Abstract
Water in healthcare environments can be a source for healthcare-associated infections (HAI). However, information on the exposure risk to opportunistic pathogens in potable water distribution systems (PWDS) is lacking. Laboratory studies characterizing the interaction of opportunistic pathogens with biofilms are needed to understand their role in water systems within healthcare facilities. A stable, repeatable, PWDS multi-species biofilm model comprising Sphingomonas paucimobilis, Methylobacterium sp., Delftia acidovorans, and Mycobacterium mucogenicum was developed in the CDC Biofilm Reactor (CBR), reaching 6 log10 CFU cm−2 within 6 days. The model was used to investigate the interaction of the opportunistic pathogen M. mucogenicum with the other species, and to determine the efficacy of monochloramine (NH2Cl) as a disinfectant against 2-week-old biofilms. Addition of 1 or 2 mg l−1 NH2Cl resulted in the same or an increased log density of viable M. mucogenicum in the biofilm while inactivating some of the Proteobacteria. Although M. mucogenicum preferentially resided in the biofilm, NH2Cl exposure caused release of viable M. mucogenicum from the biofilm into the water. Additional studies with this model should determine if sodium hypochlorite has a comparative effect and if other nontuberculous mycobacteria (NTM) respond to NH2Cl similarly.
Keywords: water distribution system biofilm, Mycobacterium mucogenicum, monochloramine, nontuberculous mycobacteria
Introduction
Potable water distribution systems (PWDS) contain a diverse microbial community of bacteria, protozoa, and fungi (Williams et al. 2004; Berry and Raskin 2006; Eichler et al. 2006; Poitelon et al. 2009; Revetta et al. 2010). Included in these communities are several genera that contain opportunistic pathogens, such as Sphingomonas and Methylobacterium (α-Proteobacteria), Ralstonia and Burkholderia (β-Proteobacteria), Legionella, Pseudomonas, and Stenotrophomonas (γ-Proteobacteria), and nontuberculous mycobacteria (NTM; Actinobacteria). Water systems, including those found in the healthcare environment, provide a site for amplification and can act as a reservoir for human pathogens that are particularly important for patients with wounds or compromised immune systems.
Although water-borne opportunistic pathogens can cause acute, chronic, or fatal healthcare-associated infections (HAI) (Anaissie et al. 2002), the majority of these organisms remain unregulated by safe drinking water guidelines (USEPA, Safe Drinking Water Act, 2006). Surveys of NTM in PWDS confirm that a wide variety of clinically relevant species are recoverable from the main distribution system and premise plumbing, including hospital water systems (Schulze-Röbbecke et al. 1992; September et al. 2004). These organisms can cause respiratory, systemic, or skin infections in the susceptible population found in acute and long term healthcare facilities (Anaissie et al. 2002; De Groote and Huitt 2006). Nonfermentative Gram-negative bacilli found in potable water, including Delftia acidovorans, Sphingomonas paucimobilis and Methylobacterium mesophilicum, sometimes cause bacteremia or pseudo-infections in the healthcare setting (Kressel and Kidd 2001; Ryan and Adley 2010; Chotikanatis et al. 2011). The combination of an immunocompromised population with direct entry routes into the body, such as surgical or central venous catheter sites, necessitates that healthcare providers control patient exposures to nonsterile potable water to reduce the incidence of HAIs attributed to water.
Mycobacterium mucogenicum, a rapidly growing NTM, has caused bacteremia associated with central venous catheters contaminated by drinking water during bathing (Kline et al. 2004; Fleming et al. 2006; Cooksey et al. 2008; Livni et al. 2008). According to a survey of drinking water from PWDS in 21 states (USA), M. mucogenicum is a common inhabitant of PWDS, occurring in >30% of samples (Covert et al. 1999). Information on quantity and virulence of most opportunistic pathogens in PWDS is lacking, which is a significant barrier to developing guidelines for preventing the transmission of opportunistic pathogens via water in the healthcare environment. Performing laboratory studies to characterize opportunistic pathogen interactions with biofilms and the response of opportunistic pathogens to disinfection is a fundamental step towards improving infection prevention protocols for healthcare water distribution systems.
PWDS biofilms have been studied in many laboratory systems with a variety of biofilm reactors, pipe loop systems, and experimental conditions (Camper et al. 1999; Murga et al. 2001; Deines et al. 2010). Some laboratory systems define biofilm inocula, while others allow undefined microbes from water to populate their systems (Martiny et al. 2005). One advantage of defining the PWDS biofilm inoculum is that experiments are repeatable and reproducible by other laboratories. This paper describes a multi-species model PWDS biofilm that contains four bacterial species common to many PWDS (Williams et al. 2004; Hong et al. 2010; Revetta et al. 2010), including three Gram-negative (Proteobacteria) species, viz. Delftia acidovorans, Sphingomonas paucimobilis, Methylobacterium sp., and the NTM M. mucogenicum. Species were chosen for the model based on their compatibility for multi-species biofilm growth and their relevance to public health as opportunistic pathogens. M. mucogenicum in particular is important as an opportunistic pathogen because it is common to many PWDS and is part of a genus known for tolerance to many disinfectants (Carson et al. 1978; Le Dantec et al. 2002).
