Abstract
The ubiquitin-proteasome proteolytic pathway is pivotal in most biological processes. Despite a great level of information available for the eukaryotic 26S proteasome—the protease responsible for the degradation of ubiquitylated proteins—several structural and functional questions remain unanswered. To gain more insight into the assembly and function of the metazoan 26S proteasome, a two-hybrid-based protein interaction map was generated using 30 Caenorhabditis elegans proteasome subunits. The results recapitulate interactions reported for other organisms and reveal new potential interactions both within the 19S regulatory complex and between the 19S and 20S subcomplexes. Moreover, novel potential proteasome interactors were identified, including an E3 ubiquitin ligase, transcription factors, chaperone proteins and other proteins not yet functionally annotated. By providing a wealth of novel biological hypotheses, this interaction map constitutes a framework for further analysis of the ubiquitin-proteasome pathway in a multicellular organism amenable to both classical genetics and functional genomics.
INTRODUCTION
As the major intracellular proteolytic mechanism in eukaryotic cells, the ubiquitin (Ub)-proteasome pathway plays a crucial role in many biological processes. Cellular proliferation, differentiation and signaling are all highly dependent upon the degradation of key proteins involved in these processes. This degradation is ATP-dependent and involves the progressive addition of activated ubiquitin to substrates by E1 (Ub-activating), E2 (Ub-carrier) and E3 (Ub-ligase) enzymes, followed by their recognition and degradation by the 26S proteasome (reviewed in Ciechanover et al., 2000).
The 26S proteasome is formed by the association of two subcomplexes, a 20S particle that constitutes the proteolytic core and a regulatory 19S particle that caps the 20S at both ends. While the crystal structure and the main enzymatic mechanisms of the 20S proteasome complex have been elucidated (Groll et al., 1997), the overall organization and function of the 19S particle are less defined despite most of the subunits having been identified and, in some cases, assigned specific functions. It appears to be composed of two structurally distinct modules called the base and the lid. The base contains nine different subunits and is thought to interact with the outer α-rings of the 20S proteasome. Six are related but nonetheless distinct ATPases of the ‘AAA’ family that have non-redundant functions based on genetic studies and are collectively involved in substrate unfolding. The yeast lid module contains eight subunits and appears to be evolutionarily related to the Cop9 signalosome and the mediator of translational initiation, eIF3, although it remains unclear whether they have common functions (reviewed in Voges et al., 1999).
Many questions concerning the 26S proteasome’s physiological organization and function remain unresolved. Although speculative, it seems likely that the 26S proteasome is only the core of a more complex and dynamic holoenzyme. In addition, the mechanism(s) of ubiquitylated substrate recognition are not well understood and the assembly process of the 26S proteasome also remains unclear. To address these questions, we performed two-hybrid analyses using 30 known subunits of the Caenorhabditis elegans 26S proteasome to more precisely define the interactions within the complex subunits and to isolate other proteins that can potentially interact with the proteasome.
RESULTS AND DISCUSSION
We identified potential C. elegans orthologs of the known 26S proteasome subunits and related proteins by BlastP using human and yeast proteins as query sequences. All predicted C. elegans proteins showed relatively high homology to their yeast and human counterparts (0.0<E-value<8e–13, Table I). Thirty predicted open reading frames (proteasome ORFs or pORFs) were successfully PCR-amplified from a highly representative worm cDNA library (Figure 1), Gateway cloned into an Entry vector and then transferred into two-hybrid Destination vectors (Hartley et al., 2000; Walhout et al., 2000a,b). This gave rise to constructs encoding fusions to either the activation domain (AD-pORFs) or the DNA binding domain (DB-pORFs) of yeast Gal4p.
Table I. Potential C. elegans proteasome subunits.
