Abstract
Large-scale, multi-site collaboration is becoming indispensable for a wide range of research and clinical activities in oncology. To facilitate the next generation of advances in cancer biology, precision oncology and the population sciences it will be necessary to develop and implement data management and analytic tools that empower investigators to reliably and objectively detect, characterize and chronicle the phenotypic and genomic changes that occur during the transformation from the benign to cancerous state and throughout the course of disease progression. To facilitate these efforts it is incumbent upon the informatics community to establish the workflows and architectures that automate the aggregation and organization of a growing range and number of clinical data types and modalities ranging from new molecular and laboratory tests to sophisticated diagnostic imaging studies. In an attempt to meet those challenges, leading health care centers across the country are making steep investments to establish enterprise-wide, data warehouses. A significant limitation of many data warehouses, however, is that they are designed to support only alphanumeric information. In contrast to those traditional designs, the system that we have developed supports automated collection and mining of multimodal data including genomics, digital pathology and radiology images. In this paper, our team describes the design, development and implementation of a multi-modal, Clinical & Research Data Warehouse (CRDW) that is tightly integrated with a suite of computational and machine-learning tools to provide actionable insight into the underlying characteristics of the tumor environment that would not be revealed using standard methods and tools. The System features a flexible Extract, Transform and Load (ETL) interface that enables it to adapt to aggregate data originating from different clinical and research sources depending on the specific EHR and other data sources utilized at a given deployment site.
Keywords: Multi-modal clinical research data warehouse, content based retrieval, decision support, machine learning, adaptable extraction, transform and load interface, large-scale multi-site collaboration
Background
Over the past decade, there has been an increased focus on the development of new methods and technologies to efficiently and reliably record, organize, and utilize patient data in complex healthcare settings. Clinical data are increasingly captured in digital form, and the volume and diversity of such data is increasing rapidly. To make best use of the growing range and number of clinical data from new laboratory tests and imaging studies, leading health care centers across the country have been making steep investments to establish enterprise-wide, data warehouses. The primary benefits that can be realized through such efforts include cost savings, efficient tracking of patient outcomes, enhanced decision support at point of care, improved prognostic accuracy and improved clinical trials matching. A significant limitation of typical data warehouse design, however, is that it relies completely upon the use of alphanumeric information to conduct searches and queries with resulting retrievals often highly dependent upon finding exact matches. Recognizing those limitations, our team has begun to design, develop and maintain an Intelligent Search and Retrieval System (IRIS) that exploits the combined use of computational imaging, genomics and data-mining capabilities. In this paper, we report on the design and implementation of IRIS, describe its current capabilities, and future development.
IRIS is designed to feature 2 modes of operation: (1) content-based search and retrieval based on image signatures and genomic profiles to facilitate efficient mining of both large and small clinical and research repositories; and (2) artificial Intelligence-based histo-genomic analysis to provide decision support for classifying subtypes of cancer and predicting disease recurrence. IRIS includes a multi-modal, Clinical & Research Data Warehouse (CRDW) that is operates in conjunction with a suite of computational and machine-learning tools to provide insight into the underlying tumor characteristics that would not be apparent by traditional methods of analysis. The system is designed to conform with all Clinical Data to Health (CD2H) program guidelines 1 and is guided by evolving approaches for establishing Learning Health Systems (LHS) 2 in which cyclical hypothesis generation and evidence evaluation become integral to improving the quality of patient care.
The Design and Key Capabilities of IRIS
Data warehouse and configurable extract, transform and load capabilities
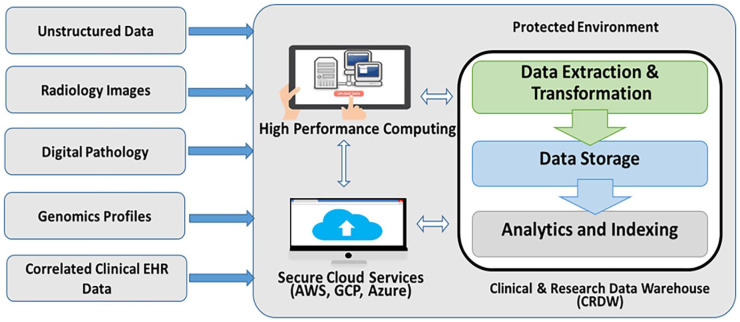
A key requirement for modern clinical and research data warehouse systems is the capacity to reliably aggregate and harmonize information originating from multiple data sources including electronic medical records, clinical trial management systems, tumor registries, biospecimen repositories, radiology and pathology archives, and next generation sequencing services. One of the key distinguishing features of the IRIS is that it is designed with a configurable extract, transform and load (ETL) interface that enables it to adapt to different clinical and research data sources depending on the environmental parameters chosen by the investigative team. See Figure 1. For example, stewards for each institution’s warehouse are free to choose which modalities (digital pathology, radiology, and genomics) that are to be included and which EHR data repositories (eg, Cerner and Epic) to draw from based on the data access guidelines and requirements at each site. Innovative solutions have already been implemented to automate the extraction of unstructured clinical information embedded in paper/text documents, including synoptic pathology reports.
