Abstract
The protein kinase TAOK3, belongs to the MAP kinase family, is one of three closely related members, namely TAOK1, TAOK2, and TAOK3. We performed a pan-cancer investigation of TAOK3 across different cancer types, including uterine carcinosarcoma, adenocarcinoma of the stomach and pancreas, and endometrial carcinoma of the uterus, to better understand TAOK3′s role in cancer. In at least 16 types of cancer, our findings indicate that TAOK3 expression levels differ considerably between normal and tumor tissues. In addition, our study is the first to identify the oncogenic role of TAOK3 locus S331 and S471 in renal clear cell carcinoma, Glioblastoma Multiforme, hepatocellular carcinoma, Lung adenocarcinoma, and Pancreatic adenocarcinoma, indicating their involvement in cancer progression. In addition, our data analysis indicates that copy number variation is the most prevalent form of mutation in the TAOK3 gene, and that there is a negative correlation between TAOK3 mRNA and DNA promoter methylation. Moreover, our analysis suggests that TAOK3 may serve as a prognostic marker for several kinds of cancer, including Colon adenocarcinoma, renal clear cell carcinoma, Lower Grade Glioma, Lung adenocarcinoma, Mesothelioma, and hepatocellular carcinoma. In addition, our research on signature cancer genes has uncovered a positive association between TAOK3 and SMAD2, SMAD4, and RNF168 in most of the malignancies we have examined. TAOK3 is also correlated with the frequency of mutations and microsatellite instability in four types of cancer. Numerous immune-related genes are closely associated with TAOK3 levels in numerous malignancies. TAOK3 expression is positively correlated with immune infiltrates, which include activated CD4 T cells, CD8 T cells, and type 2T helper cells. Our pan-cancer analysis of TAOK3 provides vital insight into its potential role across a variety of cancer types.
Keywords: TAOK3, Oncogene, Survival rate, Copy number variation, Immune cell infiltration
1. Introduction
Mitogen-activated protein kinases (MAPKs) are highly conserved and widely expressed kinases that modulate the activity of their downstream substrates by phosphorylation, resulting in either activation or deactivation. The G protein-coupled receptor (GPCR) serves as a mediator for several extracellular stimuli, including hormones, facilitating signal transduction within the cell. In the context of Saccharomyces cerevisiae, the sterile 20 (STE20) kinase serves as a mediator of G coupling proteins signals to mitogen-activated protein kinases (MAPKs) within the MAPK cascade pathway (Leberer et al., 1992, Ramer and Davis, 1993). TAO kinases have been identified as the ortholog of STE20 in mammals. The initial identification of a kinase, known as TAOK1, was accomplished in rats using a defective STE20 kinase probe to screen a cDNA library (Hutchison et al., 1998). The nomenclature “thousand and one” is derived from the fact that the TAOK1 gene is responsible for encoding a sequence of 1001 amino acids. The identification and characterization of the second TAO kinase were subsequently accomplished by the examination of related sequences with TAOK1 (Hutchison et al., 1998, Chen et al., 1999). The identification of human TAOK3, the third TAO kinase, was achieved by its association with the EPS8 (EGFR kinase substrate 8) protein in an expression library assay (Tassi et al., 1999). TAOKs play a significant role in various cellular signaling cascades, including as the stress-activated MAPK cascade involving p38/MAPK14, the SAPK cascade involving JNK, and the Hippo cascade involving Salvador-Warts-Hippo. TAOKs are also implicated in several cellular interactions that govern the responses to DNA damage, stability of the cytoskeleton, programmed cell death, and other physiological and pathological responses.
The activation of many cellular signalling pathways linked to the development of cancer is regulated by tumor-associated oncogenes (TAOKs). Disruption of kinase signalling can lead to imbalanced expression or activity, which may be associated with the occurrence of malignant transformation. Prior research conducted on cell lines has demonstrated the role of TAOKs in the regulation of the DNA damage checkpoint during the G2/M transition through the activation of p38 MAPK. The researchers observed that the inhibition of TAOKs resulted in reduced p38 activation and compromised the DNA damage response of the G2/M checkpoint (Raman et al., 2007). Furthermore, the downregulation of TAOK1 has been observed to be linked with several mitotic aberrations, leading to the loss of chromosomes in cellular systems (Liskovykh et al., 2019). Furthermore, the involvement of TAOK1 and TAOK2 in the regulation of morphological changes in response to apoptosis has been demonstrated through their mediation of the JNK pathway (Zihni et al., 2006). Another study found that adenocarcinomas of the colon exhibited decreased levels of TAOK3 in comparison to normal colon tissue (Hennig et al., 2012). Furthermore, it has been documented that TAOK3 can enhance resistance to chemotherapy drugs such as paclitaxel, epirubicin, and vinorelbine by modulating the NF-κB signalling pathway in breast cancer cell lines (Lai et al., 2020a, Lai et al., 2020b).
In this work, the role of TAOK3 was systematically studied to evaluate its association with cancer progression and development. The impact of genetic modification and mutation on TAOK3 was also analysed, as was the protein's function in immunotherapy. This article shows TAOK3 as a promising prognostic biomarker for numerous cancer types.
2. Material and methods
2.1. Gene expression analysis
In this study, we utilised data obtained from TAOK3 to examine the “Gene DE” portion of the tumour immune estimate resource, version 2 (TIMER2) web server (http://timer.cistrome.org/) (date access 10 June 2023) (Li et al., 2020). Examining TAOK3 gene expression variations in TCGA datasets between tumor and normal tissues. Furthermore, the verification of the variance in expression between tumour and normal tissues in specific tissues was conducted by the utilisation of the “Expression Analysis-Box Plots” module available on the gene expression profiling interactive analysis, version 2 (GEPIA2) web server (https://gepia2.cancer-pku.cn/ #analysis) (date access 10 June 2023) (Tang et al., 2019).
The UALCAN portal, available at https://ualcan.path.uab.edu/analysis-prot.html, was utilised to perform protein expression analysis on the Clinical Proteomic Tumour Analysis Consortium (CPTAC) dataset in a rigorous and scholarly manner (date access 15 June 2023) (Chandrashekar et al., 2017). The research focused on exploring disparities between primary tumors and healthy tissues regarding TAOK3 phosphoprotein and total protein expression. Additionally, using the UALCAN database, this study examined pathological stages of BRCA, KIRC, PAAD, and UCEC in relation to TAOK3 expression.
2.2. Survival analysis
The analysis involved examining the association between TAOK3 expression and both overall survival and disease-free survival using the Survival Map Module of the Gene Expression Profiling Interactive Analysis 2.0 (GEPIA2.0) database, accessible at http://gepia2.cancer-pku.cn/#index. (date access 20 May 2023).
2.3. Genetic and epigenetic alteration
Genetic alterations in TAOK3 were analyzed using the Cancer Genomics Dataset from the TCGA Pan-Atlas (cBioPortal) (https://cBioPortal.org) (date access 3rd August 2023) (Cerami et al., 2012, Gao et al., 2013). The section pertaining to cancer types provides an outline of the frequency of the mutant gene. Subsequently, we proceeded to ascertain the associations between copy number variations (CNVs), methylation patterns, and mRNA expression levels by utilising the GSCA database. Furthermore, the muTarget database was employed to identify the genes that have altered expression patterns in response to somatic mutations in TAOK3. (Nagy and Győrffy, 2021).
