were obtained from postoperative tissue from 19 patients with pituitary adenomas, including 10 invasive pituitary adenomas and 9 non-invasive pituitary adenomas. Firstly, protein extraction and digestion of pituitary adenoma tissue samples were carried out, and then quantitative protein analysis was performed using LC-MS/MS detection system to obtain proteomic data. This data has been deposited in the ProteomeXchange database (https://www.ebi.ac.uk/pride/, PXD039328). Pituitary adenomas gene expression dataset (GSE169498) from a GEO database (https://www.ncbi.nlm.nih.gov/geo/). There were 49 samples of invasive pituitary adenoma and 24 samples of non-invasive pituitary adenoma. In this study, all gene expression data were log2 processed (rt=log2(rt+1)) and normalized. Mitophagy-related genes were downloaded from GeneCards (https://www.genecards.org/), according to the relevance score for the top 200 mitophagy-related genes. Table 1 shows the number of pituitary adenoma samples contained in all datasets of this study.
Table 1.
The number of pituitary adenoma samples contained in all datasets of this study.
| DataSet | IPA | NIPA | Total number |
|---|---|---|---|
| Transcriptomics | 15 | 17 | 32 |
| Proteomics | 10 | 9 | 19 |
| GSE169498 | 49 | 24 | 73 |
| Total number | 74 | 50 | 124 |
Key and hub mitophagy-related invasive genes acquisition
To identify the mitophagy-related invasive genes in pituitary adenomas, we first extracted the expression profile data of mitophagy-related genes in the transcriptomics. Next, we conducted invasive correlation tests on these genes using the wilcox test method in the limma package of R software, aiming to uncover differentially expressed genes (DEGs) linked to invasive and non-invasive pituitary adenomas. In order to further determine the correlation between DEGs and mitophagy and tumor invasion, we employed the STRING database for protein-protein interaction (PPI) network mapping. Additionally, Gene Ontology(GO) enrichment analysis, WebGestalt (https://www.webgestalt.org/) Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis, and Metascape (http://metascape.org/) Disease enrichment analysis were performed to analyse the role played by these DEGs. Lastly, we utilized the Lasso regression model from the glmnet package to screen the DEGs and identify key genes associated with mitophagy. This approach enabled us to pinpoint Key mitophagy-related Invasive Genes in pituitary adenomas. We also used Cytoscape to analyze the PPI network of mitophagy-related invasive genes, and identified the top 30 of these genes as Hub mitophagy-related invasive genes through the degree algorithm in CytoHubba plug-in.
Target gene acquisition
We performed differential analysis by quantitative proteomics data of invasive and non-invasive pituitary adenomas, using student's t-test to screen for invasiveness-related proteins (|log2FC| > 1, P < 0.05). To identify the overlap between the differentially expressed proteins, Key and Hub mitophagy-related invasive genes, we utilized the Venn online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). This allowed us to pinpoint the target genes for our study. To evaluate the predictive accuracy of the target gene in assessing the invasiveness of pituitary adenomas, We identified them using transcriptomics and proteomics data. Subsequently, we conducted invasiveness correlation validation and receiver operating characteristic (ROC) analysis to assess the predictive value of this gene.
Paired protein acquisition
We employed the Venn online tool to identify the intersection of invasive differential proteins and mitophagy-related differential genes to identify mitophagy-related differential proteins. The obtained proteomics data was analyzed using the limma package. Additionally, we conducted co-expression correlation analysis between target genes and differential proteins associated with mitophagy. This analysis enabled us to screen for pairwise acting proteins of the target genes (cor>0.7 and p<0.05). We also used transcriptomics and GSE169498 to analyze the expression changes of HSPD1 and its paired proteins in invasive and non-invasive pituitary adenomas.
Invasive predictive model development and validation
Due to the substantial sample size provided by GSE169498, we selected this dataset for constructing a predictive model. The model aimed to predict the invasiveness of pituitary adenomas based on the expression patterns of target genes and paired proteins. To achieve this, pituitary adenoma invasiveness nomogram were drawn by the rms and rmda packages of R software. Additionally, we evaluated the accuracy and sensitivity of the model by utilizing both the GSE169498 and transcriptomics. The performance of the pituitary adenoma invasiveness prediction model was further assessed through the application of ROC curves, calibration plots, and decision curve analysis (DCA).
Functions exploration of invasion predictive gene
In order to gain insights into the involvement of these predictive genes in pituitary adenomas, we conducted GO enrichment analysis to elucidate the functional implications of both target genes and paired proteins at the biological process (BP), cellular component (CC), and molecular function (MF) levels. Furthermore, we employed two additional algorithms, namely Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA), to predict the potential pathways influenced by the target genes throughout the course of pituitary adenoma invasiveness. Finally, we also analyzed immune cell differences in pituitary adenoma samples from transcriptomics data using the single sample Gene Set Enrichment Analysis (ssGSEA) and CIBERSORT algorithm, as well as the correlation of target gene with immune cells.
Expression validation of invasion predictive gene
For the assessment of the relationship between target gene expression and pituitary adenoma invasiveness, we utilized postoperative pathological tissue of pituitary adenomas and performed immunohistochemical detection specifically targeting the target genes. The utilization of human tissue in these experiments was carried out after obtaining approval from the Ethics Committee of Beijing Tiantan Hospital. The classification of invasive and non-invasive subgroups of pituitary adenomas was determined based on a comprehensive evaluation of radiological presentation, pathological features, and intraoperative findings. The process of immunohistochemical staining involved initial fixation of pituitary adenoma pathology in 10% formalin, followed by embedding in conventional paraffin. Subsequently, 4μm sections were prepared and numbered for further analysis. The paraffin sections were then subjected to oven baking and dewaxing, with antigen repair using a thermal repair method. To block endogenous peroxidase activity, an immunohistochemistry kit containing hydrogen peroxide was employed. Following a 5-minute incubation in a blocking solution, we added the target gene monoclonal antibody and placed it in a wet box and took it into a 4℃ refrigerator for overnight incubation. The next day, enzyme-labeled IgG polymer was added and incubated at room temperature for 10 minutes. Finally, after DAB and hematoxylin color development, dehydration sealing. Immunohistochemical images (SlideViewer 40×) were captured using an immunohistochemical section scanner (Leica, Germany). In addition, Image J software was used to scan the immunohistochemical images of each sample under random field of view. Finally, quantitative comparison between the two groups was conducted by average optical density (AOD) value, P<0.05 was considered statistically significant.
Function experiment of HSPD1 in pituitary adenoma cells
In this study, due to the inhibition of HSPD1 expression in invasive pituitary adenomas, we completed a series of functional experiments by overexpressing HSPD1 in GH3 pituitary adenoma cells (Xiehe, China). First, we employed the plasmid transfection method to clone the HSPD1 gene onto a vector, and negative controls were transfected with blank plasmids. Post-transfection, overexpression was induced, and we validated the transfection efficiency through fluorescence labeling. Stably transfected cells were selected using puromycin. Western blot (WB) analysis served as a method to verify the expression levels of the target protein, utilizing the Recombinant Anti-Hsp60 antibody (ab190828, Abcam, US) for the WB analysis. Furthermore, to assess changes in cell proliferation and vitality, we conducted biological activity tests using the CCK8 reagent (Dojindo, Japan). In each well of a 96-well plate, 2000 cells were seeded, and after pre-culture, CCK8 reagent was added at a ratio of 100:1. The OD450 values were measured at 24h, 48h, and 72h to reflect the proliferative capacity of the cells. Clonogenic formation experiments were employed to evaluate cell proliferation and clonogenic ability. Cells were seeded at 800 cells per well in a 6-well plate. After obtaining clones with more than 50 cells, fixation was performed using 4% paraformaldehyde (Beyotime, China), followed by staining with 0.1% crystal violet (Biosharp, China) and imaging. Scratch assays were utilized to investigate cell migration and healing capabilities by assessing the distance cells traveled in the scratched area. Finally, Transwell experiments were conducted to study cell invasion and migration capabilities. We utilized Corning's Transwell plates (3422, Corning, US) coated with Matrigel (356234, BD, US) to validate changes in the invasive abilities of cells before and after transfection.