Figure 1.
Warehouse workflow.
Content based image retrieval capabilities
The extremely rich vocabulary and wide range of terms utilized throughout the medical field can often lead to inconsistencies since 2 healthcare providers or investigators may utilize different terms to refer to the same histology, diagnosis or condition. To address this limitation, a significant amount of work was undertaken by the research community over the course of nearly 30 years to develop content-based image retrieval (CBIR) methods that can quickly search through large datasets based on an objective, reproducible feature-based description of the visual content of images to provide diagnostic decision support or to facilitate efficient browsing and identification of digitized specimens exhibiting staining and histological characteristics most similar to a given query, To maximize the utility of IRIS when in the CBIR mode of operation, the user interface for the System is being developed using human-centered design to enable users to iteratively refine queries by clicking on any one of the ranked image retrievals that in-turn are used to initiate subsequent queries using the selected retrieval as the new query input. Inspired by advances in machine learning and high-performance computing, investigators are also revisiting this area of search functionality while adding the capacity of the search algorithms to integrate signatures arising from a plurality of data sources including genomics, digital pathology and/or radiology imaging studies. 3
Once a query image is loaded into IRIS, the System generates a feature vector and forwards it to the server for processing, to automatically locate and retrieve ranked sets of digitized pathology specimens and correlated molecular studies of cases from within the Warehouse that exhibit spectral and spatial profiles that most closely match that of the query. We have made sustained efforts to design, develop and optimize algorithms and methods that can quickly and reliably search through reference libraries of cases that have had their diagnosis of record and histologic type independently confirmed, to automatically identify and retrieve previously analyzed lesions which exhibit the most similar characteristics to a given query case to assist in clinical decisions and to conduct systematic comparisons of tumors within and across patient populations. One of the advantages of the CBIR approach over purely alphanumeric search strategies is it enables investigators to systematically review the data while visualizing the most relevant digitized pathology specimens. Furthermore, by utilizing a standard computational imaging toolset to both generate indices and search for matches it is possible to reduce inter- and intra-observer inconsistencies in searches and improve the objectivity with which large image repositories are interrogated.
Artificial intelligence-based analysis capabilities
The AI mode of operation for IRIS is implemented through the development of a module that uses information originating from different data types to enable decision support. IRIS utilizes digital pathology to facilitate interpretation of genomic data within the histopathologic context of disease onset and/or progression. This is accomplished by leveraging recent advances in computational imaging, machine learning and genomics that make it possible to assess combinations of clinical and pathologic data points, simultaneously. To achieve these capabilities IRIS implements workflows that perform automated detection of nuclei and delineation of tumor regions throughout the imaged specimens while generating image-based morphological features within the specimen.
As in many other similar applications that utilize machine learning and/or artificial Intelligence a significant challenge for our team in optimizing the decision support algorithms arises primarily due to limitations in assembling a sufficiently large data set to adequately train and test these methods. Other challenges that we are confronting include batch effects that are introduced during preparation of specimens and discordance among pathologists regarding the annotation of digitized specimens. Despite these drawbacks, recent studies show potential as to how AI can improve the decision-making process during cancer diagnosis while saving resources, improving reliability, and reducing patient discomfort. One of the important findings that was reported using computational technologies was recently published by New York University, USA. In this study, Coudray et al trained a large number of high-definition digitally imaged pathology glass slide specimen images (also known as Virtual Slides) using a deep learning algorithm called InceptionV3 for histopathological classification (lung cancer (adenocarcinoma and squamous cell carcinoma) and normal lung). 4 The results revealed a very high accuracy with 0.97 AUC for tissue classification. Both frozen and formalin-fixed paraffin-embedded sections were available for analysis as specimens. Furthermore, using the developed AI analysis system, 6 gene mutations, STK11, EGFR, FAT1, SETBP1, KRAS, and TP53, could be accurately predicted from the pathological images (AUC: 0.733-0.856). These results suggest that the analysis of pathological virtual slide images using AI and computational methods can enable accurate classification of lung cancer tissues and prediction of genetic mutations.