| Nomenclature | ORF or protein names | Blast results | ||||
|---|---|---|---|---|---|---|
| Subunit | H. sapiens | S. cerevisiae | C. elegans | H. sapiens – C. elegans | S. cerevisiae – C. elegans | |
| E value | E value | |||||
| 20S α-type | ||||||
| α1 | PSMA6 | Prs2 | C15H11.7 | PAS-1 | 2e–80 | 3e–57 |
| α2 | PSMA2 | Prs4 | D1054.2 | PAS-2 | 6e–83 | 2e–63 |
| α3 | PSMA4 | Prs5 | Y110A7A.14 | PAS-3 | 2e–89 | 7e–66 |
| α4 | PSMA7 | Pre6 | C36B1.4 | PAS-4 | 2e–82 | 2e–66 |
| α5 | PSMA5 | Pup2 | F25H2.9 | PAS-5 | 3e–84 | 4e–63 |
| α6 | PSMA1 | Pre5 | CD4.6 | PAS-6 | 5e–76 | 2e–62 |
| α7 | PSMA3 | Prs1 | ZK945.2 | PAS-7 | 2e–63 | 8e–47 |
| 20S β-type | ||||||
| β1 | PSMB6 | Pre3 | K08D12.1 | PBS-1 | 4e–51 | 2e–44 |
| β2 | PSMB7 | Pup1 | C47B2.4 | PBS-2 | 1e–52 | 7e–54 |
| β3 | PSMB3 | Pup3 | Y38A8.2 | PBS-3 | 2e–59 | 2e–45 |
| β4 | PSMB2 | Pre1 | T20F5.2 | PBS-4 | 2e–34 | 6e–37 |
| β5 | PSMB5 | Pre2 | K05C4.1 | PBS-5 | 3e–51 | 7e–50 |
| β6 | PSMB1 | Prs3 | C02F5.9 | PBS-6 | 2e–44 | 2e–34 |
| β7 | PSMB4 | Pre4 | F39H11.5 | PBS-7 | 1e–33 | 4e–23 |
| 19S ATPase | ||||||
| Rpt1/S7 | PSMC2 | Cim5 | C52E4.4 | RPT-1 | 0.0 | e–173 |
| Rpt2/S4 | PSMC1 | Yta5 | F29G9.5 | RPT-2 | 0.0 | e–169 |
| Rpt3/S6b | PSMC4 | Yta2 | F23F12.6 | RPT-3 | e–171 | e–142 |
| Rpt4/S10b | PSMC6 | Sug2 | F23F1.8 | RPT-4 | 0.0 | e–152 |
| Rpt5/S6a | PSMC3 | Yta1 | F56H1.4 | RPT-5 | 0.0 | e–160 |
| Rpt6/S8 | PSMC5 | Sug1 | Y49E10.1 | RPT-6 | 0.0 | e–173 |
| 19S non-ATPase | ||||||
| Rpn1/S2 | PSMD2 | Hrd2 | T22D1.9 | RPN-1 | e–168 | e–107 |
| Rpn2/S1 | PSMD1 | Sen3 | C23G10.4a | RPN-2 | e–139 | e–111 |
| Rpn3/S3 | PSMD3 | Sun2 | C30C11.2 | RPN-3 | e–93 | 2e–52 |
| Rpn4 | – | Son1 | – | – | – | – |
| Rpn5/p55 | PSMD12 | Nas5 | F10G7.8a | RPN-5 | e–114 | 8e–76 |
| Rpn6/S9 | PSMD11 | Nas4 | F57B9.10a | RPN-6 | e–109 | 8e–74 |
| Rpn7/S10a | PSMD6 | Rpn7 | F49C12.8 | RPN-7 | 2e–98 | 2e–61 |
| Rpn8/S12 | PSMD7 | Nas3 | R12E2.3 | RPN-8 | e–97 | 2e–67 |
| Rpn9/S11 | PSMD13 | Nas7 | T06D8.8 | RPN-9 | 8e–77 | 2e–28 |
| Rpn10/S5a | PSMD4 | Sun1 | B02O5.3 | RPN-10 | e–76 | 3e–35 |
| S5b | PSMD5 | – | – | – | – | – |
| Rpn11/S13 | PSMD14 | Mpr1 | K07D4.3 | RPN-11 | e–128 | e–102 |
| Rpn12/S14 | PSMD8 | Nin1 | ZK20.5 | RPN-12 | 3e–34 | 8e–13 |
| Rpn13 | - | Rpn13 | – | – | – | – |
| p28 | PSMD10 | – | – | – | – | – |
| p27 | PSMD9 | Nas2 | C44B7.1 | – | 8e–20 | e–20 |
aORF could not be cloned.