Results
Mitophagy-related invasive genes
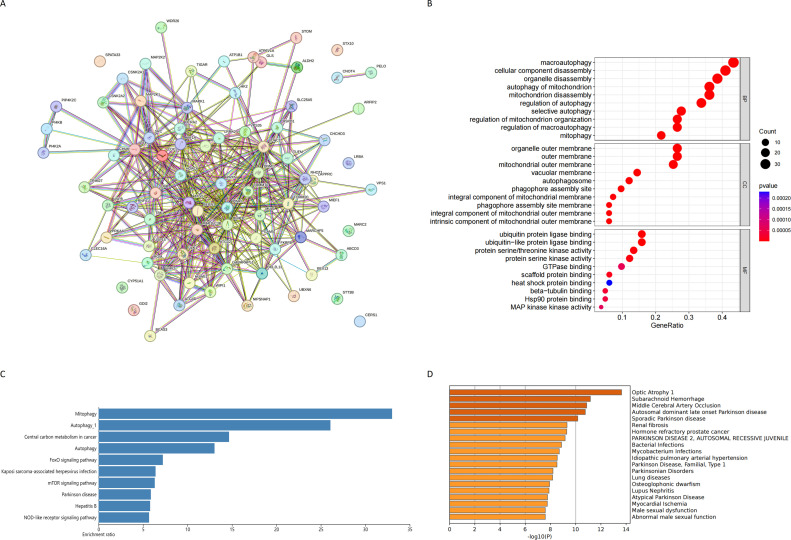
A comprehensive evaluation using differential expression validation was conducted on mitophagy-related genes in invasive and non-invasive pituitary adenomas from transcriptomics, resulting in the identification of 83 Mitophagy-related Invasive Genes. The protein-protein interaction network of these genes is depicted by the STRING database (Fig. 2A). Furthermore, GO enrichment analysis revealed the potential functions of the Mitophagy-related Invasive Genes, including macroautophagy, cellular component disassembly, organelle disassembly, autophagy of mitochondrion, mitochondrion disassembly, organelle outer membrane, outer membrane, mitochondrial outer membrane, ubiquitin protein ligase binding and ubiquitin−like protein ligase binding (Fig. 2B). Notably, the enriched pathways associated with Mitophagy–related Invasive Genes encompass Mitophagy, Autophagy_1, Central carbon metabolism in cancer, Autophagy, FoxO signaling pathway, Kaposi sarcoma-associated herpesvirus infection, mTOR signaling pathway, Parkinson disease, Hepatitis B and NOD-like receptor signaling pathway (Fig. 2C). In addition, the metscape database highlights diseases in which these genes are mainly enriched in neurological and hormonal aspects, such as Autosomal dominant late onset Parkinson disease, Sporadic Parkinson disease, Renal fibrosis, Hormone refractory prostate cancer, PARKINSON DISEASE 2, Parkinson Disease, Familial, Type 1, Parkinsonian Disorders, Male sexual dysfunction, Abnormal male sexual function (Fig. 2D).
Fig. 2.
(A) PPI network of mitophagy-related differential genes; (B) GO enrichment analysis of mitophagy-related differential genes; (C) KEGG enrichment analysis of mitophagy-related differential genes; (D) Disease enrichment analysis of mitophagy-related differential genes.
Key and hub mitophagy-related invasive genes
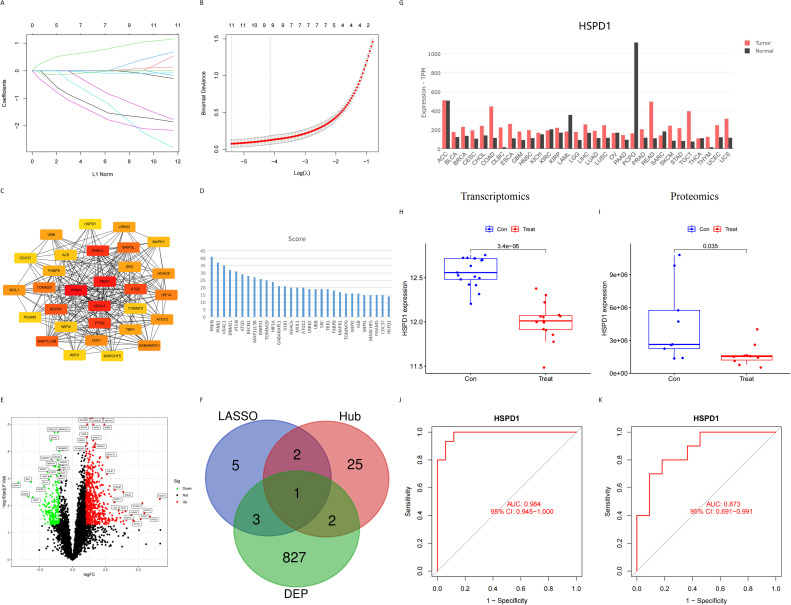
Through Lasso regression model, signature genes in mitophagy–related invasive genes were screened, and 11 Key mitophagy–related invasive genes were obtained (Fig. 3A, B). These included: PINK1, RNF41, TIGAR, VPS13D, CERS1, CHUK, PELO, SRC, PI4K2A, HSPD1 and GLS. CytoHubba plug-in was used to screen Hub mitophagy–related invasive genes. The top 30 genes with degree scores were shown in Fig. 3C and D.
Fig. 3.
(A) (B) Lasso regression analysis model of mitophagy-related differential genes; (C) PPI network of Hub mitophagy-related invasive genes; (D) The degree score ranking of Hub mitophagy-related invasive genes; (E) The Volcanic map of invasive differential proteins in proteomic data; (F) Intersection of Key and Hub mitophagy-related differential genes, invasive differential proteins; (G) Expression of HSPD1 in some normal and tumor tissues from the GEPIA database; (H) HSPD1 expression in invasive and non-invasive pituitary adenomas from transcriptomics; (I) HSPD1 expression in invasive and non-invasive pituitary adenomas from proteomics; (J) ROC curve of HSPD1 for predicting invasiveness of samples in transcriptomics; (K) ROC curve of HSPD1 for predicting invasiveness of samples in proteomics.
HSPD1 as a target gene
In proteomics, a total of 833 differentially expressed proteins (DEPs) were identified from samples of invasive and non-invasive pituitary adenomas (Fig. 3E). The Venn tool was used to obtain an intersection of 833 DEPs, 11 Key and 30 Hub mitophagy-related invasive genes, named Heat shock protein family D member 1 (HSPD1) (Fig. 3F). Through literature analysis and data summary, we finally choose HSPD1 as the focus of this study. We also demonstrated the expression of HSPD1 in some tumors and normal tissues using the GEPIA database (http://gepia.cancer-pku.cn/), which showed that HSPD1 was significantly up-regulated in colon adenocarcinoma (COAD), diffuse large B-cell lymphoma (DLBC), rectum adenocarcinoma (READ) tumors compared to normal tissues, and down-regulated in acute myeloid leukemia (LAML) and pheochromocytoma and paraganglioma (PCPG) tumors compared to normal tissues (Fig. 3G). Further analysis revealed a significant correlation between the expression of HSPD1 and the invasiveness of pituitary adenoma samples in both transcriptomics and proteomics data. Specifically, the expression of HSPD1 was significantly lower in invasive pituitary adenomas compared to non-invasive ones. This difference was supported by a p-value < 0.001 in transcriptomics (Fig. 3H) and 0.035 in proteomics data (Fig. 3I). Meanwhile, the area under the ROC curve of HSPD1 for predicting invasiveness in pituitary adenoma samples was satisfactory in both transcriptomics and proteomics data, with AUC values of 0.984 in transcriptomics (Fig. 3J) and 0.873 in proteomics (Fig. 3K).
