Abstract
Human Mediator complexes have been described as important bridging factors that enhance the effect of activators in purified systems and in chromatin. Here we report a novel basal function of a human Mediator complex. A monoclonal antibody was generated that depleted the majority of Mediator components from crude cell extracts. The removal of human Mediator abolished transcription by RNA polymerase II. This was observed on all genes tested, on TATA-containing and TATA-less promoters, both in the presence and absence of activators. To identify the relevant complex a combined biochemical and immunopurification protocol was applied. Two variants termed Mediator and basal Mediator were functionally and structurally distinguished. Basal Mediator function relies on additional constraints, which is reflected in the observation that it is essential in crude but not in purified systems. We conclude that basal Mediator is a novel general transcription factor of RNA polymerase II.
INTRODUCTION
Transcription cofactors mediate access to genes in chromatin, they help to establish, maintain or activate regulatory networks and they affect the formation and activity of basal initiation complexes. In yeast a large multiprotein complex called SRB/Mediator complex has been identified based on its ability to enhance basal and to facilitate activated transcription (Flanagan et al., 1991; Kim et al., 1994; Koleske and Young, 1994; Lee et al., 1999; Myers and Kornberg, 2000; Yudkovsky et al., 2000). Independently, several mammalian Mediator activities were discovered that supported (TRAP/SMCC, ARC, DRIP, human Srb/Mediator) or repressed (NAT) specifically the function of activators (Hampsey and Reinberg, 1999; Lemon and Tjian, 2000; Malik and Roeder, 2000). Purification and cloning of the individual polypeptides suggested that TRAP (Fondell et al., 1999; Ito et al., 1999), SMCC (Gu et al., 1999), ARC (Näar et al., 1999), DRIP (Rachez et al., 1999), NAT (Sun et al., 1998) and hSrb/Mediator (Boyer et al., 1999) are highly related, sharing many subunits. CRSP (Ryu et al., 1999) and PC2, a component of the coactivator fraction USA (Meisterernst et al., 1991), were shown to contain a limited subset of Mediator components (Malik et al., 2000). To date the individual role and the interplay of the variant Mediator forms is not well understood. TRAP/SMCC binds to several activation domains and like PC2 stimulates activator-dependent transcription in vitro in conjunction with other cofactors (Fondell et al., 1999; Ito et al., 1999; Malik et al., 2000). ARC and DRIP also bind to specific activators but enhance their effects exclusively in chromatin (Näar et al., 1999). ARC binds the coactivator CBP and contains one specific component termed ARC105/TIG-1 that appears to be absent in the other Mediator complexes (Näar et al., 1999). There are other differences in composition between the different forms, whose significance and functional importance remain elusive (Malik and Roeder, 2000).
Previous studies in mammals describe Mediator as a complex that is required for the function of regulatory activators. Here we report a critical general and basal function of human Mediator. An immunoprecipitation-based purification strategy led to the identification of two distinct Mediator forms, only one of which serves as a general RNA polymerase II transcription factor. The novel Mediator function is dependent on the context of crude physiological transcription systems.
RESULTS AND DISCUSSION
A monoclonal antibody directed against PAQ/TIG-1 depletes transcription by RNA polymerase II in nuclear extracts
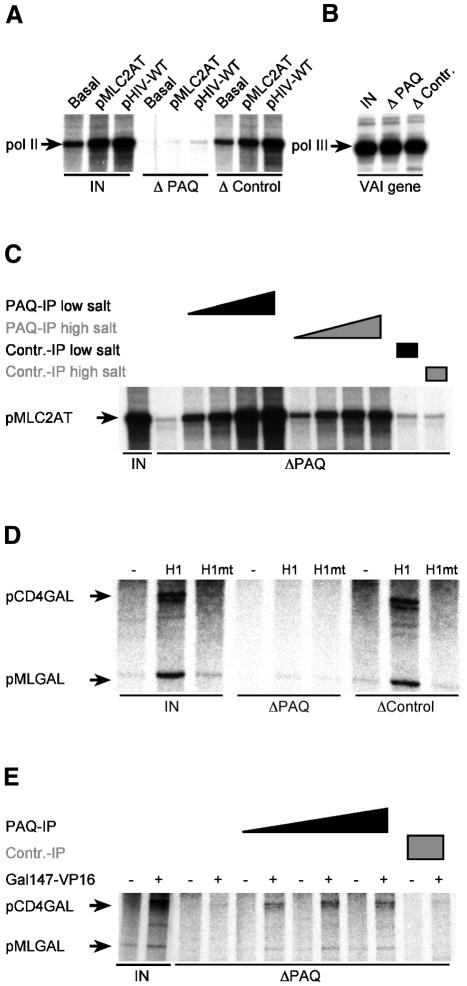