2.4. Gene enrichment analysis
Through the utilization of TIMER.2, we investigated on the correlation between TAOK3 and various cancer hallmark genes. Moreover, by employing the gene similar module in GEPIA2, we determined the top 100 targeted genes that displayed a significant association with TAOK3 across both normal tissues and cancers. We created scatter plots utilizing log2 TPM along with reporting R-value and P-value for these genes. Subsequently, using ShinyGO database (http://bioinformatics.sdstate.edu/go) (date access 20 April 2023), we visualized biological process (BP), cellular component (CC), molecular function (MF) as well as KEGG pathway enrichment analysis for this set of comparable genes procured from GEPIA2 database's top 100 entries.
2.5. Immune infiltration analysis and immune checkpoints
SangerBox, a free online application for analyzing TCGA data, was utilized in the study to measure TMB, MSI, and immune infiltrations as indicators of tumor microenvironment (date access 20 April 2023) (Shen et al., 2022). In the study, Spearman's rank correlation coefficient was utilized to determine the degree of association between 47 ICP genes and TAOK3 expression.
2.6. Statistical analyses
The Wilcoxon test was employed by TIMER to examine differential expression patterns between tumours and normal tissues. The log-rank test was utilised in the Kaplan-Meier plotter to assess the hazard ratio (HR) and log-rank P-value for the purpose of comparing survival curves. Additionally, Spearman's correlation was employed to investigate the association between gene expression levels. Statistical significance was attributed to findings with a probability coefficient below 0.05.
3. Results
3.1. TAOK3 expression level analysis shows oncogenic and tumor suppression roles in different types of cancer
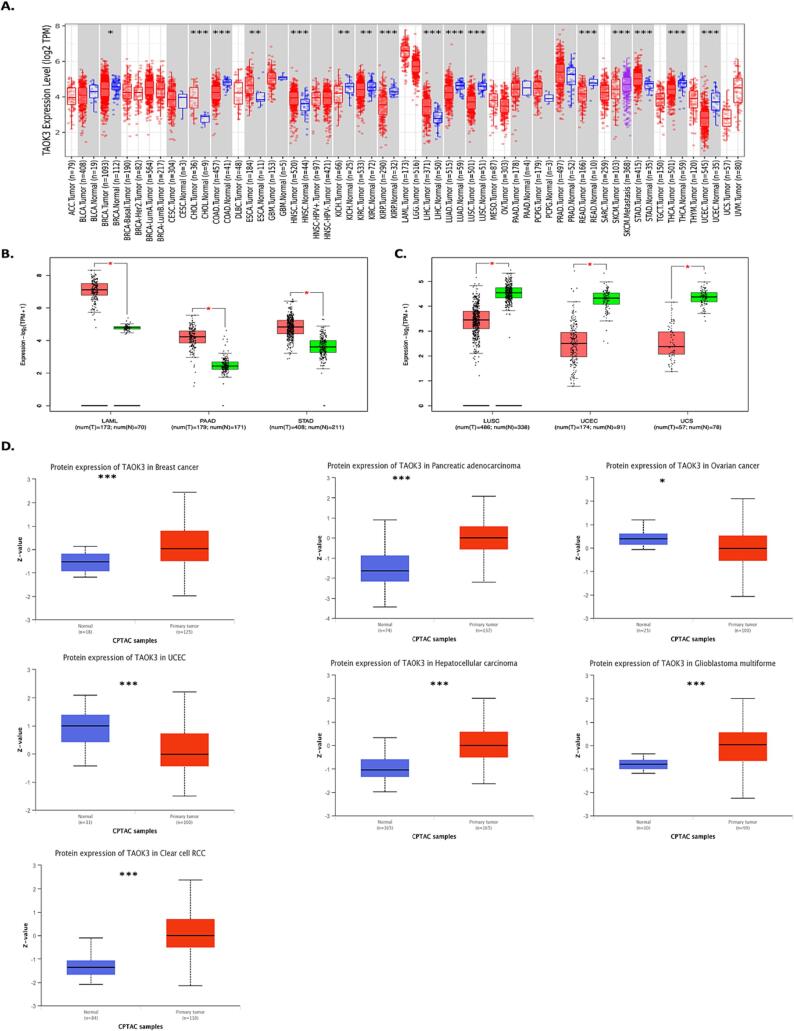
To examine the involvement of TAOK3 in cancer, we conducted an analysis of TAOK3 expression utilising the “Gene_DE” module of the TIMER2.0 web tool. This module enables the comparison of TAOK3 expression levels between tumour tissue and adjacent normal tissues. The result indicates that TAOK3 is downregulated in several types of cancer, such as invasive breast carcinoma (BRCA, p < 0.05), colon adenocarcinoma (COAD), Kidney renal papillary cell carcinoma (KIRP), Lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), Rectum adenocarcinoma (READ), Thyroid carcinoma (THCA), Uterine Corpus Endometrial Carcinoma (UCEC) (all p < 0.001). In addition, Kidney Chromophobe (KIRH) and Kidney renal clear cell carcinoma (KIRC) with p < 0.01 value for both. On the other hand, TAOK3 was upregulated in Cholangiocarcinoma (CHOL), Head and Neck squamous cell carcinoma (HNSC), Liver hepatocellular carcinoma (LIHC), Stomach adenocarcinoma (STAD) (all p < 0.001), and Esophageal carcinoma (ESCA, p < 0.01) (Fig. 1A).
Fig. 1.
mRNA and protein expression levels of TAOK3 across various tumour types and pathological stages. (A) mRNA expression levels of TAOK3 in different tumors vs corresponding controls were analyzed by TIMER 2.0 (http://timer.cistrome.org/database). * P < 0.05; ** P < 0.01; *** P < 0.001. (B)(C) Expression levels of TAOK3 as oncogenic and tumor suppressor gene different type of cancers via GEPIA2 database (http://gepia2.cancer-pku.cn). * P < 0.05; ** P < 0.01; *** P < 0.001. (D) Expression level of TAOK3 total protein between normal tissue and primary tissue of breast cancer, ovarian cancer, Pancreatic adenocarcinoma, Hepatocellular carcinoma, Glioblastoma multiforme clear cell RCC, and UCEC. * P < 0.05; ** P < 0.01; *** P < 0.001.
Using GEPIA.2 tool, we further showed that TAOK3 has oncogenic and tumor suppressor functions. The expression level of TAOK3 in Acute Myeloid Leukaemia (LAML), Pancreatic adenocarcinoma (PAAD), and STAD was higher in tumor tissue than normal, suggesting the oncogenic role of TAOK3 in these types of cancer(Fig. 1B). However, in LUSC, UCEC, and Uterine Carcinosarcoma (UCS), TAOK3 expression was lower (Fig. 1C). In the CPTAC database, the results showed higher expression of TAOK3 in BRAC, PAAD, LIHC, Glioblastoma multiforme (GMB), and KIRC. In addition, the data demonstrated a lower level of TAOK3 in ovarian cancer and UCEC (Fig. 1D).