Correlation between HSPD1 and its paired proteins
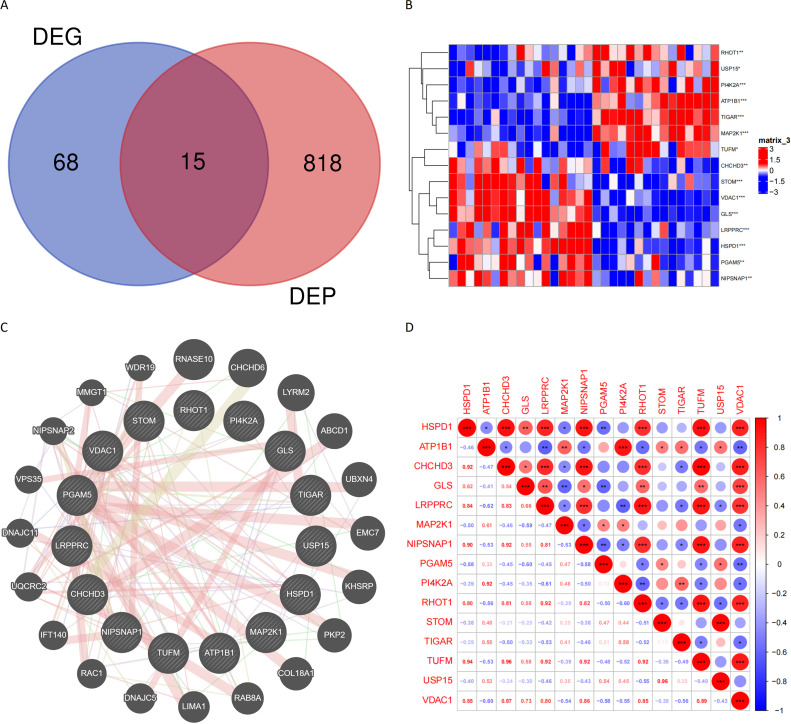
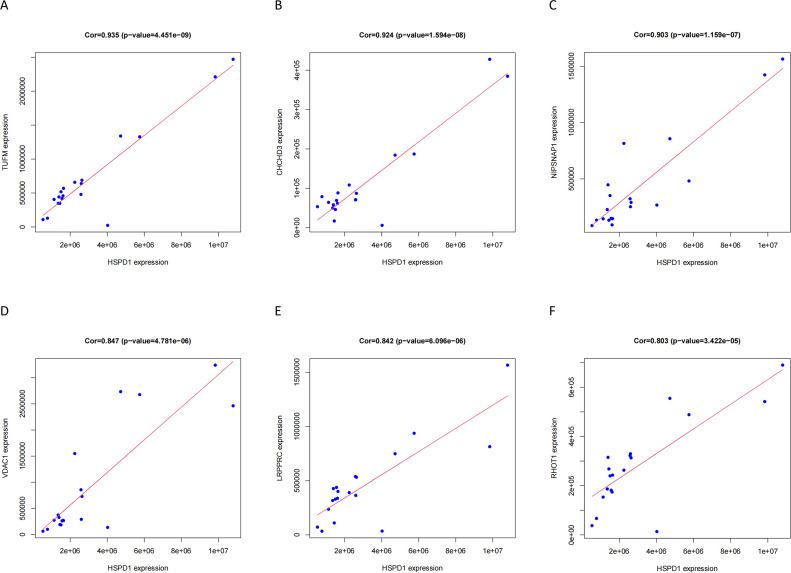
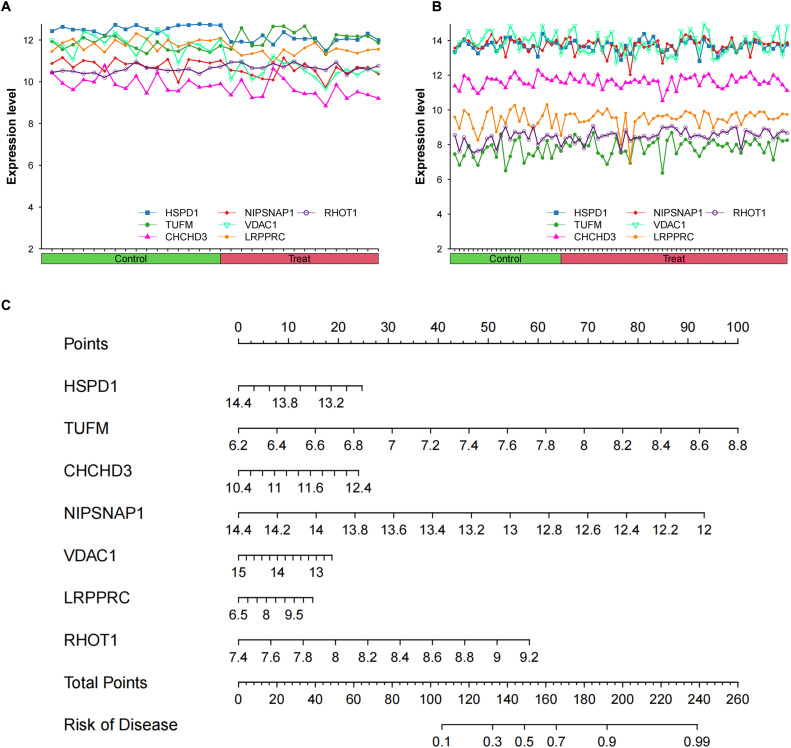
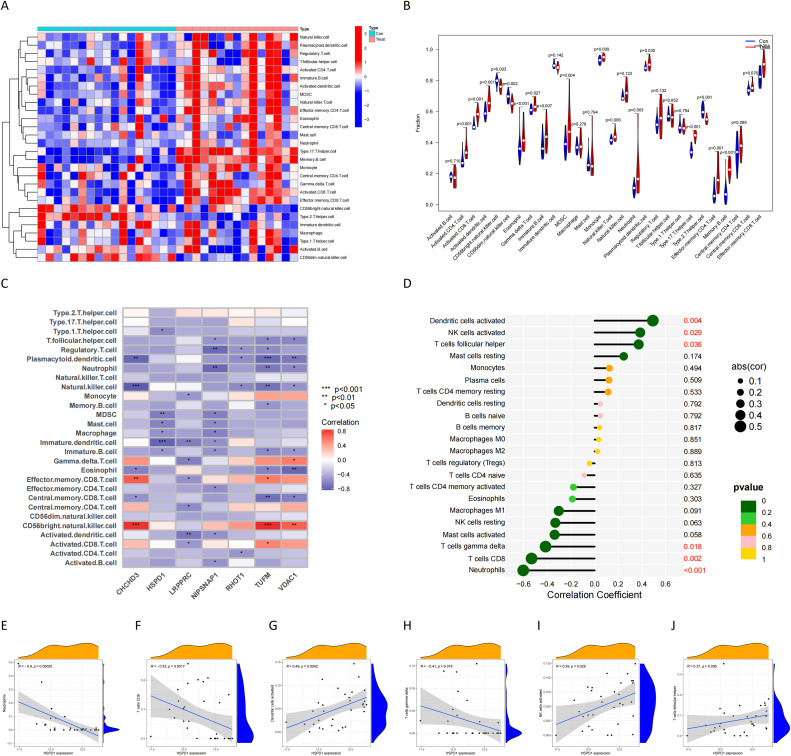
The Venn tool was utilized to identify the intersection between DEPs and Mitophagy-related Invasive Genes, resulting in the identification of 15 Mitophagy-related DEPs (Fig. 4A). The expression of these Mitophagy-related DEPs in the transcriptomics are visualized in Fig. 4B. To gain insights into the potential interactions among these DEPs and their associated proteins, the GeneMANIA database was consulted, revealing a network of interactions (Fig. 4C). A correlation heat map was constructed to illustrate the expression correlation of Mitophagy-related DEPs in the proteomics (Fig. 4D). Six HSPD1 paired proteins (TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1) were selected using the co-expression of HSPD1 with mitophagy-related DEPs in proteomics data (Cor>0.7, P<0.05, Table 2). Furthermore, the correlation curves provided evidence for the expression correlation of HSPD1 with TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1 (Fig. 5A–F). Notably, the correlation coefficients between HSPD1 and TUFM, CHCHD3, NIPSNAP1 were all > 0.9 (Fig. 5A-C), indicating a strong positive correlation. Also Fig. 6A and B demonstrate the expression changes of HSPD1 and its paired proteins in invasive and non-invasive pituitary adenomas in transcriptomics and GSE169498.
Fig. 4.
(A) Intersection of mitophagy-related differential genes and invasive differential proteins; (B) Expression profiles of mitophagy-related differential proteins in transcriptomics; (C) PPI network of mitophagy-related differential proteins in the GeneMANIA database; (D) Expression correlation of mitophagy-related differential proteins in proteomics.
Table 2.
Correlation analysis between HSPD1 and paired proteins in proteomics.
| Target | Paired proteins | Description | Cor | P-value | Rank |
|---|---|---|---|---|---|
| HSPD1 | TUFM | Tu translation elongation factor, mitochondrial | 0.935 | 4.45E-09 | 1 |
| CHCHD3 | coiled-coil-helix-coiled-coil-helix domain containing 3 | 0.924 | 1.59E-08 | 2 | |
| NIPSNAP1 | Nipsnap Homolog 1 | 0.903 | 1.16E-07 | 3 | |
| VDAC1 | voltage dependent anion channel 1 | 0.847 | 4.78E-06 | 4 | |
| LRPPRC | leucine rich pentatricopeptide repeat containing | 0.842 | 6.10E-06 | 5 | |
| RHOT1 | ras homolog family member T1 | 0.803 | 3.42E-05 | 6 |
Fig. 5.
Expression correlation between HSPD1 and its paired protein in proteomic Data.
Fig. 6.
(A) Expression changes of HSPD1 and its paired proteins in invasive and non-invasive pituitary adenomas in transcriptomics; (B) Expression changes of HSPD1 and its paired proteins in invasive and non-invasive pituitary adenomas in GSE169498; (C) A nomogram model for predicting sample invasiveness by HSPD1 and its paired protein expression in GSE169498.