We have recently identified and cloned a protein termed PAQ (PC2 associated Q-rich protein) that coeluted with the coactivator USA (Meisterernst et al., 1991). The glutamine-rich PAQ protein was identified and purified using a monoclonal antibody (1F8) directed against polyglutamine repeats (Berti et al., 2001). PAQ proved to be a close relative of TIG-1, a component initially discovered in ARC (Abraham and Solomon, 2000). Since 1F8 antibodies affected transcription in nuclear extracts we generated specific monoclonal antibodies directed against the C-terminal non-glutamine-rich regions of PAQ/TIG-1 (as there are differences to TIG-1 the protein is subsequently called PAQ). One antibody (PAQ1H7) was selected that was able to deplete PAQ from nuclear extracts. Remarkably, transcription from the adenovirus major late (ML) (pMLC2AT containing the core promoter and the USF-binding site) and the HIV-1 promoter (under Sp1-control) was abolished in these depleted nuclear extracts (ΔPAQ in Figure 1A). In contrast, extracts depleted with a non-related monoclonal rat antibody (TP14B7) of the same isotype remained fully active in transcription (Δ Control). In add-back experiments the immobilized complex restored transcription of the strong ML promoter after extensive washes with low and high salt solutions (Figure 1C). To distinguish activated from basal transcription an ML core promoter (designated Basal in Figure 1A) lacking any activator binding sites was tested in parallel and shown to be switched off upon depletion of PAQ. This suggests that PAQ or a PAQ-associated complex is required for basal transcription in the context of a crude nuclear extract. To examine whether this effect is specific for RNA polymerase II, the adeno-associated Virus VA gene, which is transcribed by RNA polymerase III, was analyzed in parallel. VA gene transcription is not affected (Figure 1B). Hence, PAQ associated proteins are critical for RNA polymerase II but not for RNA polymerase III.
Fig. 1. PAQ-associated complex is a general factor in physiological transcription systems. (A) PAQ depletion (ΔPAQ) abolishes RNA polymerase II transcription in HeLa nuclear extracts (IN), whereas depletion with control antibody (ΔControl) does not affect formation of transcripts (pol II). (B) PAQ depletion is specific for RNA polymerase II. Formation of RNA polymerase III transcripts (pol III) is not influenced. (C) Capacity of PAQ-associated complex to restore transcription in depleted nuclear extracts is resistant to high salt washes, arguing for specificity of the procedure. Titration of the respective complexes was performed in a four-fold range. (D) and (E) PAQ-associated complex is essential for TATA-less and TATA-containing promoters driven by Gal4-VP16. (D) PAQ depletion (ΔPAQ) abolishes RNA polymerase II transcription in Jurkat nuclear extracts (IN). Transcription reactions were performed with pCD4GAL (TATA-less reporter) and pMLGAL (TATA-containing reporter) as templates in the absence (–) and presence (+) of 20 ng of activators Gal-VP16:H1 (H1) and Gal-VP16:H1F442P (H1mt), respectively. (E) PAQ-associated complex restores basal and activated transcription in depleted Jurkat nuclear extracts (ΔPAQ). Titration of the complex was achieved in a three-fold range in the absence (–) and presence (+) of Gal147-VP16.
This effect is not restricted to TATA-containing promoters but is also seen on the true TATA-less CD4 promoter (Halle et al., 1997). The CD4 has basal activity near background in vitro (Figure 1D). In the presence of five upstream GAL4 binding sites and a GAL4 tethered transactivation domain the CD4 promoter template is efficiently transcribed. Here we use either the full-length activation domain of herpes simplex virus protein VP16 (consisting of amino acids 411–490; Gal147-VP16 in Figure 1E) or its subdomain VP16:H1 (amino acids 411–452; designated H1 in Figure 1D), together with a mutant of VP16:H1 (VP16:H1F442P; designated H1mt in Figure 1D) that is inactive in vivo (Regier et al., 1993; our unpublished data). This serves as a specificity control and demonstrates the physiological relevance of our crude in vitro transcription system. Also, transcription from the activated TATA-less promoter was essentially abolished upon depletion of the PAQ-associated complex (Figure 1D). The immobilized PAQ complex restored transcription, at least in part (Figure 1E). Similar results were obtained on the ML under the GAL4-VP16 control that was tested in parallel. We conclude that the antibody removes a complex that is essential for basal and GAL4-VP16-driven transcription on TATA-less and TATA-containing promoters.