Current Applications of IRIS
IRIS is currently fully functional as an automated multi-modal repository equipped with an automated ETL interface, but expansion and optimization of the System continues. To date, we have: (1) established a large and growing repository of digitized pathology specimens; (2) extracted computational features and established linkages with national tumor registry data; (3) developed software, technologies and quantitative tools based on deep-learning to support diagnostic classifications; and (4) developed data management tools that enable investigators to reliably search through large repositories to automatically retrieve targeted digitized pathology and correlated clinical data. In the next section, we describe the applications and capabilities supported by the current implementation.
IRIS data warehouse applications
The current instance of the CRDW at Rutgers enables physicians to systematically review the molecular, genomic, image-based, and correlated clinical information of patient tumors individually or as part of large cohorts to identify changes and patterns that may influence treatment decisions and potential outcomes.5,6 The CRDW core system has also yielded several peer-reviewed publications and funded grants including a collaboration with Stony Brook, Emory University and University of Kentucky to enhance the cancer registry data in Georgia, Kentucky, New Jersey and New York, with machine-learning based classifications and quantitative pathomics feature sets. Currently, the collection of cases and correlated pathology specimens from the collaboration are focused primarily on prostate cancer, lymphoma, NSCLC, melanoma, breast cancer and colorectal cancer. 7 As part of this effort, a collection of deep learning pipelines was developed and deployed to participating sites to glean salient Pathomics features from whole slide tissue images. These pipelines implement deep learning models (1) to predict distribution of tumor infiltrating lymphocytes in a tissue specimen (TIL analysis), (2) to detect and segment tumor regions (Tumor segmentation), and (3) to segment nuclei (Nucleus segmentation). The TIL analysis pipeline was trained with manually annotated image patches extracted from whole slide images from multiple cancer types. It partitions a whole slide image into image patches of 50 × 50 square microns and predicts if a patch is TIL positive, that is, the patch contains lymphocytes. The Tumor segmentation pipeline contains deep learning models for several cancer types; each model was trained with manually annotated patches extracted whole slide images from the corresponding cancer type. The current pipeline has established models for breast, lung, prostate, and pancreatic cancers. It partitions a whole slide image into patches of 88 × 88 square microns and predicts if a patch is a tumor patch, that is, the patch is in or intersects with a tumor region. The nucleus segmentation pipeline implements an instance segmentation model which was trained with an innovative training process consisting of both real and synthetic segmentation training data. In the collaborative project, the pipelines have been used to process more than 5000 whole slide images from breast, lung, and prostate cancer cases curated from SEER datasets. The imaging features from the analysis pipelines and the source whole slide images are managed in instances of a software platform, called QuIP, deployed at each collaborating site.
The CRDW has also facilitated a collaboration with the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC) at the U.S. Department of Veterans Affairs to develop and test algorithms and workflows to automate the analysis of lung adenocarcinoma. Those studies showed that combining computational nuclear signatures with traditional WHO criteria through the use of deep convolutional neural networks (CNNs) led to improved discrimination among tumor growth patterns. 8 In those studies, our team began to explore the use of nuclear signatures as a means for discriminating among different tumor growth patterns. The 2015 World Health Organization (WHO) lung cancer classification is based on histologic architectural patterns and is the current standard for rendering diagnosis and determining prognosis for patients afflicted with adenocarcinoma. However the WHO schema does not incorporate a measure of the nuclear grade exhibited in the specimen into the criteria, which may contribute additional prognostic value. Our team showed that well-differentiated tumors (lepidic pattern) tend to exhibit low nuclear grade, whereas poorly-differentiated tumors (micropapillary, solid patterns) tend to exhibit high nuclear grade. Moderately-differentiated tumors (acinar, papillary patterns) demonstrated intermediate grade nuclei with acinar pattern exhibiting the broadest distribution of nuclear grade. In those experiments we utilized transfer learning based on pre-trained state-of-the-art convolutional neural network (CNN) models to achieve multi-subtype classifications of tumor growth patterns including low-grade (LG), intermediate-grade (IG), high-grade (HG).