Furthermore, we investigated the correlation between TAOK3 and pathological stages of cancers such as of BRCA, KIRC, PAAD, and UCEC using UALCAN tool (Supplementary Fig. 1 all p < 0.05) and (Supplementary Table 1).
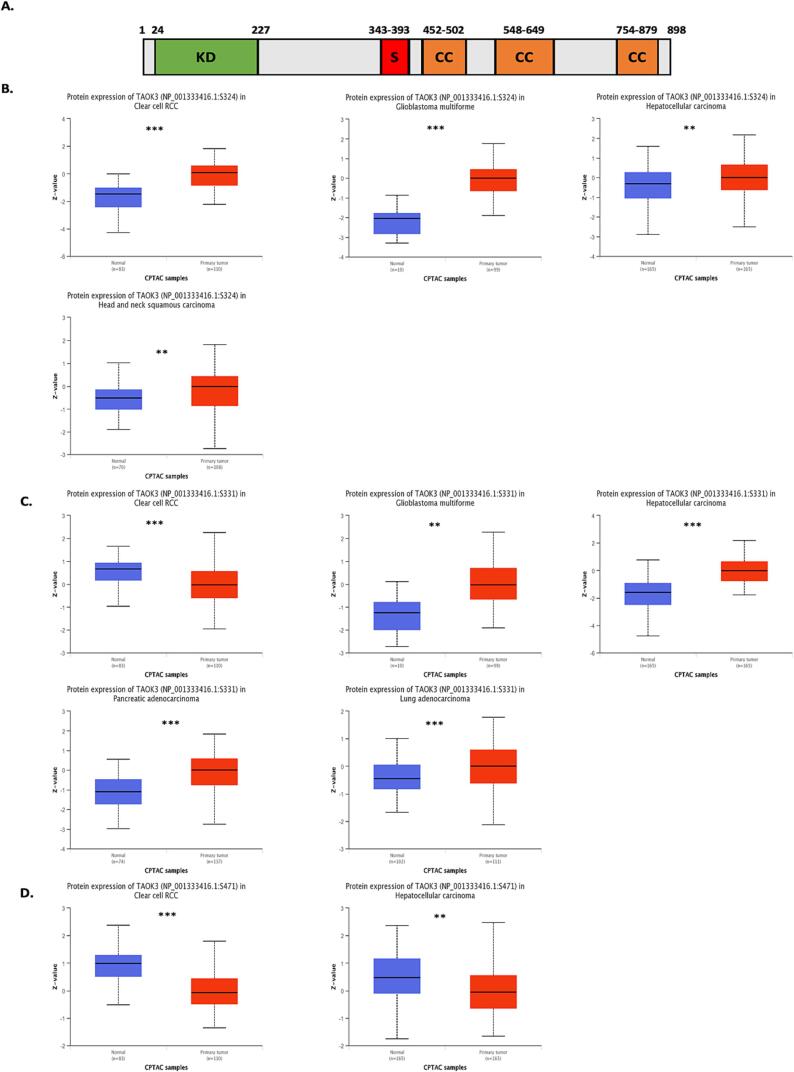
3.2. TAOK3 protein phosphorylation level correlates with cancer development
The phosphorylation level of TAOK3 was analyzed in several types of tumors using the CPTAC database (Fig. 2). A diagram displaying the significant phosphorylation sites of TAOK3, which have been associated with multiple mechanisms, was generated (Fig. 2A). The S324 locus presents a higher expression in tumor tissue versus normal one in KIRC, GMB (both p < 0.001), LIHC, and HNSC (p < 0.01) (Fig. 2B). Another TAOK3 locus is S331 which exhibits lower expression in KIRC tumors than normal (p < 0.001). However, locus 331 shows a higher expression level in GMB (p < 0.01), LIHC, LUAD, and PAAD (all p < 0.001) (Fig. 2C). Interestingly, locus S471 showed downregulation in KIRC and LIHC tumor samples compared to normal, suggesting different roles of the TAOK3 locus in cancers (Fig. 2D).
Fig. 2.
Comprehensive examination of TAOK3 phosphorylation in various cancer types. The UALCAN platform was utilised to analyse the expression level of the TAOK3 phosphoprotein in selected cancers, comparing it between normal tissue and primary tumour, based on data from the CPTAC dataset. (A) Diagram of TAOK3 phosphorylation sites. (B-D) The box plots representing various types of malignancies are presented. (E) The Impact of TAOK3 Phosphorylation on the Pathological Grade of Various Tumours.
In addition, we examined the effect of TAOK3 phosphorylation level on the tumor grade of KIRC (Supplementary Fig. 2) and (Supplementary Table 2). The data showed that locus S324 increased with tumor progression and development. However, locus S331 and S471 were downregulated in tumors compared to normal tissues.
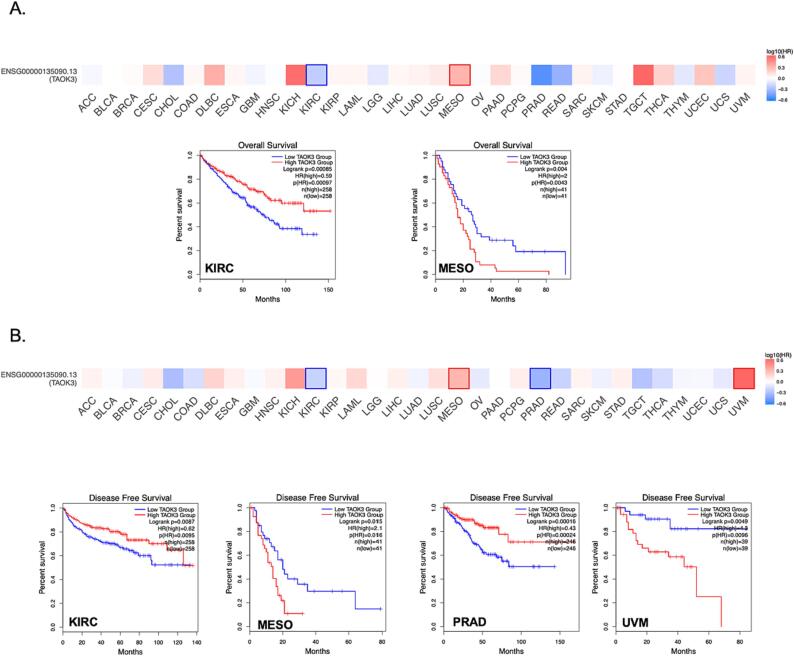
3.3. Survival analysis
To study the relationship between TAOK3 transcriptional level and the cancer patient's outcome, we performed survival analysis using GEPIA.2 tool. Overall survival (OS) heatmap for TAOK3 expression level in 33 types of cancers demonstrated that TAOK3′s higher expression is related to better overall survival in KIRC but not Mesothelioma (MESO) were higher TAOK3 levels correlated with lowered survival percentage (Fig. 3A). In addition, TAOK3 upregulation was correlated with higher disease-free survival (DFS) percentage levels in KIRC and prostate adenocarcinoma (PRAD). On the other hand, MESO and Uveal Melanoma (UVM) indicated that TAOK3′s higher expression correlates with a lower DFS percentage (Fig. 3B).