PAQ monoclonal antibody depletes the majority of Mediator components from nuclear extracts
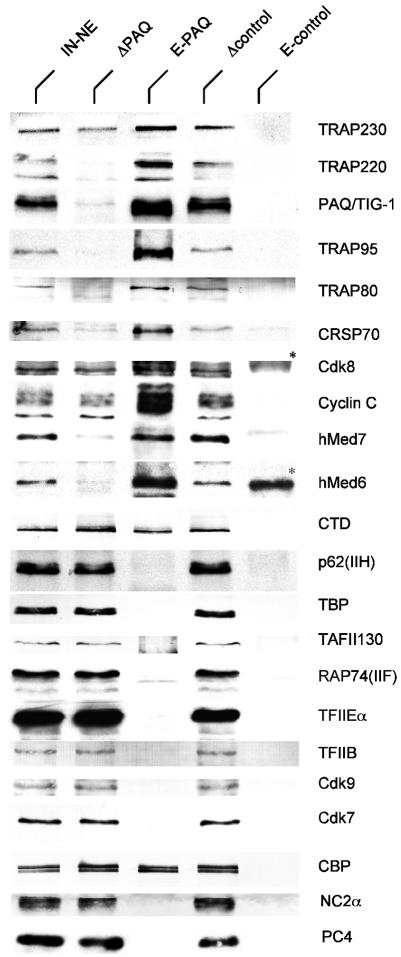
Depleted extracts and immunopurified complexes were systematically analyzed for the absence of general and accessory RNA polymerase II transcription factors (Figure 2). Depletion levels of PAQ (70–90%) correlate quantitatively to the decrease in several Mediator subunits. Examples are hMed6, hMed7 and TRAP80. The depletion of TRAP230 is less complete. We conclude that the majority of human Mediator complexes are associated with PAQ. Consistent with former studies (Näar et al., 1999) some CBP coprecipitates with PAQ antibodies. There is no indication that loss of basal transcription might be caused by codepletion of general transcription factors (GTFs). Most of the RNA polymerase II is retained in the extract. Approximately 20% of RNA polymerase II precipitates with Mediator (CTD in Figure 2). Moreover, the positive transcription elongation factors cdk7/cyclin H and cdk9/cyclin T, which antagonize negative factors DSIF and NELF (Wada et al., 1998; Yamaguchi et al., 1999; Price, 2000), are not bound to Mediator. The cyclin-kinase pair cdk8/cyclin C, which has been suggested to negatively control activated transcription via cyclin H of TFIIH (Akoulitchev et al., 2000), is in part found in the complex. The stringent washing conditions yield highly enriched complexes that, although not yet homogeneous, are free of coeluting PC4 and NC2 but contain part of the histone acetyltransferase CBP (Figure 2), the latter being in agreement with previous analyses of the ARC complex (Näar et al., 1999). These findings argue for a general basal function of a Mediator complex.
Fig. 2. PAQ-monoclonal antibody depletes the majority of Mediator components from nuclear extracts. Four per cent of input (IN-JNE), 4.4% of PAQ depleted (ΔPAQ) and mock depleted (Δcontrol) Jurkat nuclear extract together with 20% of the PAQ (E-PAQ)- or the mock precipitated (E-control) material were analyzed by western blotting. Cross-reactions of secondary antibodies with IgG-HC and IgG-LC are marked (*).
Two functionally distinct forms of human Mediator
Different from the previous studies the biochemical knockout protocol allowed for the first time the testing of Mediator function in the more physiological context of crude nuclear extracts. To our knowledge this is the strongest effect reported for human Mediator in transcription systems. In contrast to purified systems these extracts faithfully reproduce many of the transcription control mechanisms probably because they contain the negative and positive accessory and general factors in more physiological ratios. It is noteworthy that this essential basal function is seen in the absence of bona fide chromatin with templates that are not packaged into nucleosomes.