Invasive predictive model
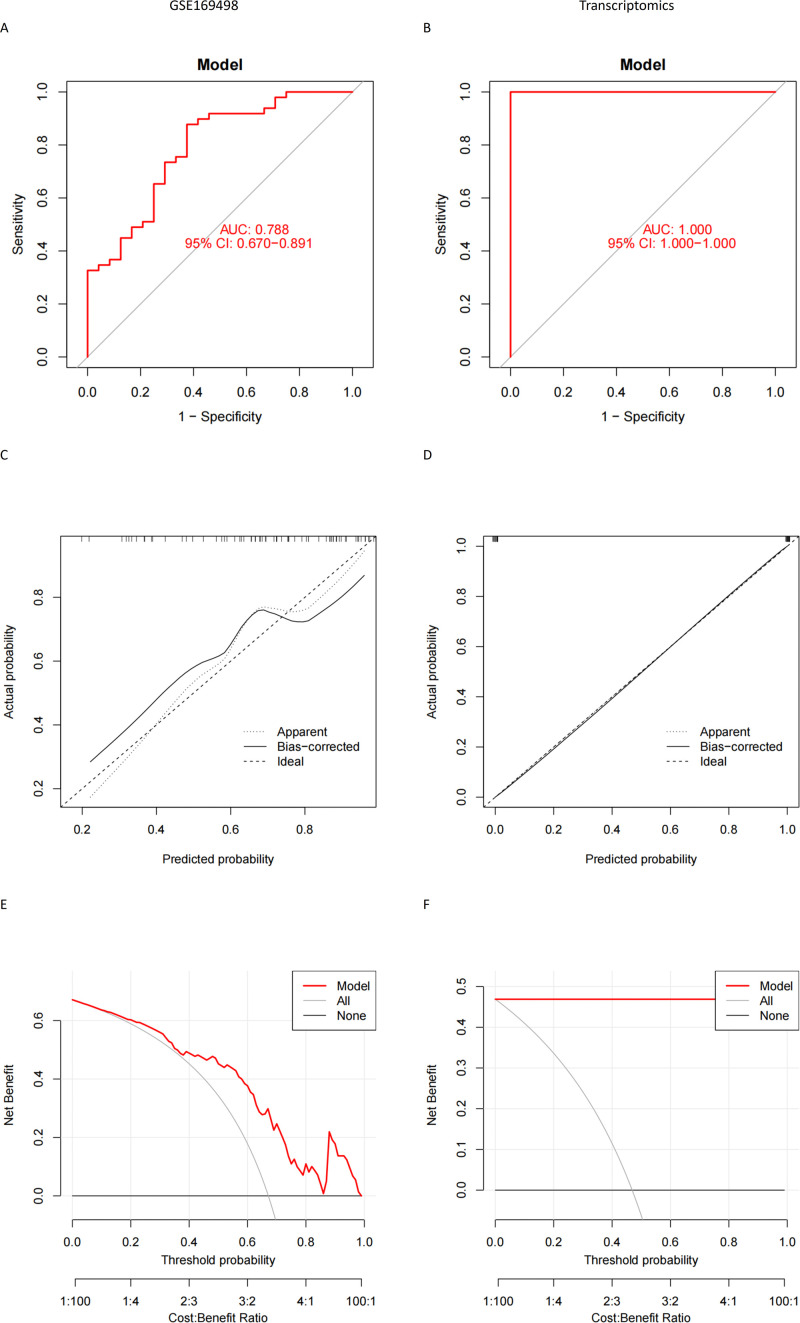
Due to the excellent predictive value of HSPD1 and its paired proteins (TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1), we employed a cohort of 73 pituitary adenoma samples from the GSE169498 dataset to construct an invasiveness prediction model based on the expression profiles of the aforementioned predictor genes (HSPD1, TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1) (Fig. 6C). The constructed prediction model was validated using both the GSE169498 and transcriptomics to ensure its reliability and robustness. Model performance evaluation involved calculating the area under ROC curve, which consistently exceeded 0.7 in both datasets (Fig. 7A, B). Calibration curves were also generated to assess the calibration accuracy of the prediction model, demonstrating a high level of calibration quality for both the GSE169498 and transcriptomics (Fig. 7C, D). In addition, DCA demonstrated excellent clinical decision-making efficiency of nomogram for both datasets (Fig. 7E, F).
Fig. 7.
(A) (B) ROC curves of the invasiveness prediction model in GSE169498 and transcriptomics; (C) (D) Calibration plot of the invasiveness prediction model in GSE169498 and transcriptomics; (E) (F) DCA analysis of invasive prediction model in GSE169498 and transcriptomics.
Potential functions of invasion predictive gene
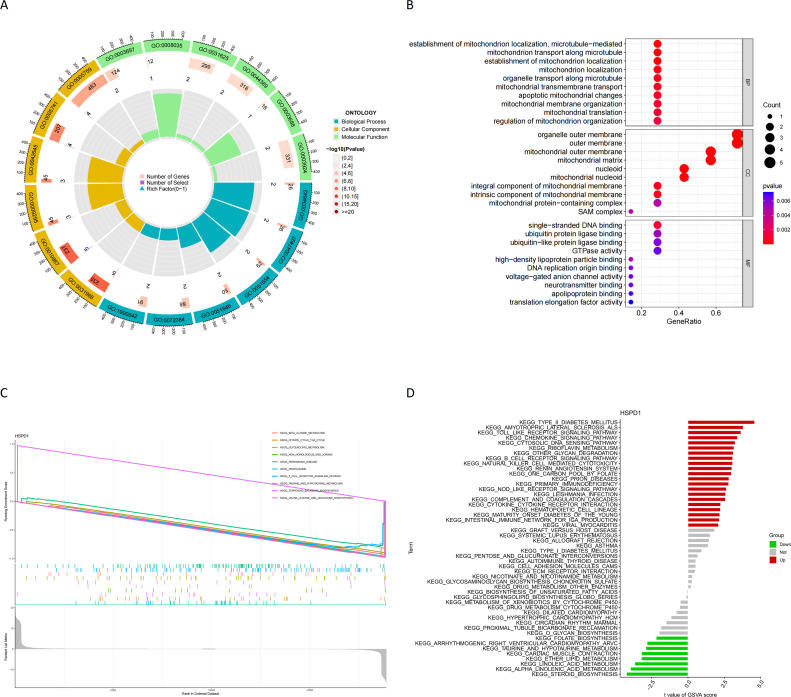
GO enrichment analysis demonstrated the potential functional roles of HSPD1 and its associated proteins. In terms of BP, the analysis revealed their involvement in (establishment of mitochondrion localization, microtubule−mediated), mitochondrion transport along microtubule, establishment of mitochondrion localization, mitochondrion localization. Regarding CC, these proteins were found to be associated with the rganelle outer membrane, outer membrane, mitochondrial outer membrane, mitochondrial matrix. In terms of MF, they exhibited single−stranded DNA binding (Fig. 8A, B). Furthermore, GSEA enrichment analysis in transcriptomics highlighted the significant pathways influenced by the downregulation of HSPD1 expression. These pathways included the beta alanine metabolism, citrate cycle TCA cycle, glycerolipid metabolism, Non homologous end joining, Parkinsons disease, proteasome, T cell receptor signaling pathway, taurine and hypotaurine metabolism, valine leucine and isoleucine biosynthesis (Fig. 8C). Additionally, GSVA enrichment analysis in GSE169498 indicated further pathways affected by the downregulation of HSPD1 expression. These pathways encompassed folate biosynthesis, arrhythmogenic right ventricular cardiomyopathy (ARVC), taurine and hypotaurine metabolism, cardiac muscle contraction, ether lipid metabolism, linoleic acid metabolism, alpha-linolenic acid metabolism, and steroid biosynthesis (Fig. 8D). The results of ssGSEA analysis in transcriptomics showed that there were significant differences, in which most of the immune cells such as CD4 + T cells, CD8 + T cells, dendritic cells, MDSC, and macrophages infiltrated the level in invasive pituitary tumors (Fig. 9A, B). HSPD1 and its paired proteins were significantly correlated with the infiltration of some immune cells, and HSPD1 expression in pituitary adenomas was significantly negatively correlated with Type 1 T helper cell, Myeloid-derived suppressor cells (MDSC), Mast cell, Macrophage, Immature dendritic cell, and Immature B cell contents (Fig. 9C). CIBERSORT analysis showed that HSPD1 expression in pituitary adenomas was significantly negatively correlated with Neutrophils, T cells CD8, T cells gamma delta, NK cells activated, and T cells follicular helper infiltration, and significantly positively correlated with Dendritic cells activated, NK cells activated, and T cells helper (Fig. 9D-J).
Fig. 8.
(A) (B) GO enrichment analysis of invasive predictive genes; (C) GSEA Enrichment analysis of differential genes between high and low HSPD1 expression groups in transcriptomics; (D) GSVA Enrichment analysis of differential genes between high and low HSPD1 expression groups in GSE169498.
Fig. 9.
(A) Heat map of immune cell composition in pituitary adenoma samples from ssGSEA; (B) Differences in immune cell composition in pituitary adenoma samples from ssGSEA; (C) Correlation between HSPD1 expression and immune cell infiltration in pituitary adenoma samples from ssGSEA. (D) Correlation analysis of HSPD1 expression and immune cell infiltration in CIBERSORT; (E-J) Correlation between HSPD1 expression and immune cell infiltration in CIBERSORT.