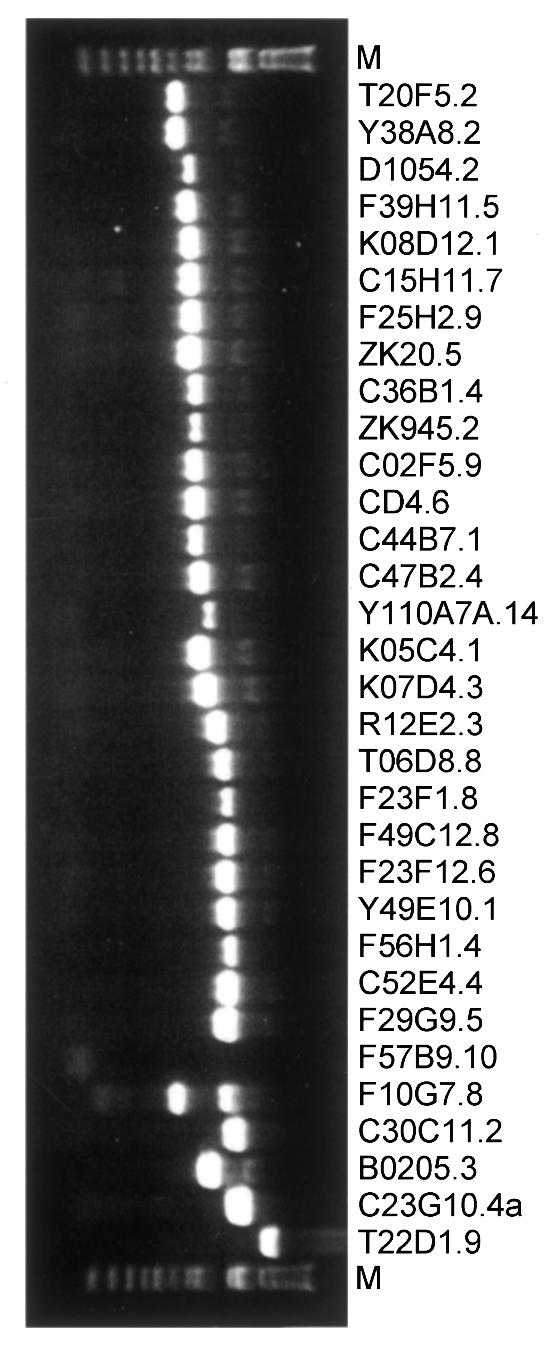
Fig. 1. PCR amplification of pORFs. PCR-amplified ORFs corresponding to potential C. elegans pORFs were analyzed by electrophoresis after organizing them by size in order to facilitate the read-out of the results (Reboul et al., 2001). PCRs were considered successful when a single band of the expected size was observed and accordingly, 30 out of 32 pORFs were successfully PCR amplified and cloned. The PCR product of Y110A7A.14 migrated as a longer fragment than its GeneFinder prediction, however this ORF was confirmed by sequencing (see Reboul et al., 2001 for an explanation). Two pORFs (F10G7.8 and F57B9.10) could not be Gateway cloned due to unsuccessful PCRs.
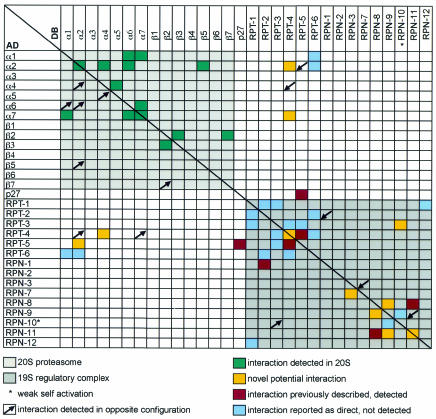
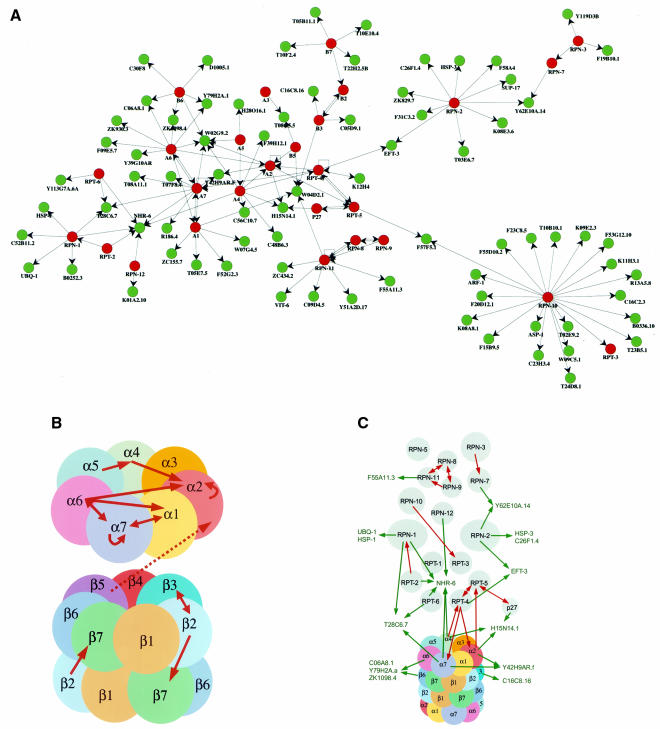
To identify potential interactions within the 26S proteasome and between proteasome subunits and other proteins, we used two complementary approaches of the yeast two-hybrid system. First, a matrix of all DB-pORFs/AD-pORFs pair-wise combinations was performed and 17 interactions were identified with two observed in both DB-X/AD-Y and DB-Y/AD-X configurations (Table II and Figure 2). Second, each of the 30 DB-pORFs was screened against a worm-AD-cDNA library using a high-throughput version of the two-hybrid system that limits the rate of false positives (Walhout et al., 2000b). Among 1254 positive AD-cDNAs recovered, retested and sequence identified, we found a total of 138 different interacting sequences or ‘interaction sequence tags’ (ISTs). The number of ISTs identified for each DB-pORF bait varied between 0 and 22 (see Supplementary data, available at EMBO reports Online). Since several ISTs were identified with more than one bait, they collectively represent 94 different genes. Links to databases (Costanzo et al., 2001; Stein et al., 2001) containing the available functional information for these genes can be found at our ‘C. elegans interactome’ website (http://vidal.dfci.harvard.edu). Sixteen potential interactors encode proteasome subunits, 11 have function(s) already assigned, 62 correspond to uncharacterized ORFs predicted from the C. elegans genome sequencing project and five were not predicted by GeneFinder (Reboul et al., 2001) (Table II). The majority of ISTs can be organized into a potential C. elegans ‘proteasome network’ using the spring layout algorithm from AGD (http://www.mpi-sb.mpg.de/AGD/) (Figure 3A).