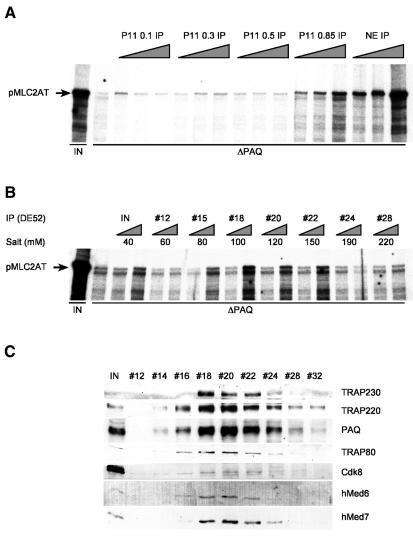
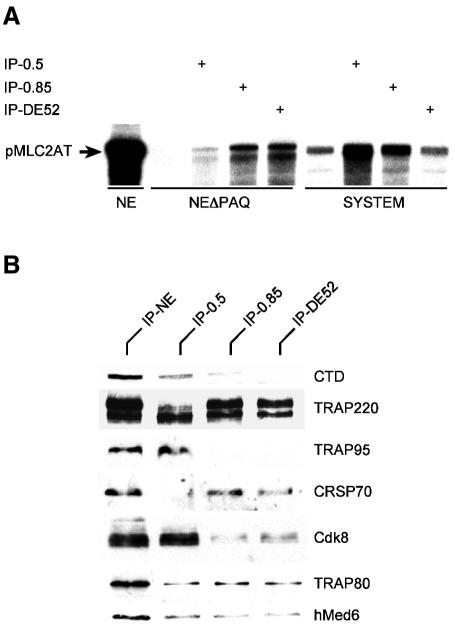
A general role of Mediator has been reported in yeast but not in mammals (Flanagan et al., 1991; Yudkovsky et al., 2000). Hence, one might ask whether the basal function is an intrinsic property of the one Mediator complex or whether PAQ antibodies coprecipitate various forms, only one of which accounts for the function. To define the relevant complex, nuclear extracts were fractionated on conventional columns. Fractions were immunoprecipitated, washed with high salt and immobilized complexes were functionally assayed in depleted nuclear extracts. The functional complex was found to elute almost exclusively in the 0.85 M KCl high salt fraction on phosphocellulose (Figure 3A). Western analysis confirmed coelution with Mediator components on a subsequent DE52 column (Figure 3C). On Superose 6 sizing columns the relevant Mediator eluted with an approximate molecular size of 1 MDa (data not shown). Despite the functional restriction roughly identical amounts of Mediator were found in the lower salt (0.5 M KCl, subsequently called Med for Mediator) and high salt fraction (0.85 M KCl, subsequently called B-Med for basal Mediator). Med contains most of the RNA polymerase II (Figure 4B). RNA polymerase II may have been responsible for moderate positive basal effects of Mediator in reconstituted systems observed previously (Kretzschmar et al., 1994; Gu et al., 1999; Rachez et al., 1999). Med-bound RNA polymerase II is functional, as is demonstrated in a purified system (SYSTEM, Figure 4A) in the presence of limiting RNA polymerase II (IP-0.5 in Figure 4A). In contrast, B-Med shows no effect on basal transcription in purified systems [compare the effects of the immunoprecipitated DE52 peak fraction (IP-DE52) in the system and the depleted extract (NEΔPAQ) in Figure 4A]. These data argue against a model in which the basal function of B-Med in nuclear extracts is caused by a huckepack RNA polymerase II molecule with specific high activity. They do provide strong evidence for a distinct and novel basal requirement of one Mediator form (B-Med) in cruder more physiological systems.
Fig. 3. PAQ-associated complex purified from the phosphocellulose (P11) 0.85 M KCl fraction restores transcription in PAQ-depleted nuclear extracts. (A) Analysis of P11 fractions for capacity to reconstitute transcription by PAQ1H7-IPs. Transcription reactions were conducted with titration of immunopurified complexes in a five-fold range. Untreated nuclear extracts (IN) and IPs (NE IP) from the latter were employed as positive controls. (B) PAQ1H7-IPs from DE52 fractions restore transcription in PAQ-depleted nuclear extracts. PAQ-IPs from input (IN) and various fractions (#) of the second DE52 column [IP(DE52)] were processed and used for add-back experiments similar to those in (A). Activity peaks at 100 mM salt (#18) on the gradient. (C) Western blot analysis of input fractions for IPs described in (B). The functional complex coelutes with the majority of Mediator components tested, except TRAP230, which exhibits a sharper elution profile.