Fig. 3.
TAOK3 gene expression linked to survival prognosis of cancers in TCGA. TCGA data analysed with GEPIA2 tool was used to perform overall survival (A) and disease-free survival (B) in correlation with TAOK3 gene expression.
3.4. Genetics and epigenetics alteration analysis
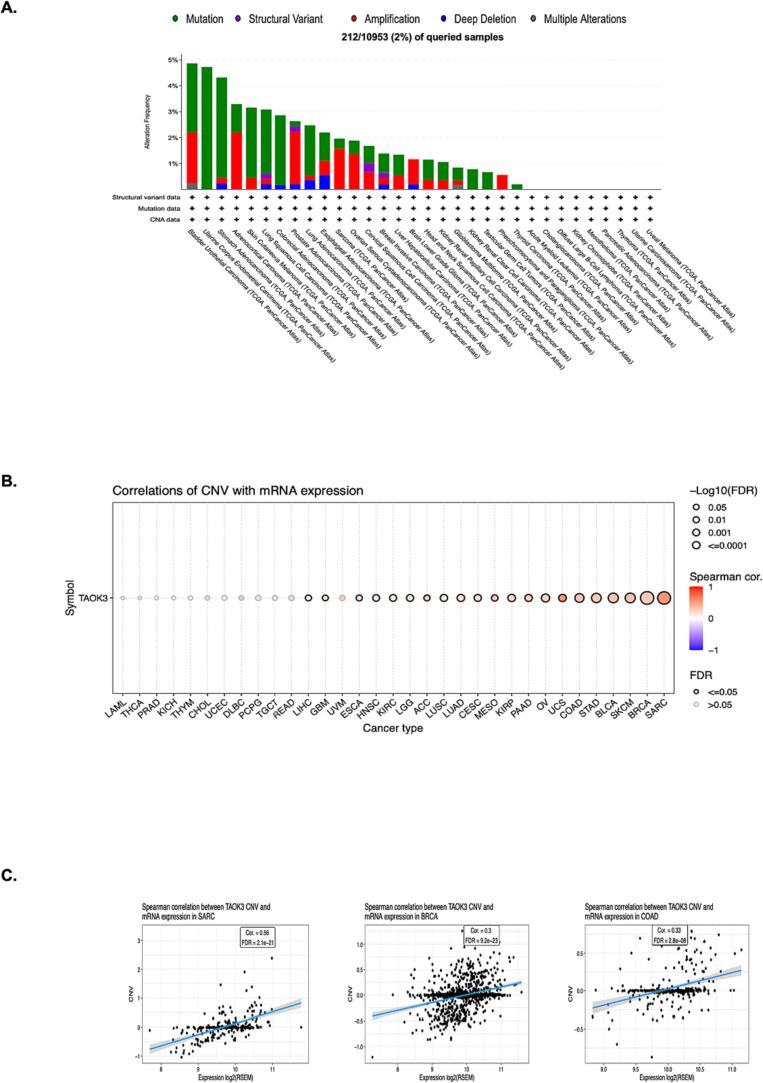
We investigated the genetic modifications of TAOK3 using the TCGA pan-cancer atlas studies, which were accessed through the online resource known as cBioPortal. The data showed 212 mutation samples of TAOK3 across 10,953 patient samples, which equals 2 %. The highest alteration frequency percentage was noted in Bladder Urothelial Carcinoma (BLCA) (4.87 %), with copy number amplification as the major type of genetic variation (1.95 %) (Fig. 4A). We noticed that TAOK3 copy number amplification was standard in most cancers, which led us to investigate further the correlation between copy number amplification and TAOK3 mRNA expression (Fig. 4B). Using GSCA (gene set cancer analysis) tool, we found a positive correlation between TAOK3 and 21 types of cancer, including Sarcoma (SARC), BRAC (both FDR<=0.0001), and COAD (FDR < 0.001) (Fig. 4B and C).
Fig. 4.
TAOK3 mutations have been analysed in a variety of cancers. (A) cBioPortal analysis of TAOK3 mutations in tumours from the TCGA. Structural variation describes a complicated rearrangement of relatively large segments through insertion, inversion, translocation, or other means. In (B) and (C), TAOK3 expression and copy number variation were examined using the GSCA method.
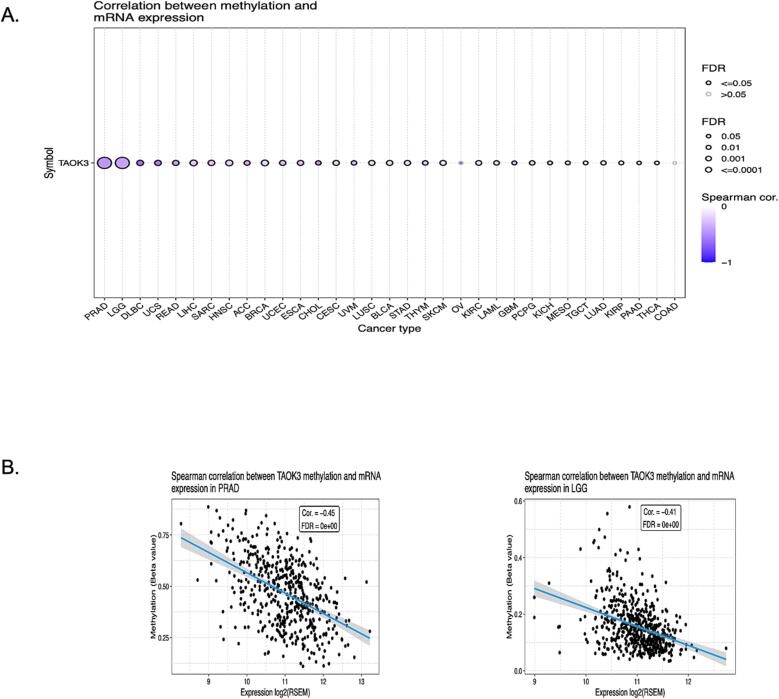
To further investigate the regulation of the TAOK3 transcriptional level, we examined the promoter DNA methylation pattern (Fig. 5). TAOK3 promoter methylation was negatively correlated with 31 types of cancer which might be the reason for the copy number amplification mutation.
Fig. 5.
The epigenetic modification of the TAOK3 gene in various tumour types. (A) Present investigation of the correlation between the expression level of TAOK3 and DNA methylation patterns across a comprehensive set of 33 cancer types. (B) The association between the expression level of TAOK3 and DNA methylation in patients with PRAD and LGG.