As a part of the precision medicine initiatives that are underway at our collaborating healthcare centers at Rutgers, Boston VA and Stony Brook, new clinical decision support algorithms, methods and strategies are being developed and tested for their capacity to improve diagnostic and prognostic accuracy and therapy planning for the care of our patients. Guided by the leadership of the learning health system (LHS) workgroup at Rutgers, the CRDW has been leveraged for a wide range of cancer and non-cancer efforts and initiatives. For example, some of the projects already underway include: (1) Comparative effectiveness of different cancer screening outreach strategies; (2) Identification of COVID-19 patient population susceptible to relapse after treatment with Paxlovid; (3) Identification of high risk patient for opioid overdose or relapse; and (4) Improving capacity to identify those patients presenting to the emergency department seemingly unrelated urgent care who are most likely to eventually receive diagnosis of cancer to ultimately develop an intervention to streamline navigation and treatment.
Artificial intelligence-Based analysis applications
In a set of parallel efforts, our team has been investigating the potential of utilizing a combination of genomic and computational imaging signatures to characterize prostate cancer and survival models to study their correlations in prostate cancer. The results of the study show that integrating image biomarkers from CNN with a recurrence network model, called long short-term memory (LSTM) and genomic pathway scores, is more strongly correlated with disease recurrence than using standard clinical markers and image-based features. 9 In addition, 5 survival models were assessed in the context of other prostate clinical prognostic factors, including primary and secondary Gleason patterns, prostate-specific antigen levels, age, and clinical tumor stages. The highest hazard ratio for predicting prostate cancer recurrence was based on Cox regression using an elastic net penalty. Based on knowledge gained from these studies, the team built a unified system using whole-slide histology images and corresponding genomic data through deep neural networks to identify the most salient computational biomarkers. The experimental results showed that the computational biomarkers extracted by this approach resulted in a hazard ratio of 5.73 and C-index of 0.74, which were higher than those using standard clinical prognostic factors and other image-based features. Given the clinical impact that predicting disease recurrence in patients with Gleason score 7 prostate cancer can have on treatment and care, continues to optimize the workflow to fully investigate the potential use of integrated histo-genomic signatures for these purposes. Once optimized, we plan to integrate these capabilities into the decision support module of IRIS.
Alignment and analytical pipelines
Most recently, in an effort to improve user experience and reproducibility, we developed a shell script to orchestrate alignment and analysis pipelines in Linux containers. This script requires the following arguments: directory of tumor detection outputs, directory of TIL detection outputs, a comma-separated values (CSV) table with the survival information for each case, and the path to an output directory. The alignment and survival analyses are implemented in R and are contained in a Linux container with R version 4.2.1. The shell script downloads this versioned Linux container and runs it with either Apptainer/Singularity or Docker, depending on which is available on the host machine. All code is version controlled with Git and is available publicly on GitHub. The shell script is available at https://github.com/SBU-BMI/tumor-til-survival-analysis and the alignment and analysis pipelines are available at https://github.com/SBU-BMI/til_align .
The following analyses are implemented by the above container. Lymphocyte and tumor detection pipelines generate predictions at patch sizes that are optimized for each task. Subsequent calculation of lymphocyte invasion into tumor-containing patches requires rescaling and alignment. Once overlaid, the lymphocyte and tumor probability heatmaps are thresholded based on the thresholds derived from individual training (Inception Lymphocyte: 0.1, ResNet Cancer: 0.5). For each Whole Slide Image, we then calculate what percentage of predicted tumor-containing patches are also predicted to be lymphocyte-containing patches (Percent Invasion). For descriptive statistics of invasion, Percent Invasion is applied directly. For downstream survival analysis, a patient’s Percent Invasion is scaled by the standard deviation of Percent Invasion within the dataset (Scaled Invasion). This calculation is the same as previously reported. 10
Following alignment, we implement a standardized analytical pipeline to interrogate tumor-TIL invasion characteristics. Patients are classified as TIL-high or TIL-low by binning around the mean invasion value for the dataset. We then generate survival analyses for both categorical invasion (TIL-High vs TIL-Low) and the impact of continuous lymphocyte invasion (Scaled Invasion). This allows us to quickly understand the correlation of lymphocyte invasion with patient outcomes in a stable way across a multitude of cohorts. These same pipelines will also be made available as part of the IRIS toolset.