Fig. 4. Two forms of PAQ associated Mediators. (A) Purified B-Med (IP-DE52; see text) exhibits basal activity only in crude but not in purified transcription systems. Immunopurified Mediator from P11 0.5 M KCl fraction (IP-0.5), P11 0.85 M KCl (IP-0.85) and DE52 0.1 M KCl fraction (IP-DE52), respectively, was tested either in a depleted (NEΔPAQ) nuclear extract or in a purified system (SYSTEM) containing TBP (in the absence of TAFs). (B) B-Med purified from the DE52 0.1 M KCl fraction (IP-DE52) contains neither RNA polymerase II (CTD) nor TRAP95. The presence of CRSP70 and the absence of TRAP95 distinguish B-Med from the P11 0.5 M KCl derived complex (IP-0.5). The amount of cdk8 is substantially reduced.
Both Mediator complexes are large, containing many polypeptides that might be differentially bound or modified, which complicates the analysis. Having a powerful IP procedure in hand, here we have initially focused on two questions. First, can we detect features that allow us to clearly distinguish the two forms? If so, is B-Med identical to one of the Mediator forms described earlier? Western analysis demonstrated qualitative and quantitative differences between Med and B-Med. As one prominent feature B-Med lacks TRAP95 and contains CRSP70, while Med contains TRAP95 and lacks CRSP70 (Figure 4B). Furthermore, we noted that Med and B-Med contain different ratios of distinct forms of TRAP220 (Figure 4B). B-Med, when compared with Med, further contains sub-stoichiometric concentrations of cdk8/cyclinC, TRAP100, TRAP230 and TRAP240 (Figure 4B and data not shown). These differences are quantitative and not qualitative and, although reminiscent of PC2 and CRSP descriptions, their physiological relevance needs to be established. B-Med most closely resembles ARC, although there are differences (Malik et al., 2000). Nonetheless, to finally establish a relationship, testing the corresponding complexes in a depleted nuclear extract will be critical. Furthermore, it is presently not clear whether the observed differences are relevant for the alternative function. In any event, they clearly establish structural differences between Med and B-Med and define possible functional surfaces whose functional importance will be investigated in the future. Taken together we report a novel general function of a major form of one specific Mediator complex (B-Med). This function is detectable only through the combination of a biochemical depletion procedure in a crude system and the purification of the relevant complex through conventional chromatography combined with an immunopurification protocol. This may have been a unique opportunity to detect such a general function. Unraveling the molecular details will allow the testing of the resulting molecular hypothesis in cells and in living animals.
METHODS
In vitro transcription reactions. Extracts, templates and the reconstituted transcription system have been described [(Werten et al., 1998) and references therein]. Transcription reactions were conducted with 10 ng (pMLGAL), 20 ng (pCD4GAL) and 100 ng (pHIVWT, pMLC2AT) of linearized templates as described in the figure legends.
Antibodies. Monoclonal antibodies against the C-terminal part (amino acids 291–746) of PAQ were generated in rats using GST fusion proteins. PAQ1H7 was used in this investigation. Antisera were purchased from Santa-Cruz if not further specified. Mouse monoclonal antibodies directed against the CTD of RNA polymerase II (8WG16) and against the poly(Q) motif (1F8) were used.
Immunodepletion of nuclear extracts. PAQ1H7 was immobilized on protein G–Sepharose (Pharmacia) at a concentration of 2 mg/ml, washed with PBS (pH 7.4) and equilibrated in buffer C [20 mM Tris–HCl pH 7.3 (RT), 10% glycerol, 0.1% NP-40, 0.2 mM EDTA, 1 mM PMSF, 0.1 mM benzamidine, 0.25 mM DTT] containing 0.15 M KCl and 200 ng/µl BSA (Roche) on ice. One microliter nuclear extracts containing 6–12 mg/ml protein were brought to 0.15 M KCl, 0.1% NP-40, 1 mM PMSF, 0.1 mM benzamidine, 1 µg/µl aprotinin, leupeptin and pepstatin A, respectively, followed by incubation with 200 µl PAQ1H7 protein G–Sepharose resin at 6°C for 2–3 h. Controls were performed with TP14B7 antibody resin prepared in the same way.