3.5. TAOK3 mutation and hallmark genes of cancer analysis
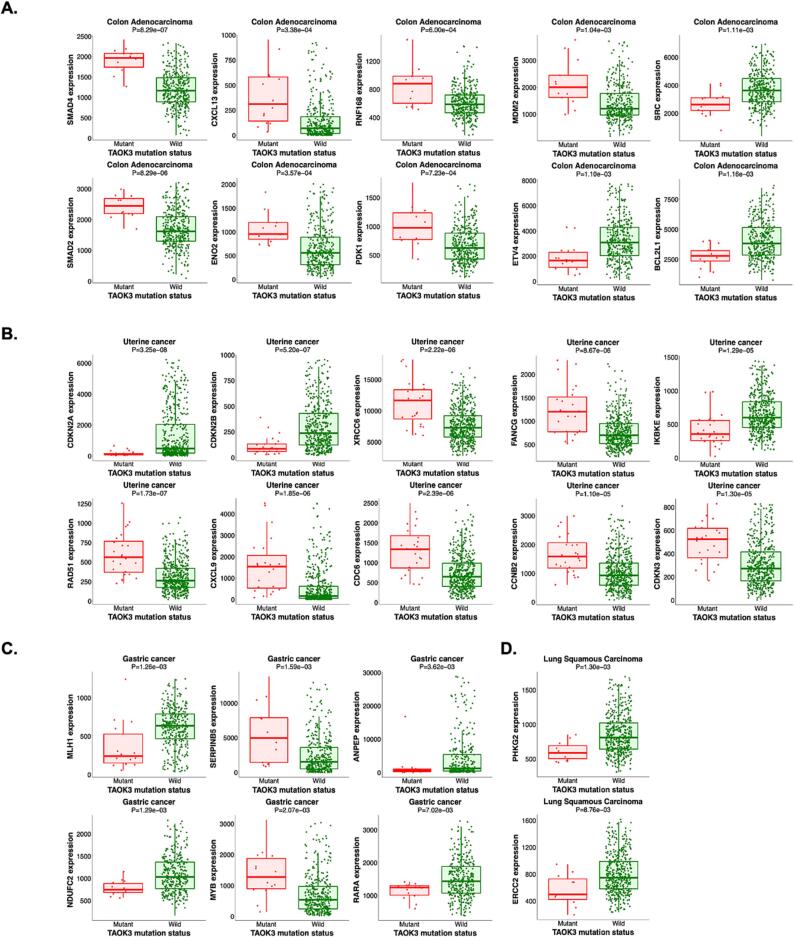
To further explore TAOK3 mutation, we investigated the correlation of TAOK3 mutation with hallmark cancer genes via muTarget platform. We analyzed the effect of TAOK3 mutation on all types of cancer, but we could only obtain data from 4 types of Cancers, including COAD, uterine cancer, gastric cancer, and lung squamous carcinoma (Fig. 6). We found 16 genes to be upregulated in response to TAOK3 mutation, including SMAD4, SMAD2, MDM2, RNF168, CXCL13, ENO2, PDK1, XRCC6, FANCG, RAD51, CXCL9, CDC6, CCNB2, CDKN3, SERPINB5, and MYB. On the other hand, some genes were downregulated following TAOK3 mutation, such as SRC, ETV4, BCL2L1, CDKN2A, CDKN2B, IKBKE, MLH1, NDUFC2, ANPEP, RARA, PHKG2, and ERCC2.
Fig. 6.
TAOK3 mutation impact on Cancer Hallmark genes. Expression level of cancer hallmark genes in colon adenocarcinoma, uterine cancer, gastric cancer, and lung squamous carcinoma.
3.6. Enrichment analysis of TAOK3 and correlated hallmark cancer genes
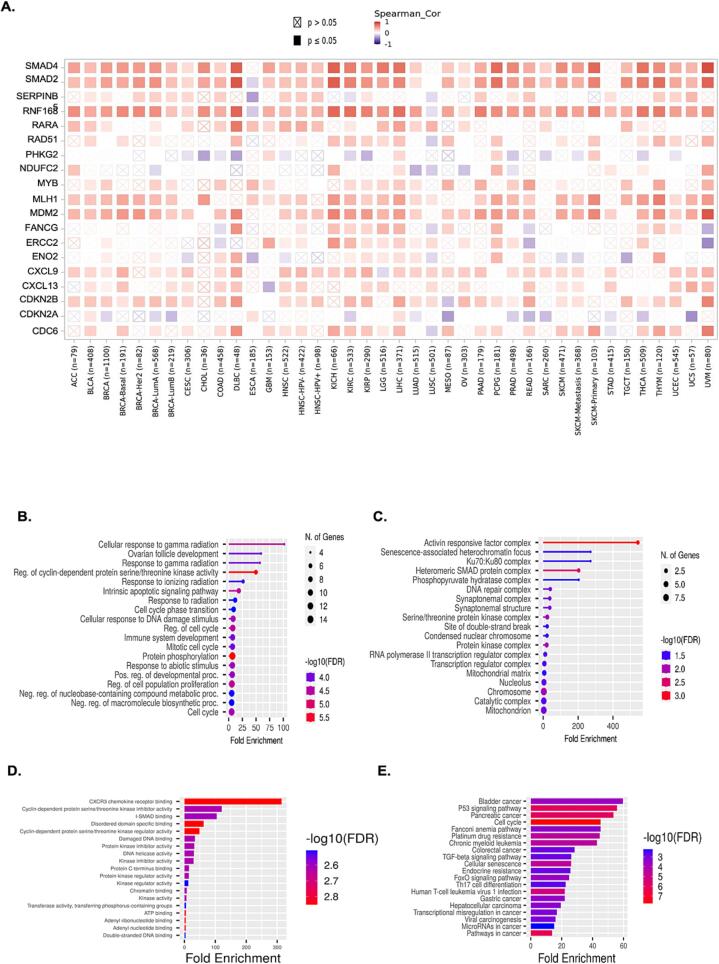
To investigate the correlation of TAOK3 with hallmark cancer genes affected by TAOK3 mutation, we used the TIMER2.0 web tool to identify this correlation. The heatmap showed that SMAD4, SMAD2, and RNF168 correlate with TAOK3 positively in 37 types of cancers (Fig. 7. A). In addition, a gene ontology analysis of the 28 genes that correlated with TAOK3 was conducted, including biological process, cell component, and molecular function (Fig. 7B, C, and D). The data showed that the cellular response to gamma radiation and CXCR3 chemokine receptor binding are the biological and molecular mechanisms that TAOK3 might play a role in cancer progression and treatment. Furthermore, function analysis for these genes has shown that bladder cancer, P53 signaling pathway, Pancreatic cancers, and cell cycle are the top four related mechanisms to TAOK3 and its correlated genes.
Fig. 7.
Conducting an enrichment analysis of the co-expression genes associated with the TAOK3 gene. (A) Heat map illustrating the link between TAOK3 and the cancer signature genes across various cancer types was generated using TIMER 2.0, a publicly accessible database (http://timer.cistr ome.org/database). (B-D) The cancer hallmark genes connected to TAOK3 were subjected to gene ontology analysis using the shining GO tool. (E) KEGG pathway analysis to investigate the association between TAOK3 expression and cancer signature genes.