Conclusions and Future Work
Key innovations in IRIS can be summarized as follows: (1) algorithms, tools and analytic pipelines that enable investigators to systematically interrogate large-scale repositories based on computational imaging signatures and genomic markers to facilitate clinical assessment and translational research—this capability enables systematic examination of relationships and correlations among the morphologic and genomic characteristics of cancers and clinical patient outcomes in large and diverse datasets; (2) a scalable system that supports automated aggregation and indexing of multi-modal data including genomics and digital pathology images; (3) a novel, multi-stage, hierarchical searching algorithm that enables fast content-based image retrieval and pre-sorting of large datasets of digital WSIs; (4) deep learning analytic pipelines that carry out integrated analyses of genomic and computational pathology biomarkers to predict survival and recurrence; and (5) methods to perform multi-site performance studies while receiving dynamic feedback from tumor boards.
To test and further optimize the performance of the content-retrieval algorithms we plan to leverage the consortium that has been established among investigators at Stony Brook, Rutgers and cancer registries at Georgia, Kentucky, New Jersey and New York and utilize precision, recall, and average precision metrics with relevance feedback, as well as SamMatch, average precision, mean average precision and median average precision. Classification performance of the lung tumor growth patterns and the non-tumor (NT) tissue will be conducted using confusion matrices with rows representing ground-truth and columns representing computed results in order for true positive (TP), true negative (TN), false positive (FP) and false negative (FN) counts and F1-scores to be reported. In addition, cross-validation will be used to assess classification accuracy with labeled (training) data partitioned into a training and a validation (test) set prior to open-set evaluations. Further, we will utilize mixed sets of cases from de-identified extracts including digitized pathology and genomic profiles from the VA Corporate Data Warehouse and the GDC data set of which none were used in the training set to enable our team to conduct “open-set,” prospective performance. The algorithms and methods will also be evaluated by direct comparison of expert decisions with those rendered by IRIS.
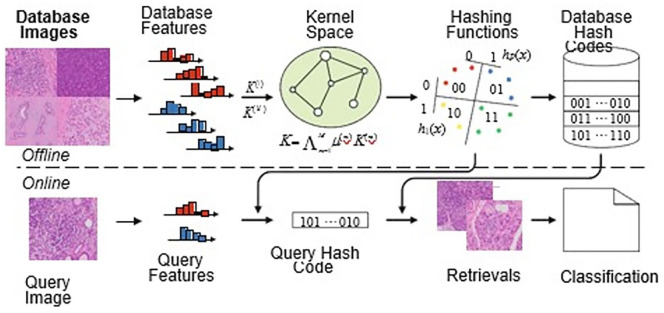
To improve efficiencies and further challenge and evaluate IRIS, we will implement and compare performance of the search and retrieval strategies with those based on binary vectors. Specifically, we will explore the use of hashing techniques including joint kernel-based supervised hashing (JKSH) to encode the high-dimensional feature vectors extracted from the computational imaging signatures and genomic profiles so that they can be encapsulated into short binary vectors. Hashing-based retrieval approaches are an active area of research that is gaining popularity in the medical imaging community due to their exceptional efficiency and scalability. A joint kernel function can be constructed as a linear combination of the kernels for individual features. A series of hashing functions can then be constructed based upon the characteristics of the kernel. As part of an offline process, a supervised optimization algorithm will be utilized to learn the kernel weights and hashing functions based upon the cases within a given reference library. Individual queries can be classified according to a weighted majority vote of the retrievals. The next step of the algorithm requires transformation of the binary vectors from each of the different modalities into a single vector. Note that, this approach could conceivably include additional modalities including CT or MRI. The flexibility of the workflow of the system is shown in Figure 2.
Figure 2.
The flowchart of the designed 2 step CBR system.
Depending on the performance results we will either implement the best of breed approach or we may adopt a hybrid search and retrieval strategy. In either case, we will subsequently integrate online learning to the System so that we can efficiently fine-tune the algorithms as new cases are added to the “ground-truth” Warehouse.