Inhibition of HSPD1 expression in invasive pituitary adenomas
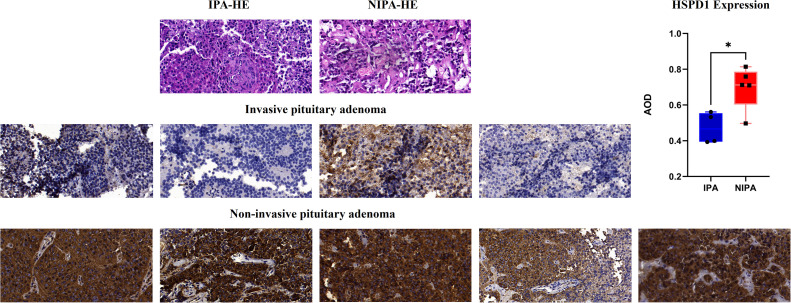
All postoperative pathological tissues were verified as pituitary adenomas by our pathology department (Fig. 10). Immunohistochemical staining was employed to evaluate the expression of HSPD1 in pituitary adenomas. The findings revealed that HSPD1 was mainly expressed in the cytoplasmic fraction of pituitary adenoma cells. In addition, Image J analysis results indicated that the expression average optical density (AOD) value of HSPD1 in IPA was significantly lower than that in NIPA. Specifically, the expression of HSPD1 was significantly higher in non-invasive pituitary adenomas compared to invasive ones, which aligns with the analysis of transcriptomics and proteomics data (Fig. 10). This suggests that inhibition of HSPD1 expression might serve as a predictive marker for the invasive advancement of pituitary adenomas.
Fig. 10.
HE and immunohistochemical staining of invasive and noninvasive pituitary adenomas (SlideViewer 40×).
HSPD1 affects the function of pituitary adenoma cells
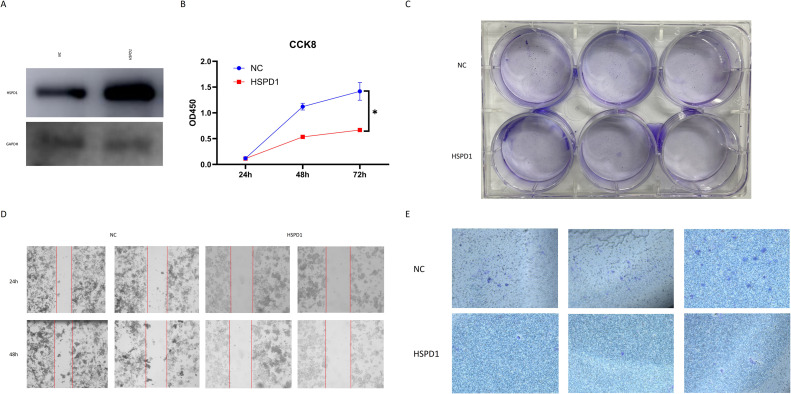
WB results confirmed the overexpression of HSPD1 in GH3 cells after plasmid transfection (Fig. 11A). The results of CCK8 and clonal formation experiments showed that the number of cells in the HSPD1 overexpression group was significantly reduced compared with that in the negative control (NC, Blank plasmid) group, indicating that the proliferation activity of cells was significantly reduced by HSPD1 overexpression (Fig. 11B, C). The results of the scratch test showed that the wound healing in NC group was significantly faster than that in the HSPD1 overexpression group, which indicated that the HSPD1 overexpression weakened the migration ability of pituitary adenoma cells (Fig. 11D). The results of transwell assay showed that the number of transwell chamber cells in NC group was significantly higher than that in HSPD1 overexpression group, indicating that HSPD1 overexpression caused a decrease in the invasive characteristics of pituitary adenomas (Fig. 11E). These functional experiments all showed that HSPD1 overexpression reduced the proliferation, migration and invasion of pituitary adenoma cells, illustrating the diagnostic and therapeutic potential of HSPD1 in invasive pituitary adenomas.
Fig. 11.
(A) HSPD1 expression in pituitary adenoma cells of overexpression group and NC group; (B) CCK8 assay of pituitary adenoma cells in HSPD1 overexpression and NC groups; (C) Colony formation assay of pituitary adenoma cells in HSPD1 overexpression and NC groups; (D) Scratch tests of pituitary adenoma cells in HSPD1 overexpression and NC groups; (E) Transwell assay of pituitary adenoma cells in HSPD1 overexpression and NC groups.
Discussion
Invasive pituitary adenomas have the capability to infiltrate neighboring brain tissue, blood vessels, and bones, making surgical intervention challenging and complete resection often unattainable, regardless of the surgical approach used (transsphenoidal or craniotomy). Postoperative endocrine therapy, radiotherapy, or chemotherapy can offer some benefits to patients; however, disease progression and recurrence pose significant threats to patient safety [16,17]. To address this current dilemma, it is crucial to gain a comprehensive understanding of the mechanisms underlying the invasiveness of pituitary adenomas and identify effective targets for inhibition.
Mitochondria are the main sites of aerobic respiration in eukaryotic cells. Mitophagy, a selective form of autophagy, eliminates dysfunctional or surplus mitochondria through lysosomal degradation pathways, thereby regulating mitochondrial quality control and enabling cells to meet metabolic demands while safeguarding against the detrimental effects of damaged mitochondria [18,19]. Deficiencies in mitophagy have been implicated in various human diseases, including cancer, neurodegenerative diseases, cardiovascular diseases, and liver diseases [20,21]. Depending on the type and stage of the tumor mitochondrial autophagy can exhibit multiple roles. In the early stage of tumors, mitochondrial autophagy can control the mitochondrial population in a healthy and stable state, reduce intracellular oxidative and genotoxic stress, and inhibit tumorigenesis. During tumor progression, however, mitochondrial autophagy is enhanced, reducing oxidative stress within tumor cells and providing circulating substrates required for metabolism. Programmed cellular metabolic rearrangements are manifested by increased rates of tumor cell glycolysis and lactate production even under aerobic conditions to ensure tumor cell survival, proliferation, and maintenance [22,23]. The process greatly enhances the malignancy of tumors. Mitophagy plays a role in facilitating this metabolic switch to a glycolytic phenotype and contributes to tumor progression by regulating metabolic program rearrangements [24]. Mitophagy is strongly associated with drug resistance, representing a key mechanism leading to the failure of apoptotic pathway activation. Inhibition of autophagy has been shown to sensitize cancer cells to the cytotoxicity of anticancer agents, thus influencing treatment outcomes [9,10]. Furthermore, mitophagy can promote the invasive potential and metastatic ability of cancer cells by promoting epithelial-mesenchymal transition (EMT) activation [25]. Whelan et al. found that mitophagy supports the EMT-mediated transformation of low CD44-expressing keratinocytes into high CD44-expressing keratinocytes by regulating oxidative stress and Parkin-dependent mitochondrial clearance [26]. Marín-Hernández et al., in their study, revealed that cancer cells undergo EMT activation and increased invasiveness in hypoxia and hypoglycemia conditions, accompanied by mitophagy activation and impaired mitochondrial function [27]. In conclusion, studying the mechanism of mitochondrial autophagy in aggressive pituitary tumors will help us to deepen our understanding of the disease and provide a more accurate scientific basis for therapeutic strategies.
In this study, we focused on HSPD1, a core gene associated with mitophagy in invasive pituitary adenomas. Genomics and proteomics analysis demonstrated that HSPD1 expression was significantly down-regulated in invasive pituitary tumors compared to non-invasive pituitary tumors. In light of this association, we postulated that HSPD1 may be related to the invasiveness of pituitary adenomas. Six protein molecules (TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1) significantly associated with HSPD1 expression were subsequently selected by co-expression correlation analysis, and a nomogram model associated with the risk of pituitary adenoma invasion was constructed together with HSPD1. The model demonstrated that the risk of pituitary adenoma invasion was inversely correlated with HSPD1 expression. The reliability of the model was then evaluated using ROC curve analysis, calibration curve analysis, and decision curve analysis. Subsequently, we compared the immune microenvironment between the two groups, and the results showed that there were significant differences, in which most of the immune cells such as CD4 + T cells, CD8 + T cells, dendritic cells, MDSC, and macrophages infiltrated the level in invasive pituitary tumors. In addition, we also found that HSPD1 expression was negatively correlated with mast cells, macrophages, immature B cells, immature dendritic cells, and MDSC, so we speculated that HSPD1 down-regulation may promote the infiltration of a variety of immune cells by regulating mitophagy and then increase the invasiveness of pituitary tumors.