3.7. Correlation between TAOK3 gene expression and TMB, MSI, and ICP genes
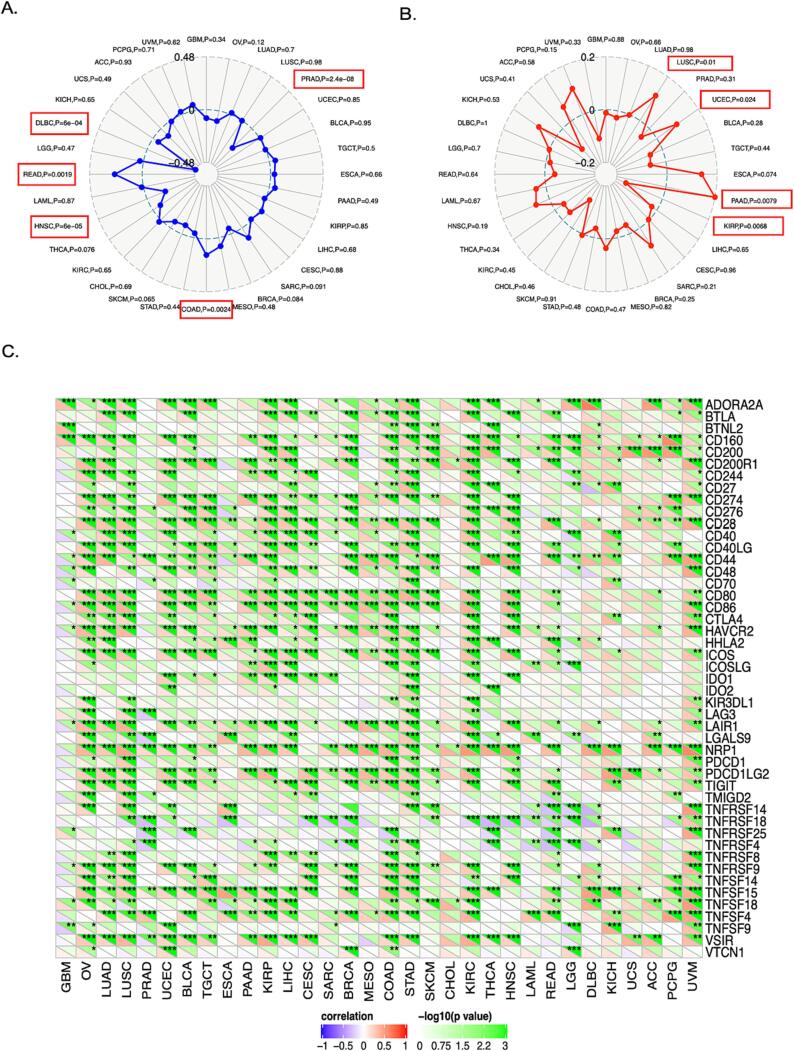
To investigate the effect of TAOK3 on cancer immunotherapy, we explored the gene expression in correlation with Tumour mutational burden (TMB), Microsatellite instability (MSI), and Immune checkpoints (ICP). The MSI data showed that TAOK3 gene expression correlated negatively with PRAD, HNSC, and DLBC but was positively correlated with COAD and READ (Fig. 8A). In addition, TMB data demonstrated that TAOK3 gene expression correlated positively with LUSC, PAAD, and UCEC, while TAOK3 expression was negatively correlated with KIRP (Fig. 8B). Looking at ICP correlation with TAOK3 gene expression, we found that the ICP correlates positively with TAOK3 via almost 35 genes in 32 types of cancers except for such as TNFRSF4, 18, and 25, which correlates negatively.
Fig. 8.
The associations between the expression of TAOK3 and several aspects of immunity, such as immunological marker sets, tumour mutational burden (TMB), and microsatellite instability (MSI), in the context of cancer. (A) The radar map illustrates the association between the expression of TAOK3 and MSI. (B). The radar map illustrates the link between the expression of TAOK3 and the TMB. The black number indicates the range, while the blue and red curve represents the correlation coefficient. The present study investigates the potential correlation between the expression of TAOK3 and sets of immunological markers. The black number indicates the range, whereas the red curve indicates the correlation coefficient. *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.8. Correlation between gene expression and immune cells infiltration
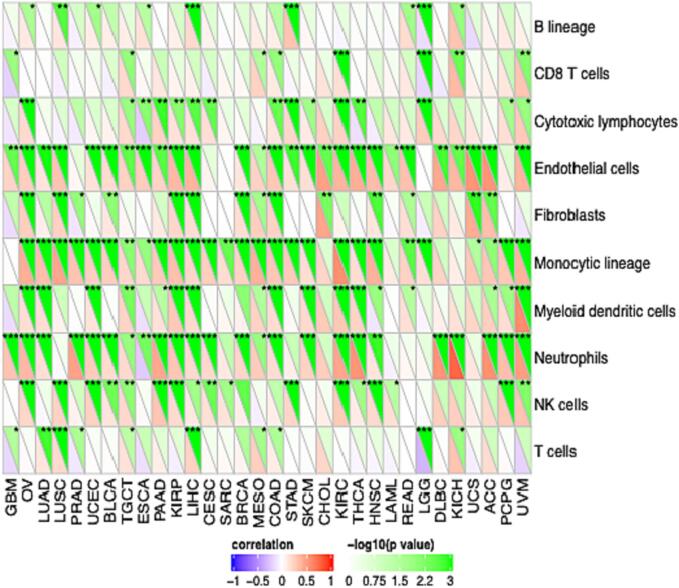
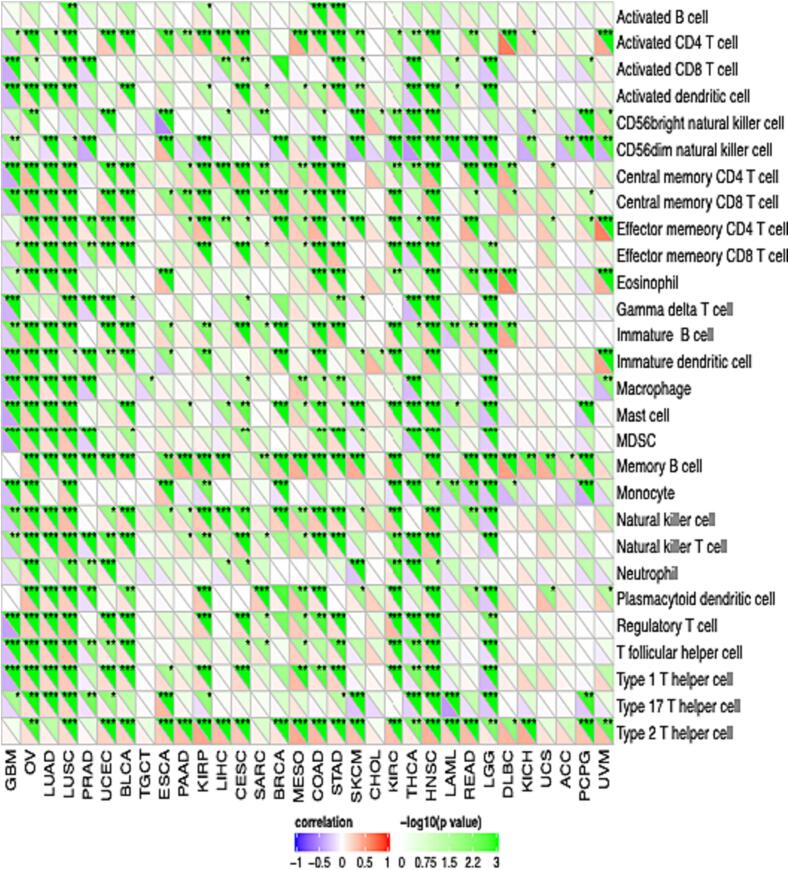
In consequence, we analyzed the immune cell infiltration correlation with TAOK3 expression. Most immune cells correlate positively with TAOK3, especially neutrophils, endothelial cells, and monocytic lineage (Fig. 9). In addition, we looked at the immune pathway that correlates with TAOK3 gene expression. We noticed that TAOK3 correlated positively with type 2T helper cells and memory B cells in almost all cancers. However, it was negatively correlated with CD54dim neutral killer cells and monocyte. In addition, we explored that OV, LUSC, PRAD, UCEC, and BLCA were correlated positively with almost all immune cells, but GMB was negatively correlated (Fig. 10).