“Efficient validation and testing require multicenter assessments involving multiple pathologists and datasets.” 11 As an Enterprise Healthcare System, the VA has access to large, varied datasets that facilitate the creation of pipelines to further the goals of the Cancer Moonshot Initiative. In addition, the Boston VA Bioinformatics team offers expertise in machine learning, on premise scalable infrastructure at the VA-CRRC Martinsburg Data Center, and a growing collaboration with the Applied Proteogenomics OrganizationaL Learning and Outcomes (APOLLO) network. Through a collaboration between Rutgers and Boston VA we will utilize these data sets that include a range of different histopathology preparations and inter-observer variability across the VA Healthcare system to optimize these algorithms.
Through this collaboration the team will be able to expand the cancers under consideration to include: squamous cell carcinoma of the larynx, squamous cell carcinoma of the trachea, adenocarcinoma of the trachea, salivary gland-type tumors of the trachea, adenosquamous carcinoma of the lung, large cell carcinoma of the lung, salivary gland-type tumors of the lung, sarcomatoid carcinoma of the lung, and typical and atypical carcinoid of the lung.
Footnotes
Author Contributions: DJF and JHS contributed to overall design, concept, and oversight. DJF, JHS and TK wrote and edited the manuscript. RG provided expertise in diagnostic and digital pathology. EB, JRK, LATH and TK implemented and optimized the analytic pipelines. WC developed key methods, tools, workflows and databases. ND and SA provided support for development of algorithms and data analysis. AS, BQ, ED, GH, KW, TI and MJS oversaw access to registries and compliance components of the project.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported, in part, by UH3-CA225021, U24-CA215109, UG3-CA225021, U24-CA180924-05, and 5UL1TR003017 grants from the National Institutes of Health and generous private support to Stony Brook from Bob Beals and Betsy Barton. Additional support was provided through funding from the U.S. Department of Veterans Affairs - Boston Healthcare System through contract, IPA-RU-092920. This work leveraged resources from XSEDE, which is supported by NSF ACI-1548562 grant, including the Bridges system (NSF ACI-1445606) at the Pittsburgh Supercomputing Center. Services, results and/or products in support of the research were generated by Rutgers Cancer Institute of New Jersey Biomedical Informatics Shared Resource NCI-CCSG 7P30CA072720-24.
ORCID iD: Luke Austin Torre-Healy  https://orcid.org/0000-0002-9513-4620
https://orcid.org/0000-0002-9513-4620
References
- 1. Payne PRO, Embi PJ, Cimino JJ. Clinical research informatics. In: Shortliffe EH, Cimino JJ. eds. Biomedical Informatics: Computer Applications in Health Care and Biomedicine. Springer International Publishing; 2021;913-940. [Google Scholar]
- 2. Platt JE, Raj M, Wienroth M. An analysis of the learning health system in its first decade in practice: scoping review. J Med Internet Res. 2020;22:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vanguri RS, Luo J, Aukerman AT, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer. 2022;3:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coudray N, Moreira A, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. medRxiv. 2017;24:1559-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirshfield KM, Tolkunov D, Zhong H, et al. Clinical actionability of comprehensive genomic profiling for management of rare or refractory cancers. Oncologist. 2016;21:1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foran DJ, Chen W, Chu H, et al. Roadmap to a comprehensive clinical data warehouse for precision medicine applications in oncology. Cancer Inform. 2017;16:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foran DJ, Durbin EB, Chen W, et al. An expandable informatics framework for enhancing central cancer registries with digital pathology specimens, computational imaging tools, and advanced mining capabilities. J Pathol Inf. 2022;13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaldana F, Qi X, Ren J, et al. Analysis of Lung Adenocarcinoma Based on Nuclear Features and WHO Subtype Classification Using Deep Convolutional Neural Networks on Histopathology Images. Presented at the Newport, Rhode Island, 2018. [Google Scholar]
- 9. Ren J, Karagoz K, Gatza ML, et al. Recurrence analysis on prostate cancer patients with Gleason score 7 using integrated histopathology whole-slide images and genomic data through deep neural networks. J Med Imaging. 2018;5:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le H, Gupta R, Hou L, et al. Utilizing automated breast cancer detection to identify spatial distributions of tumor-infiltrating lymphocytes in invasive breast cancer. Am J Pathol. 2020;190:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida H, Kiyuna T. Requirements for implementation of artificial intelligence in the practice of gastrointestinal pathology. World J Gastroenterol. 2021;27:2818-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]