Heat shock protein family D member 1 (HSPD1), also known as HSP60, is an ATP-dependent heat shock protein primarily localized in the mitochondrial matrix and expressed in various locations outside mitochondria, such as the outer mitochondrial surface, intracellular vesicles, and nuclei [28,29]. HSPD1 facilitates the degradation of misfolded or denatured proteins, influencing immune system activation, regulating mitophagy, and promoting apoptosis [30]. Downregulation of HSPD1 can result in the accumulation of misfolded proteins, leading to the recruitment of PRKN/Parkin, ubiquitin, and opsin (OPTN) to mitochondria, ultimately triggering mitophagy [31]. Abnormal HSPD1 expression and subcellular localization have been implicated in numerous diseases, including neurodegenerative diseases, cardiovascular diseases, inflammatory diseases, and various cancers [32], [33], [34], [35], [36]. While HSPD1 plays a vital role in cancer development, it has been shown to promote tumor progression, induce apoptosis, and enhance invasiveness in different types of tumors [37]. Beatrice et al. found that HSPD1 is highly expressed in non-small cell lung cancer tissues and associated with poor patient prognosis. It could serve as a potential marker to target mitochondrial metabolism [38]. Wu et al. reported that HSPD1 promotes multiple myeloma cell proliferation by regulating energy rearrangement and protein synthesis [39]. Additionally, high HSPD1 expression is associated with poor prognosis in tumors such as gastric cancer, pancreatic cancer, and head and neck squamous cell carcinoma [40], [41], [42]. However, HSPD1 also inhibits tumor progression. Duan et al. reported that HSPD1 inhibits ovarian cancer cell proliferation via the mitochondrial 3-oxoacyl-ACP synthase (OXSM)-mediated lipoic acid (LA) synthesis pathway [43]. It has also been reported that HSPD1 oxidation mediated by PKCδ-mtROS is able to activate MAPK and prevent cell cycle in hepatocellular carcinoma, thereby inducing cell differentiation and inhibiting cell invasion and migration [44]. Moreover, HSPD1 oxidation disrupts the balance of mitochondrial ROS and triggers mitophagy activation to maintain normal cell morphology. Thus, HSPD1 may influence tumor progression through its impact on mitophagy. Our experiments also verified that HSPD1 was significantly associated with pituitary adenoma cell proliferation, migration and invasion.
The other six protein molecules that constitute the model are all important regulatory molecules for mitochondrial activity. CHCHD3 localizes predominantly to the inner mitochondrial membrane (IM), facing the intermembrane space (IMS), and was originally reported as a substrate for cAMP-dependent protein kinase (PKA) [45]. Darshi et al found that CHCHD3 plays an important role in maintaining mitochondrial structure, cristae morphology and mitochondrial function, and may also be involved in protein import and/or assembly through SAM complexes [46]. CHCHD3 knockdown can severely affect the normal metabolism of cells due to severe defects in mitochondrial morphology. NIPSNAP1 is a modulator of calcium channels and is mainly located in the IMM. Some reports have found that NIPSNAP1 is able to bind to LC3/GABARAP which is involved in the regulation of mitophagy [47]. Abudu et al then found that NIPSNAP1/2 was able to recruit proteins involved in selective autophagy, such as autophagy receptor and ATG8 protein, which are essential proteins for mitophagy [48]. LRPPRC is an autophagy suppressor gene that is mainly located in mitochondria. LRPPRC has been found to regulate the ROS/HIF1-α pathway possibly by affecting autophagy, which in turn plays a role in removing cellular debris and damaging mitochondria [49]. LRPPRC has also been associated with the progression of various tumors such as lung, gastric, colon and breast cancers, and down-regulation of LRPPRC can promote apoptosis and inhibit cell proliferation and invasion in lung adenocarcinoma and lymphoma cells [50,51]. Song et al demonstrated that LRPPRC can regulate retinoblastoma activation, invasion and glycolysis by mediating autophagy inhibition and ROS/HIF1-α cell migration [52]. TUFM, CDAC1, and RHOT1 are also involved in the regulation of mitophagy. Among them, RHOT1 is not only a substrate for PINK1-PRKN-dependent degradation, but may also promote PRKN contact with its potential substrates, thereby promoting mitophagy [53]. The self-antagonistic characteristics of TUFM are able to avoid excessive mitochondrial degradation and thus more finely regulate mitophagy [54].
In this study, we observed significant downregulation of the mitophagy-related gene HSPD1 in invasive pituitary adenomas, suggesting its potential role in influencing the invasiveness of these tumors through modulating mitophagy. Subsequently, we identified six protein molecules significantly associated with both HSPD1 expression and mitophagy and constructed a predictive nomogram model for assessing the invasiveness risk of pituitary adenomas. The model demonstrated good accuracy which was confirmed. Through immune correlation analysis we hypothesize that HSPD1 inhibition may promote immune cell infiltration by regulating mitophagy which in turn increases pituitary adenoma invasiveness. Although the specific mechanism by which mitophagy contributes to the invasive behavior of pituitary adenomas remains unclear, targeting mitophagy holds promise as a therapeutic approach for these tumors.
Conclusion
We have identified the downregulation of HSPD1 as a promising predictive marker for invasive pituitary adenomas, indicating that its suppression may contribute to the progression of pituitary adenomas towards invasiveness. HSPD1 exhibits a significant association with mitochondrial autophagic processes, and its co-expression with TUFM, CHCHD3, NIPSNAP1, VDAC1, LRPPRC and RHOT1 demonstrates a strong predictive value for assessing the invasiveness of pituitary adenomas. These findings suggest that factors involved in the mitophagy process may play a critical role in regulating the progression of pituitary adenomas. The differential expression of these factors may represent a key mechanism underlying the invasive behavior of these tumors. Further investigations are warranted to elucidate the precise mechanism by which the mitophagy process contributes to the invasive progression of pituitary adenomas. Such studies will provide valuable insights for the development of targeted therapeutic interventions.
Ethical approval
This study was approved by the ethics committee of Beijing Tiantan Hospital, Capital Medical University (KY2022-172-03), and patients provided written informed consent to participate in this study.
Availability of data and material
Transcriptomic data will be uploaded to a public database prior to publication of the manuscript. Proteomics has been uploaded to the ProteomeXchange database (PXD039328). GSE169498 is publicly available in the GEO database.
Funding
This work was supported by the Capital clinical characteristic diagnosis and treatment technology research and translational application topic (2-1-1-860-13).
CRediT authorship contribution statement
Yu Zhang: Conceptualization, Data curation, Formal analysis, Methodology, Resources, Writing – original draft, Writing – review & editing. Xin Ma: Data curation, Methodology. Congyu Liu: Data curation, Formal analysis, Writing – review & editing. Zhixu Bie: Data curation, Formal analysis, Methodology. Gemingtian Liu: Data curation, Formal analysis, Writing – review & editing. Pinan Liu: Funding acquisition, Project administration, Supervision, Writing – review & editing. Zhijun Yang: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
There is no conflict of interest to declare in this manuscript.
Acknowledgments
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.101886.
Contributor Information
Pinan Liu, Email: pinanliu@ccmu.edu.cn.