Table II. IST results.
| Bait | Prey | ||||
|---|---|---|---|---|---|
| Subunit | Gene name | Gene name | Hits | Ma | Comments |
| α1/PAS-1 | C15H11.7 | ZK945.2 | 108 | + | proteasome subunit α7 [consistent with 20S crystal structure (Groll et al., 1997)] |
| T05E7.5 | 22 | unknown | |||
| W04D2.1 | 3 | putative actin-binding protein | |||
| W07G4.5 | 1 | unknown | |||
| ZC155.7 | 1 | unknown | |||
| F52G2.3 | 1 | unknown | |||
| α2/PAS-2 | D1054.2 | H15N14.1 | 10 | ortholog of yeast protein Sec18, member of the AAA ATPase family | |
| W02G9.2 | 3 | member of the kelch motif family | |||
| D1054.2 | 2 | + | proteasome subunit α2 | ||
| F56H1.4 | 1 | proteasome subunit RPT-5 | |||
| Y42H9AR.f | 1 | strong similarity to D. melanogaster CG7809 and human DKFZP434D156 | |||
| α3/PAS-3 | Y110A7A.14 | T08G5.5 | 1 | unknown, similarity with human TGF-β receptor associated protein -1 (TRAP-1) | |
| α4/PAS-4 | C36B1.4 | C48B6.3 | 12 | unknown | |
| W02G9.2 | 6 | member of the kelch motif family | |||
| F39H12.1 | 4 | unknown | |||
| C48D5.1 | 1 | NHR-6 | |||
| C56C10.7 | 1 | unknown | |||
| H15N14.1 | 1 | ortholog of yeast protein Sec18, member of the AAA ATPase family | |||
| D1054.2 | – | + | proteasome subunit α2 (M. Groll, personal communication) | ||
| F23F1.8 | – | + | proteasome subunit RPT-4 | ||
| α5/PAS-5 | F25H2.9 | C36B1.4 | 8 | + | proteasome subunit α4 [consistent with 20S crystal structure (Groll et al., 1997)] |
| W02G9.2 | 6 | member of the kelch motif family | |||
| C38D4.6 | 2 | PAL-1 | |||
| H28016.1 | 1 | ATP synthetase α subunit, putative ortholog of human ATP5A1 | |||
| α6/PAS-6 | CD4.6 | ZK945.2 | 145 | + | proteasome subunit α7 [consistent with 20S crystal structure (Groll et al., 1997)] |
| F09E5.7 | 32 | unknown | |||
| D1054.2 | 7 | + | proteasome subunit α2 (M. Groll, personal communication) | ||
| Y79H2A.1 | 6 | unknown | |||
| ZK1098.4 | 5 | member of the initiation factor 2 subunit protein family | |||
| Y39G10AR | 3 | no GeneFinder prediction | |||
| W02G9.2 | 2 | member of the kelch motif family | |||
| ZK930.3 | 2 | unknown | |||
| C06A8.1 | 2 | member of the methylenetetrahydrofolate reductase protein family | |||
| C15H11.7 | 1 | proteasome subunit α1 |
aM: interaction detected in the matrix experiment.