Fig. 9.
The correlation between genes expression and immune infiltration levels in pan-cancer. The correlation between immune cells including B cells, T cells, CD8 + T cells, neutrophils, macrophages, dendritic cells and TAOK3 expression through the SangerBox 3.0 tools (http://sangerbox.com/Index).
Fig. 10.
Correlation analysis between abundance of tumor-infiltrating lymphocytes and TAOK3 expression in different cancer types.
4. Discussion
GCKs, also known as Group C kinases, belong to a subclass family of Set 20 kinases. Their primary function is to regulate stress signal transmission, leading to cell cycle arrest and apoptosis. This regulation is achieved through the modulation of two key signalling cascades, namely the JNK and p38 MAPK pathways, as well as the tumor-suppressive Hippo pathway (Kyriakis and Avruch, 2012, Miller et al., 2019). One of the subfamilies of the germinal centre kinase (GCK) family is known as Thousand and One kinase (TAOKs), which encompasses three distinct kinase proteins: TAOK1, TAOK2, and TAOK3. TAOKs play a significant role in various cellular signalling pathways, such as the p38/MAPK14 stress-activated MAPK cascade, the JNK/SAPK cascade, and the Salvador–Warts–Hippo cascade. TAOKs are also implicated in several cellular interactions that govern the responses to DNA damage, integrity of the cytoskeleton, programmed cell death, and other physiological and pathological responses (Miller et al., 2019). This study aimed to investigate the role of TAOK3 in cancer by a comprehensive pan-cancer analysis. Previous research has indicated that TAOK3 exhibits both oncogenic and tumour suppressor characteristics in several studies.
The oncogenic role of TAOK3 in breast cancer was initially identified by an analysis of a Kaplan Meier dataset. This dataset revealed that breast cancer patients who received adjuvant treatment had a worse rate of recurrence-free survival, which was found to be correlated with the high expression level of TAOK3 (Lai et al., 2020a, Lai et al., 2020b). In addition, TAOK3 was found to induce drug resistance to microtubule-targeted drugs. Furthermore, it has been observed that TAOK3 can confer resistance to microtubule-targeted medicines (such as paclitaxel, epirubicin, and vinorelbine) in breast cancer cell lines through the activation of the NF-κB signalling pathway (20). The expression of TAOK3 in pancreatic cancer cells exhibited a positive correlation with the development of cancer stem cell-enriched spheroids. Conversely, the reduction of TAOK3 expression resulted in a decrease in the ability of stem cells to form spheroids and increased the sensitivity of cells to gemcitabine therapy (Bian et al., 2019a, Bian et al., 2019b). Furthermore, it has been determined that TAOK3 functions as a gene that responds to androgens in cells affected by prostate cancer (Romanuik et al., 2009). The analysis demonstrated a correlation between the expression of TAOK3 and the likelihood of recurrence following androgen restriction therapy in individuals with prostate cancer (Bii et al., 2018). In line with these findings, we found that TAOK3 protein expression was increased in BRCA, PAAD, LIHC, and KIRC. Furthermore, this research demonstrated the effect of TAOK3 overexpression on pathological levels, with TAOK3 protein levels increasing with cancer progression in BRCA, KIRC, and PAAD.
However, in colorectal cancer, microarray analysis showed that TAOK3 downregulated in colon adenocarcinoma compared to the normal colon suggesting its role as tumour suppressor (Hennig et al., 2012). On the other hand, TAOK3 mRNA levels were found to be downregulated in COAD, LUSC, UCEC, LUAD, KIRP and UCS cancer tissues compared to their normal tissue counterparts. Taken together, these findings suggest that TAOK3 has both oncogenic and tumour suppressor roles in different cancers.
TAOK3 is a serine/threonine protein kinase that plays a role in several cellular processes, including cell proliferation, differentiation, and apoptosis. TAOK3 can be phosphorylated at multiple sites by various kinases and the specific effects of TAOK3 phosphorylation can vary depending on the site and the context of the phosphorylation. TAOK3 exhibits a dual association with the SAPK/JNK pathway, as demonstrated by its ability to suppress SAPK/JNK activity in COS7 cells through the human epidermal growth factor (Tassi et al., 1999). However, a separate study provided evidence that the overexpression of TAOK3 resulted in the activation of SAPK/JNK in NIH3T3 cells (Zhang et al., 2000). Moreover, the activation of JNK1/2, caspase-9, and PARP showed a negative correlation with the expression of TAOK3 in Hela cells during apoptosis (MacKeigan et al., 2005). Furthermore, the activation of JNK was observed in the mouse brain with defective TAOK3, as compared to the control group (Kapfhamer et al., 2012). Hence, the perplexity in regulating the TAOK3-JNK pathway may arise from variances in cellular environment. However, further research is necessary to examine the correlation between TAOK3 and JNK. Furthermore, the elimination of TAOK1/2/3 in HEK293A cells leads to a substantial reduction in the phosphorylation of YAP/TAZ and their confinement inside the cytoplasm (Plouffe et al., 2016). However, the significance of the TAOK3 locus has not been extensively examined, making our findings the first to investigate it. The level of phosphorylation of TAOK3 at S324 locus is significantly higher in KIRC, GBM, LIHC and HNSC cancer tissues compared to their adjacent normal tissues. Interestingly, the level of TOAK3 phosphorylation at S324 locus significantly increased with the progression of clear cell RCC cancer stages but not the phosphorylation of TAOK3 at S331 and S471 locus, suggesting a pathological function of S324 phosphorylation of TAOK3 in several cancers including clear cell RCC. Functional studies investigating the role of TAOK3 in cancer progression are needed to confirm our findings.