Zhijun Yang, Email: zhijunyang@ccmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Di Ieva A., Rotondo F., Syro L.V., Cusimano M.D., Kovacs K. invasive pituitary adenomas–diagnosis and emerging treatments. Nat. Rev. Endocrinol. 2014;10(7):423–435. doi: 10.1038/nrendo.2014.64. JulEpub 2014 May 13. PMID: 24821329. [DOI] [PubMed] [Google Scholar]
- 2.Delgado-López P.D., Pi-Barrio J., Dueñas-Polo M.T., Pascual-Llorente M., MC G.B. Recurrent non-functioning pituitary adenomas: a review on the new pathological classification, management guidelines and treatment options. Clin. Transl. Oncol. 2018;20(10):1233–1245. doi: 10.1007/s12094-018-1868-6. OctEpub 2018 Apr 5. PMID: 29623588. [DOI] [PubMed] [Google Scholar]
- 3.Nie D., Fang Q., Li B., et al. Research advances on the immune research and prospect of immunotherapy in pituitary adenomas. World J. Surg. Oncol. 2021;19(1):162. doi: 10.1186/s12957-021-02272-9. Published 2021 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosseinkhan N., Honardoost M., Emami Z., Cheraghi S., Hashemi-Madani N., Khamseh M.E. A systematic review of molecular alterations in invasive non-functioning pituitary adenoma. Endocrine. 2022;77(3):500–509. doi: 10.1007/s12020-022-03105-9. SepEpub 2022 Jun 16. PMID: 35711030. [DOI] [PubMed] [Google Scholar]
- 5.Tatsi C., Stratakis C.A. invasive pituitary tumors in the young and elderly. Rev. Endocr. Metab. Disord. 2020;21(2):213–223. doi: 10.1007/s11154-019-09534-8. JunPMID: 31912365. [DOI] [PubMed] [Google Scholar]
- 6.Fleseriu M., Popovic V. The journey in diagnosis and treatment, from pituitary adenoma to invasive pituitary tumors. Rev. Endocr. Metab. Disord. 2020;21(2):201–202. doi: 10.1007/s11154-020-09561-w. JunPMID: 32488740. [DOI] [PubMed] [Google Scholar]
- 7.Onishi M., Yamano K., Sato M., Matsuda N., Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3) doi: 10.15252/embj.2020104705. Feb 1Epub 2021 Jan 13. PMID: 33438778; PMCID: PMC7849173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poole L.P., Macleod K.F. Mitophagy in tumorigenesis and metastasis. Cell Mol. Life Sci. 2021;78(8):3817–3851. doi: 10.1007/s00018-021-03774-1. AprEpub 2021 Feb 13. PMID: 33580835; PMCID: PMC8259496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulikov A.V., Luchkina E.A., Gogvadze V., Zhivotovsky B. Mitophagy: Link to cancer development and therapy. Biochem. Biophys. Res. Commun. 2017;482(3):432–439. doi: 10.1016/j.bbrc.2016.10.088. Jan 15Epub 2017 Feb 3. PMID: 28212727. [DOI] [PubMed] [Google Scholar]
- 10.Vara-Perez M., Felipe-Abrio B., Agostinis P. Mitophagy in cancer: a tale of adaptation. Cells. 2019;8(5):493. doi: 10.3390/cells8050493. May 22PMID: 31121959; PMCID: PMC6562743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praharaj P.P., Panigrahi D.P., Bhol C.S., Patra S., Mishra S.R., Mahapatra K.K., Behera B.P., Singh A., Patil S., Bhutia S.K. Mitochondrial rewiring through mitophagy and mitochondrial biogenesis in cancer stem cells: a potential target for anti-CSC cancer therapy. Cancer Lett. 2021;498:217–228. doi: 10.1016/j.canlet.2020.10.036. Feb 1Epub 2020 Nov 10. PMID: 33186655. [DOI] [PubMed] [Google Scholar]
- 12.Dai K., Radin D.P., Leonardi D. PINK1 depletion sensitizes non-small cell lung cancer to glycolytic inhibitor 3-bromopyruvate: Involvement of ROS and mitophagy. Pharmacol. Rep. 2019;71(6):1184–1189. doi: 10.1016/j.pharep.2019.08.002. DecEpub 2019 Aug 14. PMID: 31669882. [DOI] [PubMed] [Google Scholar]
- 13.Liu C., Wu Z., Wang L., Yang Q., Huang J., Huang J. A mitophagy-related gene signature for subtype identification and prognosis prediction of hepatocellular carcinoma. Int. J. Mol. Sci. 2022;23(20):12123. doi: 10.3390/ijms232012123. Oct 12PMID: 36292980; PMCID: PMC9603050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C., Lu S., Wang X.Z., Wang C.C., Wang L., Liang S.P., Luo T.F., Wang Z.C., Piao M.H., Chi G.F., Ge P.F. FOXO3a protects glioma cells against temozolomide-induced DNA double strand breaks via promotion of BNIP3-mediated mitophagy. Acta Pharmacol. Sin. 2021;42(8):1324–1337. doi: 10.1038/s41401-021-00663-y. AugEpub 2021 Apr 20. PMID: 33879840; PMCID: PMC8285492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T., Xu T., Wang Y., Zhou Y., Yu D., Wang Z., He L., Chen Z., Zhang Y., Davidson D., Dai Y., Hang C., Liu X., Yan C. Cannabidiol inhibits human glioma by induction of lethal mitophagy through activating TRPV4. Autophagy. 2021;17(11):3592–3606. doi: 10.1080/15548627.2021.1885203. NovEpub 2021 Feb 25. PMID: 33629929; PMCID: PMC8632311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raverot G., Burman P., McCormack A., Heaney A., Petersenn S., Popovic V., Trouillas J., Dekkers O.M. European society of endocrinology. European society of endocrinology clinical practice guidelines for the management of invasive pituitary tumours and carcinomas. Eur. J. Endocrinol. 2018;178(1):G1–G24. doi: 10.1530/EJE-17-0796. JanEpub 2017 Oct 18. PMID: 29046323. [DOI] [PubMed] [Google Scholar]
- 17.Dworakowska D., Grossman A.B. invasive and malignant pituitary tumours: state-of-the-art. Endocr. Relat. Cancer. 2018;25(11):R559–R575. doi: 10.1530/ERC-18-0228. Nov 1PMID: 30306782. [DOI] [PubMed] [Google Scholar]
- 18.Tatsuta T., Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27(2):306–314. doi: 10.1038/sj.emboj.7601972. Jan 23PMID: 18216873; PMCID: PMC2234350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgakopoulos N.D., Wells G., Campanella M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 2017;13(2):136–146. doi: 10.1038/nchembio.2287. Jan 19PMID: 28103219. [DOI] [PubMed] [Google Scholar]
- 20.Redmann M., Dodson M., Boyer-Guittaut M., Darley-Usmar V., Zhang J. Mitophagy mechanisms and role in human diseases. Int. J. Biochem. Cell Biol. 2014 Aug;53:127–133. doi: 10.1016/j.biocel.2014.05.010. Epub 2014 May 16. PMID: 24842106; PMCID: PMC4111979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weil R., Laplantine E., Curic S., Génin P. Role of optineurin in the mitochondrial dysfunction: potential implications in neurodegenerative diseases and cancer. Front. Immunol. 2018;9:1243. doi: 10.3389/fimmu.2018.01243. Jun 19PMID: 29971063; PMCID: PMC6018216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. Mar 4PMID: 21376230. [DOI] [PubMed] [Google Scholar]
- 23.Ohshima K., Morii E. Metabolic reprogramming of cancer cells during tumor progression and metastasis. Metabolites. 2021;11(1):28. doi: 10.3390/metabo11010028. Jan 2PMID: 33401771; PMCID: PMC7824065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naik P.P., Birbrair A., Bhutia S.K. Mitophagy-driven metabolic switch reprograms stem cell fate. Cell Mol. Life Sci. 2019;76(1):27–43. doi: 10.1007/s00018-018-2922-9. JanEpub 2018 Sep 28. PMID: 30267101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerra F., Guaragnella N., Arbini A.A., Bucci C., Giannattasio S., Moro L. Mitochondrial dysfunction: a novel potential driver of epithelial-to-mesenchymal transition in cancer. Front. Oncol. 2017;7:295. doi: 10.3389/fonc.2017.00295. Dec 1PMID: 29250487; PMCID: PMC5716985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan K.A., Chandramouleeswaran P.M., Tanaka K., Natsuizaka M., Guha M., Srinivasan S., Darling D.S., Kita Y., Natsugoe S., Winkler J.D., Klein-Szanto A.J., Amaravadi R.K., Avadhani N.G., Rustgi A.K., Nakagawa H. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36(34):4843–4858. doi: 10.1038/onc.2017.102. Aug 24Epub 2017 Apr 17. PMID: 28414310; PMCID: PMC5570661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marín-Hernández Á., Gallardo-Pérez J.C., Hernández-Reséndiz I., Del Mazo-Monsalvo I., Robledo-Cadena D.X., Moreno-Sánchez R., Rodríguez-Enríquez S. Hypoglycemia enhances epithelial-mesenchymal transition and invasiveness, and restrains the warburg phenotype, in hypoxic HeLa cell cultures and microspheroids. J. Cell Physiol. 2017;232(6):1346–1359. doi: 10.1002/jcp.25617. JunEpub 2016 Sep 30. PMID: 27661776. [DOI] [PubMed] [Google Scholar]
- 28.Cheng M.Y., Hartl F.U., Martin J., Pollock R.A., Kalousek F., Neupert W., Hallberg E.M., Hallberg R.L., Horwich A.L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337(6208):620–625. doi: 10.1038/337620a0. Feb 16PMID: 2645524. [DOI] [PubMed] [Google Scholar]
- 29.Dubrez L., Causse S., Borges Bonan N., Dumétier B., Garrido C. Heat-shock proteins: chaperoning DNA repair. Oncogene. 2020;39(3):516–529. doi: 10.1038/s41388-019-1016-y. JanEpub 2019 Sep 20. PMID: 31541194. [DOI] [PubMed] [Google Scholar]