Fig. 2. Caenorhabditis elegans proteasome interaction mapping: two-hybrid interactions between 26S proteasome subunits. The data shown here was obtained for all pair-wise combinations by compiling both the matrix (DB-pORF/AD-pORF) and the screens (DB-pORF/AD-cDNAs). See Supplementary data for a detailed description of the comprehensive two-hybrid screens.
Fig. 3. Proteasome subunit/subunit interactions and new potential interactors. Circles and arrows represent proteins and two-hybrid interactions (from DB-X to AD-Y), respectively. (A) Comprehensive two-hybrid protein network of the C. elegans 26S proteasome. Red circles represent proteasome subunits and green circles represent other potential interactors. (B) Two-hybrid interactions between α and β subunits of the 20S. The α and β rings were separated and each subunit colored differently for clarity. (C) A new model for the 26S proteasome. The 19S subunits used in this study were colored in gray and interactions detected between 26S proteasome subunits are shown with red arrows. Novel potential interactors forming clusters with proteasome subunits and proteins previously characterized for which a model of interaction with the proteasome could be envisaged are shown in green.
Similar to other functional genomic approaches, large-scale two-hybrid interaction mapping can generate both false negatives (sensitivity) and false positives (specificity) (Walhout et al., 2000b). The latter issue was of particular concern since proteasome subunits are abundant and a priori, prone to interact with many proteins. To assess this, we took advantage of the fact that the structure of the 20S proteasome is known (Groll et al., 1997). We detected 14 interactions between the 20S proteasome subunits (Figures 2 and 3B). Among them, eight (α1/α7, α4/α5, α6/α7, α7/α1, α7/α6, β2/β3, β3/β2 and β2/β7) were expected since the pairs involved are direct neighbors in the complex. In addition, the 20S structure suggests that α2 is in contact with β5 and that its N-terminal tail interacts with the N-terminal tail of α4 and with an α6 domain (M. Groll, personal communication). Indeed, β5/α2, α4/α2 and α6/α2 interactions were detected in our study (Figures 2 and 3B). Thus, at least 80% (11/14) of the observed interactions between 20S subunits are concordant with the crystal structure of the complex. The remaining three interactions are more difficult to reconcile. However, the α6/α1 interaction suggests that the N-termini of the corresponding partners are in contact, while the detection of two homodimeric interactions (α2/α2 and α7/α7) might be explained by bridging partners known to occur in some two-hybrid interactions. Finally, the average number of potential interactors per bait in the context of the proteasome map is similar to that observed for other interaction projects [4.6 compared with an overall average of ∼5.0 (Walhout et al., 2000b; S. Boulton and M. Vidal, in preparation)]. Altogether, the specificity of the screens performed appears optimal and thus the resulting proteasome interaction map is likely to give rise to reliable hypotheses.
We next examined whether the map provides any information on the organization of the 19S subcomplex and how it assembles with the 20S proteasome. Four direct interactions previously reported (Ferrell et al., 2000; Uetz et al., 2000; Cagney et al., 2001; Hartmann-Petersen et al., 2001) and involving 19S regulatory subunits and the modulator p27 protein were detected [RPT-5/C44B7.1(p27), RPT-5/RPT-4, RPN-8/RPN-11 and RPT-2/RPN-1] (Figures 2 and 3C). In addition, six novel potential interactions (RPT-3/RPN-10, RPT-4/RPT-4, RPN-3/RPN-7, RPN-8/RPN-9, RPN-9/RPN-11 and RPN-11/RPN-11) were observed (Figures 2 and 3C). Although it is known that the 19S/20S association is reversible, ATP dependent and probably regulated by phosphorylation (Voges et al., 1999; Verma et al., 2000), little information is available concerning their contact points (Ferrell et al., 2000; Satoh et al., 2001). Interestingly, we detected four potential interactions between two 19S ATPases and three 20S α-subunits (RPT-4/α2, α2/RPT-5, α4/RPT-4 and RPT-4/α7) (Figures 2 and 3C). Although they score relatively weakly in the two-hybrid system, these interactions represent an important step in understanding how the 20S and 19S particles assemble.