Affinity purification. Fractions were adjusted to 0.15 M KCl, 0.1% NP-40, 1 mM PMSF, 1 mM DTT, 0.1 mM benzamidine and each with 1 µg/µl aprotinin, leupeptin and pepstatin A followed by a preclearing step with TP14B7 antibody resin at 6°C for 30 min. PAQ-Mediator was purified by incubation with one-tenth column volume (CV) PAQ1H7 resin for 3 h at 6°C. Beads were precipitated gently and washed with 100 CV buffer C either at low salt (0.15 M KCl) or high salt (1 M KCl) conditions, indicated in the figure legends and used directly for in vitro transcription reactions or analyzed by SDS–PAGE followed by western blotting.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Bob Roeder, Roger Kornberg and Len Freedman for antibodies. We thank Shona Murphy for the VA plasmid and B. Günzler for help. This work was supported by grants from the HFSP and the DFG (SFB190) to M.M. and by a Pionier-grant (NWO-MW 900-98-142) to H.Th.M.T.
REFERENCES
- Abraham S. and Solomon, W.B. (2000) A novel glutamine-rich putative transcriptional adaptor protein (TIG-1), preferentially expressed in placental and bone-marrow tissues. Gene, 255, 389–400. [DOI] [PubMed] [Google Scholar]
- Akoulitchev S., Chuikov, S. and Reinberg, D. (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature, 407, 102–106. [DOI] [PubMed] [Google Scholar]
- Berti L. et al. (2001) Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a Mediator subunit. Genomics, 74, 320–332. [DOI] [PubMed] [Google Scholar]
- Boyer T.G., Martin, M.E., Lees, E., Ricciardi, R.P. and Berk, A.J. (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature, 399, 276–279. [DOI] [PubMed] [Google Scholar]
- Flanagan P.M., Kelleher, R.J., Sayre, M.H., Tschochner, H. and Kornberg, R.D. (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature, 350, 436–438. [DOI] [PubMed] [Google Scholar]
- Fondell J.D., Guermah, M., Malik, S. and Roeder, R.G. (1999) Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl Acad. Sci. USA, 96, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Malik, S., Ito, M., Yuan, C.X., Fondell, J.D., Zhang, X., Martinez, E., Qin, J. and Roeder, R.G. (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell, 3, 97–108. [DOI] [PubMed] [Google Scholar]
- Halle J.-P., Haus-Seuffert, P., Woltering, C., Stelzer, G. and Meisterernst, M. (1997) A conserved tissue-specific structure at a human T cell receptor β chain core promoter. Mol. Cell. Biol., 17, 4220–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. and Reinberg, D. (1999) RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev., 9, 132–139. [DOI] [PubMed] [Google Scholar]
- Ito M. et al. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Bjorklund, S., Li, Y., Sayer, M.H. and Kornberg, R.D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell, 77, 599–608. [DOI] [PubMed] [Google Scholar]
- Koleske A.J. and Young, R.A. (1994) An RNA polymerase II holoenzyme responsive to activators. Nature, 368, 466–469. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Stelzer, G., Roeder, R.G. and Meisterernst, M. (1994) RNA Polymerase II cofactor PC2 facilitates activation of transcription by GAL4-AH in vitro. Mol. Cell. Biol., 14, 3927–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.C., Park, J.M., Min, S., Han, S.J. and Kim, Y.J. (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B. and Tjian, R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Malik S. and Roeder, R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Malik S., Gu, W., Wu, W., Qin, J. and Roeder, R.G. (2000) The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell, 5, 753–760. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Roy, A.L., Lieu, H.M. and Roeder, R.G. (1991) Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell, 66, 981–993. [DOI] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg, R.D. (2000) Mediator of transcriptional regulation. Annu. Rev. Biochem., 69, 729–749. [DOI] [PubMed] [Google Scholar]
- Näar A.M., Beaurang, P.A., Zhou, S., Abraham, S., Solomon, W. and Tjian, R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- Price D.H. (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol., 20, 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C. et al. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Regier J.L., Shen, F. and Triezenberg, S.J. (1993) Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl Acad. Sci. USA, 90, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Zhou, S., Ladurner, A.G. and Tjian, R. (1999) The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature, 397, 446–450. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang, Y., Cho, H., Rickert, P., Lees, E., Lane, W. and Reinberg, D. (1998) NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell, 2, 213–222. [DOI] [PubMed] [Google Scholar]
- Wada T. et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev., 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werten S., Stelzer, G., Goppelt, A., Langen, F.M., Gros, P., Timmers, H.T., Van der Vliet, P.C. and Meisterernst, M. (1998) Interaction of PC4 with melted DNA inhibits transcription. EMBO J., 17, 5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., Hasegawa, J. and Handa, H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell, 97, 41–51. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish, J.A. and Hahn, S. (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature, 408, 225–229. [DOI] [PubMed] [Google Scholar]