- 30.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013;14(10) doi: 10.1038/nrm3658. Oct630-42Epub 2013 Sep 12. PMID: 24026055; PMCID: PMC4340576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan C., Gong L., Chen L., Xu M., Abou-Hamdan H., Tang M., Désaubry L., Song Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16(3):419–434. doi: 10.1080/15548627.2019.1628520. MarEpub 2019 Jun 16. PMID: 31177901; PMCID: PMC6999623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng R., Liu Z., Tang H., Zhang W., Chen Y., Xu R., Chen L., Song J., Liu X., Deng H. HSP60 silencing promotes Warburg-like phenotypes and switches the mitochondrial function from ATP production to biosynthesis in ccRCC cells. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101218. JunEpub 2019 May 14. PMID: 31112866; PMCID: PMC6526248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusk M.S., Damgaard B., Risom L., Hansen B., Ostergaard E. Hypomyelinating leukodystrophy due to HSPD1 mutations: a new patient. Neuropediatrics. 2016;47(5):332–335. doi: 10.1055/s-0036-1584564. OctEpub 2016 Jul 12. PMID: 27405012. [DOI] [PubMed] [Google Scholar]
- 34.Wachoski-Dark E., Zhao T., Khan A., Shutt T.E., Greenway S.C. Mitochondrial protein homeostasis and cardiomyopathy. Int. J. Mol. Sci. 2022;23(6):3353. doi: 10.3390/ijms23063353. Mar 20PMID: 35328774; PMCID: PMC8953902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enomoto H., Mittal N., Inomata T., Arimura T., Izumi T., Kimura A., Fukuda K., Makino S. Dilated cardiomyopathy-linked heat shock protein family D member 1 mutations cause up-regulation of reactive oxygen species and autophagy through mitochondrial dysfunction. Cardiovasc. Res. 2021;117(4):1118–1131. doi: 10.1093/cvr/cvaa158. Mar 21PMID: 32520982. [DOI] [PubMed] [Google Scholar]
- 36.Parma B., Wurdak H., Ceppi P. Harnessing mitochondrial metabolism and drug resistance in non-small cell lung cancer and beyond by blocking heat-shock proteins. Drug Resist. Updat. 2022;65 doi: 10.1016/j.drup.2022.100888. DecEpub 2022 Oct 28. PMID: 36332495. [DOI] [PubMed] [Google Scholar]
- 37.Yun C.W., Kim H.J., Lim J.H., Lee S.H. Heat shock proteins: agents of cancer development and therapeutic targets in anti-cancer therapy. Cells. 2019;9(1):60. doi: 10.3390/cells9010060. Dec 24PMID: 31878360; PMCID: PMC7017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parma B., Ramesh V., Gollavilli P.N., Siddiqui A., Pinna L., Schwab A., Marschall S., Zhang S., Pilarsky C., Napoli F., Volante M., Urbanczyk S., Mielenz D., Schrøder H.D., Stemmler M., Wurdak H., Ceppi P. Metabolic impairment of non-small cell lung cancers by mitochondrial HSPD1 targeting. J. Exp. Clin. Cancer Res. 2021;40(1):248. doi: 10.1186/s13046-021-02049-8. Aug 7PMID: 34364401; PMCID: PMC8348813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X., Guo J., Chen Y., Liu X., Yang G., Wu Y., Tian Y., Liu N., Yang L., Wei S., Deng H., Chen W. The 60-kDa heat shock protein regulates energy rearrangement and protein synthesis to promote proliferation of multiple myeloma cells. Br. J. Haematol. 2020;190(5):741–752. doi: 10.1111/bjh.16569. SepEpub 2020 Mar 10. PMID: 32155663. [DOI] [PubMed] [Google Scholar]
- 40.Li X.S., Xu Q., Fu X.Y., Luo WS. Heat shock protein 60 overexpression is associated with the progression and prognosis in gastric cancer. PLOS One. 2014;9(9) doi: 10.1371/journal.pone.0107507. Sep 10PMID: 25207654; PMCID: PMC4160299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai Y.P., Yang M.H., Huang C.H., Chang S.Y., Chen P.M., Liu C.J., Teng S.C., Wu K.J. Interaction between HSP60 and beta-catenin promotes metastasis. Carcinogenesis. 2009;30(6):1049–1057. doi: 10.1093/carcin/bgp087. JunEpub 2009 Apr 15. PMID: 19369584. [DOI] [PubMed] [Google Scholar]
- 42.Zhou C., Sun H., Zheng C., Gao J., Fu Q., Hu N., Shao X., Zhou Y., Xiong J., Nie K., Zhou H., Shen L., Fang H., Lyu J. Oncogenic HSP60 regulates mitochondrial oxidative phosphorylation to support Erk1/2 activation during pancreatic cancer cell growth. Cell Death Dis. 2018;9(2):161. doi: 10.1038/s41419-017-0196-z. Feb 7PMID: 29415987; PMCID: PMC5833694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Y., Yu J., Chen M., Lu Q., Ning F., Gan X., Liu H., Ye Y., Lu S., Lash GE. Knockdown of heat shock protein family D member 1 (HSPD1) promotes proliferation and migration of ovarian cancer cells via disrupting the stability of mitochondrial 3-oxoacyl-ACP synthase (OXSM) J. Ovarian. Res. 2023;16(1):81. doi: 10.1186/s13048-023-01156-8. Apr 22PMID: 37087461; PMCID: PMC10122320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandal J.P., Shiue C.N., Chen Y.C., Lee M.C., Yang H.H., Chang H.H., Hu C.T., Liao P.C., Hui L.C., You R.I., Wu W.S. PKCδ mediates mitochondrial ROS generation and oxidation of HSP60 to relieve RKIP inhibition on MAPK pathway for HCC progression. Free Radic. Biol. Med. 2021;163:69–87. doi: 10.1016/j.freeradbiomed.2020.12.003. Feb 1Epub 2020 Dec 8. PMID: 33307168. [DOI] [PubMed] [Google Scholar]
- 45.Xie J., Marusich M.F., Souda P., Whitelegge J., Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581(18):3545–3549. doi: 10.1016/j.febslet.2007.06.052. Jul 24Epub 2007 Jun 27. PMID: 17624330. [DOI] [PubMed] [Google Scholar]
- 46.Darshi M., Mendiola V.L., Mackey M.R., Murphy A.N., Koller A., Perkins G.A., Ellisman M.H., Taylor S.S. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 2011;286(4):2918–2932. doi: 10.1074/jbc.M110.171975. Jan 28Epub 2010 Nov 16. PMID: 21081504; PMCID: PMC3024787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigbolt K.T., Zarei M., Sprenger A., Becker A.C., Diedrich B., Huang X., Eiselein S., Kristensen A.R., Gretzmeier C., Andersen J.S., Zi Z., Dengjel J. Characterization of early autophagy signaling by quantitative phosphoproteomics. Autophagy. 2014;10(2):356–371. doi: 10.4161/auto.26864. FebEpub 2013 Nov 21. PMID: 24275748; PMCID: PMC5396084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Princely Abudu Y., Pankiv S., Mathai B.J., Håkon Lystad A., Bindesbøll C., Brenne H.B., Yoke Wui Ng M., Thiede B., Yamamoto A., Mutugi Nthiga T., Lamark T., Esguerra C.V., Johansen T., Simonsen A. NIPSNAP1 and NIPSNAP2 act as "eat me" signals for mitophagy. Dev. Cell. 2019;49(4):509–525.e12. doi: 10.1016/j.devcel.2019.03.013. May 20Epub 2019 Apr 11. PMID: 30982665. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q., Cai S., Guo L., Zhao G. Propofol induces mitochondrial-associated protein LRPPRC and protects mitochondria against hypoxia in cardiac cells. PLOS One. 2020;15(9) doi: 10.1371/journal.pone.0238857. Sep 8PMID: 32898195; PMCID: PMC7478836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui J., Wang L., Ren X., Zhang Y., Zhang H. LRPPRC: a multifunctional protein involved in energy metabolism and human disease. Front. Physiol. 2019;10:595. doi: 10.3389/fphys.2019.00595. May 24PMID: 31178748; PMCID: PMC6543908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H.Y., Ma Y.D., Zhang Y., Cui J., Wang ZM. Elevated levels of autophagy-related marker ULK1 and mitochondrion-associated autophagy inhibitor LRPPRC are associated with biochemical progression and overall survival after androgen deprivation therapy in patients with metastatic prostate cancer. J. Clin. Pathol. 2017;70(5):383–389. doi: 10.1136/jclinpath-2016-203926. MayEpub 2016 Sep 27. PMID: 27679555. [DOI] [PubMed] [Google Scholar]
- 52.Song K., Li B., Chen Y.Y., Wang H., Liu K.C., Tan W., Zou J. LRPPRC regulates metastasis and glycolysis by modulating autophagy and the ROS/HIF1-α pathway in retinoblastoma. Mol. Ther. Oncolytics. 2021;22:582–591. doi: 10.1016/j.omto.2021.06.009. Jun 24PMID: 34589577; PMCID: PMC8450181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safiulina D., Kuum M., Choubey V., Hickey M.A., Kaasik A. Mitochondrial transport proteins RHOT1 and RHOT2 serve as docking sites for PRKN-mediated mitophagy. Autophagy. 2019;15(5):930–931. doi: 10.1080/15548627.2019.1586260. MayEpub 2019 Mar 4. PMID: 30806158; PMCID: PMC6526870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin J., Chen K., Chen W., Yao Y., Ni S., Ye M., Zhuang G., Hu M., Gao J., Gao C., Liu Y., Yang M., Zhang Z., Zhang X., Huang J., Chen F., Sun L., Zhang X., Yu S., Chen Y., Jiang Y., Wang S., Yang X., Liu K., Zhou H.M., Ji Z., Deng H., Haque M.E., Li J., Mi L.Z., Li Y., Yang Y. Paradoxical mitophagy regulation by PINK1 and TUFm. Mol. Cell. 2020;80(4):607–620.e12. doi: 10.1016/j.molcel.2020.10.007. Nov 19Epub 2020 Oct 27. PMID: 33113344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic data will be uploaded to a public database prior to publication of the manuscript. Proteomics has been uploaded to the ProteomeXchange database (PXD039328). GSE169498 is publicly available in the GEO database.