TAOK3 alteration could be involve in cancer progression and development. It has been shown that TAOK3 overexpression is contributing at breast cancer progression through inducing drug-resistance, a process that is regulated through upregulation of NF-KB singaling (Lai et al., 2020a, Lai et al., 2020b). Inhibition of TAOK3 genetically or pharmacologically, reduced the survival of breast cancer cells and increased their sensitivity to different chemotherapies (Lai et al., 2020a, Lai et al., 2020b). Additionally, TAOK3 signaling has been shown to regulate survival of cancer stem cells (Bian et al., 2019a, Bian et al., 2019b). When TAOK3 is absent, there is a decrease in colony formation, expression of stem cell markers, cancer cell invasion and an increased sensitivity to the cytotoxic effects of gemcitabine (Bian et al., 2019a, Bian et al., 2019b, Iizuka et al., 2021). Conversely, when TAOK3 is overexpressed, there is an increase in stem cell characteristics such as tumour initiation and the cancer invasion and metastases (Bian et al., 2019a, Bian et al., 2019b, Iizuka et al., 2021). Ultimately and taking in consideration the potential oncological role of TAOK3 in several cancers, the development of a potent and specific inhibitor for TAOK 3 could offer a successful treatment for a wide range of malignancies and pathogenesis associated with TAOK3. Our study has identify that TAOK3 genetic alterations correlated with the higher expression of other genes in SMAD4, SMAD2, CXCL13, RNF168, MDM2 and ENO2 in COAD and RAD51, CXXL9, CDC6, CCNB2 and CDKN3 in uterine cancer. Most of these genes involve in regulating several signaling pathways including cell cycle progression, TGF-beta signaling pathway and differentiation of T cells. Collectively, these signaling pathways contribute at cancer cell proliferation and cell survival and ultimately the progression of several cancers.
5. Institutional Review Board Statement
Not applicable.
6. Informed Consent Statement
Not applicable.
CRediT authorship contribution statement
Glowi Alasiri: Conceptualization, Software, Validation, Writing – original draft, Writing – review & editing. Bahauddeen Alrfaei: Methodology. Ali M. Alaseem: Writing – review & editing. Osama A. AlKhamees: Methodology. Jehad A. Aldali: Writing – review & editing. Ala M. Aljehani: Methodology. Abdulaziz Alfahed: Writing – review & editing. Mohammad Azhar Aziz: Conceptualization. Ghadir Almuhaini: . Mana M. Alshehri: Conceptualization, Software, Validation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (1209)”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2023.101942.
Contributor Information
Glowi Alasiri, Email: gaalasiri@imamu.edu.sa.
Ali M. Alaseem, Email: amalaseem@imamu.edu.sa.
Osama A. AlKhamees, Email: Oakhamees@imamu.edu.sa.
Jehad A. Aldali, Email: jaaldali@imamu.edu.sa.
Ala M. Aljehani, Email: amaljehani@imamu.edu.sa.
Ghadir Almuhaini, Email: Muhainig@ksau-hs.edu.sa.
Mana M. Alshehri, Email: Mana_Alshehri@dfci.Harvard.edu, Alshehrima3@ngha.med.sa, Mana_Alshehri@dfci.Harvard.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bian Y., Teper Y., Griner L.A.M., et al. Target deconvolution of a multikinase inhibitor with antimetastatic properties identifies TAOK3 as a key contributor to a cancer stem cell-like phenotype. Mol. Cancer Ther. 2019;18:2097–2110. doi: 10.1158/1535-7163.MCT-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y., Teper Y., Mathews Griner L.A., et al. Target deconvolution of a multikinase inhibitor with antimetastatic properties identifies TAOK3 as a key contributor to a cancer stem cell-like phenotype. Mol. Cancer Ther. 2019;18:2097–2110. doi: 10.1158/1535-7163.MCT-18-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bii V.M., Collins C.P., Hocum J.D., et al. Replication-incompetent gammaretroviral and lentiviral vector-based insertional mutagenesis screens identify prostate cancer progression genes. Oncotarget. 2018;9:15451. doi: 10.18632/oncotarget.24503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.Cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hutchison M., Cobb M.H. Isolation of the protein kinase TAO2 and identification of its mitogen-activated protein kinase/extracellular signal-regulated kinase kinase binding domain. J. Biol. Chem. 1999;274:28803–28807. doi: 10.1074/jbc.274.40.28803. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig E.E., Mikula M., Rubel T., et al. Comparative kinome analysis to identify putative colon tumor biomarkers. J. Mol. Med. 2012;90:447–456. doi: 10.1007/s00109-011-0831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison M., Berman K.S., Cobb M.H. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 1998;273:28625–28632. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]
- Iizuka S., Quintavalle M., Navarro J.C., et al. Serine-threonine kinase TAO3-mediated trafficking of endosomes containing the invadopodia scaffold TKS5alpha promotes cancer invasion and tumor growth. Cancer Res. 2021;81:1472–1485. doi: 10.1158/0008-5472.CAN-20-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D., King I., Zou M.E., et al. JNK pathway activation is controlled by Tao/TAOK3 to modulate ethanol sensitivity. PLoS One. 2012;7:e50594. doi: 10.1371/journal.pone.0050594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis J.M., Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol. Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- Lai T.-C., Fang C.-Y., Jan Y.-H., et al. Kinase shRNA screening reveals that TAOK3 enhances microtubule-targeted drug resistance of breast cancer cells via the NF-κB signaling pathway. Cell Commun. Signal. 2020;18:1–14. doi: 10.1186/s12964-020-00600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T.C., Fang C.Y., Jan Y.H., et al. Kinase shRNA screening reveals that TAOK3 enhances microtubule-targeted drug resistance of breast cancer cells via the NF-kappaB signaling pathway. Cell Commun. Signal. 2020;18:164. doi: 10.1186/s12964-020-00600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., et al. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Fu J., Zeng Z., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskovykh M., Goncharov N.V., Petrov N., et al. A novel assay to screen siRNA libraries identifies protein kinases required for chromosome transmission. Genome Res. 2019;29:1719–1732. doi: 10.1101/gr.254276.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKeigan J.P., Murphy L.O., Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- Miller C.J., Lou H.J., Simpson C., et al. Comprehensive profiling of the STE20 kinase family defines features essential for selective substrate targeting and signaling output. PLoS Biol. 2019;17:e2006540. doi: 10.1371/journal.pbio.2006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Á., Győrffy B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer. 2021;148:502–511. doi: 10.1002/ijc.33283. [DOI] [PubMed] [Google Scholar]
- Plouffe S.W., Meng Z., Lin K.C., et al. Characterization of Hippo pathway components by gene inactivation. Mol. Cell. 2016;64:993–1008. doi: 10.1016/j.molcel.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M., Earnest S., Zhang K., et al. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–2014. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer S.W., Davis R.W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanuik T.L., Wang G., Holt R.A., et al. Identification of novel androgen-responsive genes by sequencing of LongSAGE libraries. BMC Genomics. 2009;10:1–19. doi: 10.1186/1471-2164-10-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Song Z., Zhong X., et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1:e36. doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Kang B., Li C., et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi E., Biesova Z., Di Fiore P.P., et al. Human JIK, a novel member of the STE20 kinase family that inhibits JNK and is negatively regulated by epidermal growth factor. J. Biol. Chem. 1999;274:33287–33295. doi: 10.1074/jbc.274.47.33287. [DOI] [PubMed] [Google Scholar]
- Zhang W., Chen T., Wan T., et al. Cloning of DPK, a novel dendritic cell-derived protein kinase activating the ERK1/ERK2 and JNK/SAPK pathways. Biochem. Biophys. Res. Commun. 2000;274:872–879. doi: 10.1006/bbrc.2000.3244. [DOI] [PubMed] [Google Scholar]
- Zihni C., Mitsopoulos C., Tavares I.A., et al. Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1. J. Biol. Chem. 2006;281:7317–7323. doi: 10.1074/jbc.M513769200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.