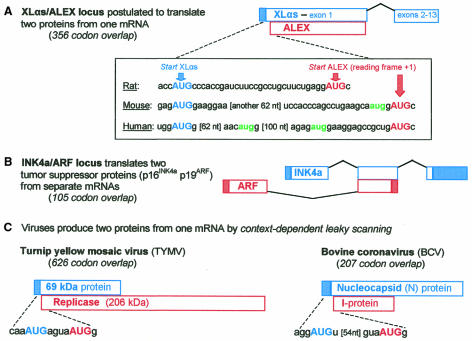
Given the roominess of the mammalian genome, the discovery of a gene that saves space by being translated in two reading frames over a distance of 356 codons is unexpected. Klemke et al. (2001) have found that the rat XLαs gene, previously shown to encode a G protein α subunit, encodes a second protein—dubbed ALEX—in the +1 reading frame. The start codon for ALEX (AUGALEX) is 32 nt downstream from AUGXLαs (Figure 1A). Even among viruses, where overlapping genes are employed to compress the genome to a packageable size, such a long dual-coding domain is rare. The 105 codon overlap in the INK4A/ARF locus is the only other extensive dual-coding sequence identified thus far in mammals (Sharpless and DePinho, 1999). Whereas the two proteins produced from the INK4A/ARF locus are translated from separate mRNAs (Figure 1B), only one mRNA has been found for the XLαs/ALEX locus.
Fig. 1. Dual-coding mRNAs from eukaryotic cells and viruses. Possible mechanisms for translating the cellular mRNAs (A, B) are discussed in the text. In the mouse XLαs/ALEX transcript, translation of XLαs might start upstream from the indicated point; see DDBJ/EMBL/GenBank entry AJ245739. With viral mRNAs (C), leaky scanning allows access to the downstream start codon not only when the first AUG resides in a poor context (TYMV; Weiland and Dreher, 1989) but also, to a limited extent, when the first AUG resides in a good but not perfect context, as in BCV. In the latter case, the low-level translation of I-protein was suppressed when the N-protein start site was changed from aggAUGu to the more optimal aggAUGg (Senanayake and Brian, 1997). Coding (open boxes) and noncoding domains (filled boxes) are not drawn to scale.
Klemke et al. (2001) showed that antibodies against the cDNA-derived ALEX polypeptide detect the corresponding endogenous protein in rat PC12 cells, proving that the second open reading frame (ORF) is expressed. Conservation of overlapping ORFs in the human and murine XLαs genes supports the hypothesis that functional proteins are produced from both reading frames. The most intriguing finding is that XLαs and ALEX proteins appear to interact, as evidenced by cross immunoprecipitation. When a functional assay for ALEX becomes available, it will be interesting to determine whether interaction of XLαs and ALEX as nascent polypeptides confers some advantage. A positive result would suggest a rationale for the unusual gene structure.
What is not clear is how ribosomes gain access to the second initiation site. Comparison with viruses (Figure 1C) suggests that the structure of XLαs/ALEX mRNA in rats might be compatible with a leaky scanning mechanism. The first AUG is in a good but not perfect context (like BCV, it has A in position –3 but lacks G in position +4) and therefore a small fraction of ribosomes might reach AUGALEX by leaky scanning. This explanation is tenable because the observed translation of ALEX was low, as seen by comparing its yield to that of XLαs when the dual-coding mRNA was tested in vitro. The yield of ALEX from the dual-coding mRNA was also much lower than from a shortened transcript that lacks the XLαs start codon.
The postulated structure of XLαs/ALEX mRNA in humans and mice is not compatible with leaky scanning, however. Unlike the rat transcript, the human and mouse mRNAs (Figure 1A) have additional AUG codons between the XLαs and ALEX start sites and these intervening start codons would block access to AUGALEX. Of course, the puzzle of how ALEX gets translated from a downstream position could be solved simply by invoking direct internal initiation, but the operation of that mechanism with cellular mRNAs remains uncertain (Kozak, 2001).
Further speculation about how ALEX gets translated is premature until we are sure that a single mRNA is indeed the normal template for synthesis of both proteins. Translation has not yet been tested with the human and mouse transcripts. In rats, where the dual-coding mRNA supports only low level translation of ALEX, a better translated mRNA might yet be found. Only one mRNA was detected in rats by northern blotting but that assay does not always resolve transcripts with small differences near the 5‘ end.
Comparison of ALEX protein structure as predicted from the cDNAs of rats, humans and mice reveals strong conservation of the C-terminal domain but the predicted N-terminal domains are completely different in sequence and length. In rats, the similarity of proteolytic digestion products between the endogenous and cDNA-derived proteins supports the view that ALEX initiates at the point shown in Figure 1A. The endogenous protein has not yet been analyzed in mice and humans, however, and in those cases translation of ALEX could conceivably initiate within an upstream exon, as occurs with the INK4A/ARF tumor suppressor genes (Figure 1B). It is interesting that the key functional domain of the ARF protein derives from the unique upstream exon rather than from the dual-coding exon (Sharpless and DePinho, 1999), but there is no reason to think that what is true of INK4A/ARF applies to XLαs/ALEX. In any event, future experiments that explore how translation of ALEX is initiated will also confirm the structure of the protein.
Regardless of whether the two proteins turn out to be translated by a novel mechanism from a single mRNA or by the conventional scanning mechanism from separate (yet to be identified) mRNAs, the economical structure of the XLαs/ALEX locus is a remarkable discovery.
REFERENCES
- Klemke M., Kehlenbach, R.H. and Huttner, W.B. (2001) Two overlapping reading frames in a single exon encode interacting proteins—a novel way of gene usage. EMBO J., 20, 3849–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2001) New ways of initiating translation in eukaryotes? Mol. Cell. Biol., 21, 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake S.D. and Brian, D.A. (1997) Bovine coronavirus I-protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res., 48, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho, R.A. (1999) The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev., 9, 22–30. [DOI] [PubMed] [Google Scholar]
- Weiland J.J. and Dreher, T.W. (1989) Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res., 17, 4675–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]