We also expected that our results might shed light on the existence of a putative proteasome holoenzyme and on the mechanism(s) of ubiquitylated substrate recognition. However, because of possible false positives, the two-hybrid interactions between proteasome proteins and other proteins should be viewed cautiously. Therefore, we classified the 78 non-proteasome subunit interactors according to their increasing likelihood of biological relevance. The first class (58/78) comprises those that can interact with a single proteasome subunit and either have been assigned a function outside of the proteasome pathway or remain uncharacterized in any organism (Table II). While it is difficult to formulate hypotheses concerning these interactors, additional functional genomic approaches currently ongoing for C. elegans as well as new biological or biochemical information might eventually reveal any relevant functional association to the proteasome (Sternberg, 2001; Vidal, 2001).
A second class of 12 members was defined with interactors that associate with at least two proteasome subunits while showing no obvious functional link to the Ub-proteasome pathway (excluding PAL-1; see Methods). By linking multiple proteins, these interactors form ‘interaction clusters’ (for example α6/C06A8.1/β6, α6/Y79H2A.a/β6, α6/ZK1098.4/β6, α2/Y42H9AR.f/α7, and RPN-2/Y62E10A.14/RPN-7) (Figure 3C and Table II), which suggests a higher likelihood of biological relevance (Walhout et al., 2000b). Interestingly, two interactions associate with three proteasome subunits: T28C6.7 with α7, RPT-6 and RPN-1, and H15N14.1 with α4, α2 and p27. H15N14.1 is the likely ortholog of Sec18p, a member of the ‘AAA’ ATPase family. Another family member, Cdc48p, has been implicated in the Ub-proteasome pathway (Ghislain et al., 1996). These two facts add credence to H15N14.1 being a genuine proteasome interactor. Strikingly, this class of interactors also includes NHR-6 which is able to bind to six different proteasome subunits (RPN-1, RPT-2, RPT-6, RPN-12, α4 and α7) (Figure 3C). Since NHR-6 is not a spurious two-hybrid interactor (Walhout et al., 2000b), it could represent a new proteasome subunit. However, based on its primary sequence, NHR-6 is predicted to be a nuclear hormone receptor and further analyses will be necessary to understand its functional link with the proteasome.
Proteins in the third class (seven members) have orthologs that can be directly or indirectly linked to the degradation pathway and are thus more likely to generate or consolidate meaningful biological hypotheses. The C. elegans EFT-3 protein interacts with both RPN-2 and RPT-4 and is an ortholog of the mammalian elongation factor EF-1α, which is required for Ub-dependent proteasomal degradation of certain substrates (Gonen et al., 1994). Moreover, its yeast ortholog (Tef1p) has recently been demonstrated to be a proteasome interactor (Verma et al., 2000). EFT-3 thus appears a likely proteasome cofactor and our identifying the potential proteasome subunits involved could prove useful in further assessing its function in proteolysis. F55A11.3 and the chaperone heat shock protein HSP-3 (potential interactors of RPN-11 and RPN-2, respectively) are likely orthologs of the yeast proteins Hrd1p and Kar2p, respectively, which are important for endoplasmic reticulum (ER)-associated protein degradation (Plemper and Wolf, 1999). This process appears to take place on both sides of the ER membrane with proteasomal degradation of aberrant proteins occurring on the cytosolic side. Since Hrd1p contains a RING finger motif localized in the cytosol and has recently been shown to display E3 ligase activity (Bays et al., 2001), it is possible that the RPN-11/F55A11.3 interaction is involved in recruiting the proteasome to the ER membrane for the rapid removal of abnormal ER proteins. Interestingly, other E3s have recently been shown to interact with the proteasome (Xie and Varshavsky, 2000), suggesting that E3s could play an important role in targeting substrates to the proteasome. Another likely interactor is the cytosolic chaperone HSP-1 that was found to interact with the 19S RPN-1 subunit (Figure 3C and Table II). It was recently shown that four different yeast orthologs of HSP-1 co-purify with the yeast 19S regulatory complex (Verma et al., 2000) and chaperones appear necessary for the degradation of many substrates both in yeast and in higher eukaryotes (Ciechanover et al., 2000; McClellan and Frydman, 2001). These observations together with the RPN-1/HSP-1 interaction suggest that cytosolic extra-proteasomal chaperones could play a role not only in the folding/unfolding steps preceding substrate degradation, but also in the targeting of these substrates to the proteasome.
A central question concerning the proteasome is how it recognizes its ubiquitylated substrates. The 19S RPN-10 subunit might be involved in this process, however, since this subunit is dispensable for yeast viability (Van Nocker et al., 1996), it is commonly accepted that other proteasome subunits and/or bridging proteins might be important for substrate recognition. Our interaction map provides information on the possible identity of these other Ub-binding subunits. Importantly, the interaction between UBQ-1 (linear poly-Ub protein) and RPN-1 suggests that the latter is involved in Ub recognition. Furthermore, C16C8.16 and C26F1.4, which interact with β3 and RPN-2, respectively, contain Ub-like domains. Since such domains are present in proteins such as PLIC and BAG, which can serve as functional links between the ubiquitylation machinery and the proteasome (Kleijnen et al., 2000; Lüders et al., 2000), it is possible that these two proteasome subunits are also involved in substrate recognition. In this context, it is noteworthy that based on structure predictions, RPN-1 and RPN-2 possess a domain that could accommodate denatured proteins and perhaps ubiquitylated substrates (Lupas et al., 1997).
The work described here reinforces the usefulness of the two-hybrid strategy to map subunit interactions within multimeric metazoan complexes. The results not only shed light on the intra- and inter-organization of the 19S and 20S sub-complexes and their assembly into the active 26S proteasome, but also identify new potential substrates or cofactors involved in proteasome regulation, function and liaison with the ubiquitin machinery. Moreover, they elaborate how certain proteins already known to play a role in the Ub-proteasome pathway may function. Finally, many of the interactions described here might serve as potential targets for drug discovery efforts aimed at finding novel inhibitors of proteasome functions (Vidal and Endoh, 1999; Wright et al., 2000).
METHODS
Gateway cloning of C. elegans pORFs.
The C. elegans pORF sequences were obtained by BlastP at NCBI (http://www.ncbi .nlm.nih.gov/blast/) or from WormBase (http://www.wormbase.org), PCR amplified from a C. elegans cDNA library (worm-AD-cDNA, Walhout et al., 2000a,b) and recombinationally cloned into the pDONR201 vector (Gateway, Invitrogen). These Entry clones were then transferred into Gateway two-hybrid Destination vectors, pAD-Dest and pDB-Dest, to give DB-pORF and AD-pORF constructs (Hartley et al., 2000; Walhout et al., 2000a,b).
Transformation of DB-pORFs into yeast cells.
The 30 DB-pORFs were transformed into Mav103 yeast cells and transformants were selected on minimal media lacking leucine, then tested for self-activation (Walhout et al., 2000b). All DB-pORFs were re-amplified from individual yeast colonies for sequence verification using an Applied Biosystems (ABI) protocol. All pORF sequence tags (OSTs, Reboul et al., 2001) are available at http://worfdb.dfci.harvard.edu.
Matrix experiment.
Each AD-pORF plasmid was transformed into each DB-pORF-containing haploid Mav103 yeast strain in order to generate a matrix of 900 (30 × 30) DB-pORF/AD-pORF cotransformants. Briefly, transformation-competent yeast cells expressing each DB-pORF were prepared and transformed with each AD-pORF plasmid in U-bottom 96-well plates (QIAGEN) using 20 µl of competent cells per reaction. After transformation, the cells were resuspended in 8 µl of H2O and spotted onto minimal media lacking both leucine and tryptophan to select for cotransformants. After 18 h, the colonies were replica-plated onto selective medium to analyze for the two-hybrid interaction phenotypes as described previously (Walhout et al., 2000b). For each yeast cotransformant exhibiting at least one two-hybrid interaction phenotype, both DB-pORF and AD-pORF inserts were PCR amplified directly from yeast colonies and subsequently sequence verified using an ABI protocol with appropriate primers. These ISTs are available at http://vidal.dfci.harvard.edu.
Large-scale two-hybrid screening.
Competent yeast cells were prepared for each DB-pORF, transformed with 30 µg of worm AD-cDNA library and plated onto minimal selective media lacking leucine, tryptophan and histidine and containing 3-aminotriazole. Positive colonies from each screen were PCR-amplified and re-introduced into fresh yeast cells containing the bait DB-pORF to re-test for two-hybrid interaction phenotypes (Walhout et al., 2000b). Those giving rise to more than one PCR product or that failed to re-test positively were systematically eliminated. The remaining positive AD-cDNA inserts were then sequenced and identified (see below). As a control, the DB-pORF insert was PCR amplified from three random positives of each screen and sequenced for verification. PAL-1 is repeatedly obtained with unrelated proteins (Walhout et al., 2000b) and was disregarded as a likely false positive. We also verified that each predicted ORF sequence encoding a potential interactor was in the same translational frame as the Gal4p-AD-encoding sequence.
Bioinformatic and database analyses.
A local ACeDB database (Durbin and Thierry-Mieg, 1994), called WISTdb (Worm Interaction Sequence Tags database) was built based on a selected subset of the WormBase data and used in conjunction with the Acembly software (http://alpha.crbm.cnrs-mop.fr/acembly/) to align all insert sequences with the C. elegans genome for identification and to verify their translational frames.
Supplementary data.
Supplementary data are available at EMBO reports Online.
Supplementary Material
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ray Deshaies, Stan Fields and Jean Thierry-Mieg for sharing unpublished results, Marcel Dorée and Andrew Goldsborough for reading the manuscript, and Michael Groll for his help in analyzing several interactions within the 20S proteasome. A.D. was supported by a fellowship from the Ligue Nationale Contre le Cancer. This work was supported by an ATIPE program (French CNRS) and grants from Association pour la Recherche sur le Cancer and Fondation pour la Recherche Médicale awarded to O.C., as well as by grants 5R01HG01715-02 (NHGRI), P01CA80111-02 (NCI), 7 R33 CA81658-02 (NCI) and 232 (MGRI) awarded to M.V.
REFERENCES
- Bays N.W., Gardner, R.G., Seelig, L.P., Joazeiro, C.A. and Hampton, R.Y. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biol., 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Cagney G., Uetz, P. and Fields, S. (2001) Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiolog. Genomics, in press. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Orian, A. and Schwartz, A.L. (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays, 97, 2497–2502. [DOI] [PubMed] [Google Scholar]
- Costanzo M.C., et al. (2001) YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res., 29, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. and Thierry-Mieg, J. (1994) The ACeDB genome database. In Suhai, S. (ed.), Computational Methods in Genome Research. Plenum Press, New York, NY.
- Ferrell K., Wilkinson, C.R., Dubiel, W. and Gordon, C. (2000) Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci., 25, 83–88. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Dohmen, R.J., Lévy, F. and Varshavsky, A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J., 15, 4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Gonen H., Smith, C.E., Siegel, N.R., Kahana, C., Merrick, W.C., Chakraburtty, K., Schwartz, A.L. and Ciechanover, A. (1994) Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain Nα-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl Acad. Sci. USA, 91, 7648–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H.D. and Huber, R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Hartley J.L., Temple, F.T. and Brasch, M.A. (2000) DNA cloning using in vitro site-specific recombination. Genome Res., 10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R., Tanaka, K. and Hendil, K.B. (2001) Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys., 386, 89–94. [DOI] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih, A.H., Zhou, P., Kumar, S., Soccio, R.E., Kedersha, N.L., Gill, G. and Howley, P.M. (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- Lüders J., Demand, J. and Höhfeld, J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem., 275, 4613–4617. [DOI] [PubMed] [Google Scholar]
- Lupas A., Baumeister, W. and Hofmann, K. (1997) A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome (anaphase-promoting complex). Trends Biochem. Sci., 22, 195–196. [DOI] [PubMed] [Google Scholar]
- McClellan A.J. and Frydman, J. (2001) Molecular chaperones and the art of recognizing a lost cause. Nature Cell Biol., 3, 51–53. [DOI] [PubMed] [Google Scholar]
- Plemper R.K. and Wolf, D.H. (1999) Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci., 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Reboul J. et al. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- Satoh K., Sasajima, H., Nyoumura, K.I., Yokosawa, H. and Sawada, H. (2001) Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry, 40, 314–319. [DOI] [PubMed] [Google Scholar]
- Stein L., Sternberg, P., Durbin, R., Thierry-Mieg, J. and Spieth, J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P.W. (2001) Working in the post-genomic C. elegans world. Cell, 105, 173–176. [DOI] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Van Nocker S., Sadis, S., Rubin, D.M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D. and Vierstra, R.D. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol., 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Chen, S., Feldman, R., Shieltz, D., Yates, J., Dohmen, J. and Deshaies J.R. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell, 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M. (2001) A biological atlas of functional maps. Cell, 104, 333–339. [DOI] [PubMed] [Google Scholar]
- Vidal M. and Endoh, H. (1999) Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol., 17, 374–381. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl, P. and Baumeister, W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S. and Vidal, M. (2000a) Gateway recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol., 328, 575–592. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N. and Vidal, M. (2000b) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science, 287, 116–122. [DOI] [PubMed] [Google Scholar]
- Wright J., Hillsamer, V.L., Gore-Langton, R.E. and Cheson, B.D. (2000) Clinical trials referral resource. Current clinical trials for the proteasome inhibitor PS-341. Oncology (Huntingt.), 14, 1589–1597. [PubMed] [Google Scholar]
- Xie Y. and Varshavsky, A. (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl Acad. Sci. USA, 97, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.