The objectives of this study were (1) to develop a repeatable and relevant multispecies PWDS biofilm model system containing M. mucogenicum and three other opportunistic pathogens, (2) to evaluate the survival of M. mucogenicum within PWDS biofilm after disinfection with monochloramine in either batch or continuous flow conditions, and (3) to examine the release of M. mucogenicum from PWDS biofilm into water.
Materials and methods
Cultures
D. acidovorans (ATCC 15668; American Type Culture Collection, Manassas, VA), S. paucimobilis (CDC SP-01), M. mucogenicum (CDC MM-01), and Methylobacterium sp. (CDC ME-01) were grown from frozen stocks on R2A agar (Becton Dickinson Diagnostic Systems, Sparks, MD), incubated at ambient room temperature for 7 days, and passaged no more than two times before inoculation into biofilm. The CDC isolates were identified by multiple methods, including morphological characteristics on R2A agar. M. mucogenicum CDC MM-01, an environmental isolate from hospital water associated with an outbreak of central line associated blood stream infections, was identified by fluorescence high-performance liquid chromatography analysis of mycolic acids and hsp65, 16S rRNA, and rpoB gene sequencing (Cooksey et al. 2008). S. paucimobilis CDC SP-01 is an environmental isolate from a 10 μg ml−1 Fentanyl Citrate in 0.9% Sodium Chloride IV Bag involved in a separate hospital outbreak. Methylobacterium sp. CDC ME-01 is an environmental isolate from water in an outpatient dialysis facility. Identification of both S. paucimobilis CDC SP-01 and Methylobacterium sp. CDC ME-01 was confirmed by 16S rRNA gene sequencing.
Potable water
Autoclaved DeKalb County (Georgia) municipal drinking water (treated surface water containing free chlorine as the secondary disinfectant; DeKalb County Public Works Department; EPA ID: GA0890001), collected over the 7 month period of the study, was used for all experiments. The mean water quality parameters measured in water leaving the treatment plant include temperature, 188C (standard deviation, SD = 2.5); total organic carbon (TOC), 0.79 mg l−1 (SD = 0.11); dissolved O2, 14 mg l−1 (SD = 0.38); alkalinity, 21 mg l−1 (SD = 1.7); hardness (total), 13 mg l−1; nitrate, 0.49 mg l−1 (SD = 0.09); free chlorine, 1.40 mg l−1 (SD = 0.14); and chloramines, 0.03 mg l−1 (SD = 0.01). pH equaled 8.85 (SD = 0.49), as measured in autoclaved water in the laboratory. Free chlorine in the autoclaved drinking water was measured using the FAS-DPD method (APHA 2005) to confirm that no residual levels were present prior to experimentation.
Inocula
For all assays and model experiments, single species inocula were prepared in either autoclaved drinking water (for the paired growth assays) or Butterfield Buffer (Becton, Dickinson and Company, Sparks, MD; Donlan et al. 2005) to a concentration equivalent to 0.10 OD600 (Hach DR/4000U spectrophotometer, Hach Company, Loveland, CO). For the paired growth assays, the single species suspensions were stored overnight in an environmental chamber (Caron 6030, CARON Products & Services, Inc, Marietta, OH) at 25°C, to allow the bacteria to acclimate to the water. The absorbance at 600nm was measured and adjusted to 0.10 OD600 if necessary the next day, before use in the assay. An aliquot of each inoculum was diluted and plated onto R2A, to determine the concentration of each species. The mean cell density of each species suspension was 8.66 log10 CFU ml−1 (SD = 0.16), 8.81 (SD = 0.16), 8.60 (SD = 0.15) and 8.22 (SD = 0.15) for D. acidovorans, S. paucimobilis, M. mucogenicum, and Methylobacterium sp., respectively (n = 13).
Paired growth assay
Preliminary paired-growth biofilm assays were performed in 96-well polystyrene plates by adding overnight suspensions of each possible pairing of the four model species at a 1:1 proportion, for a final concentration of 105 CFU ml−1 per species. Controls of single species in autoclaved drinking water were also run. The plates were incubated in an environmental chamber at 25°C and 46% humidity for 72 h. To sample the biofilm, plates were rinsed three times in 250 μl well−1 of phosphate buffered saline (PBS, pH 7.2) before extracting the biofilm by pipetting vigorously for 10 s to resuspend the cells in 250 μl well−1 of PBS with 0.02% Tween® 80 (PBST). Biofilm organisms were enumerated by 1:10 serially diluting suspensions and spread-plating onto R2A agar.
CDC Biofilm Reactor (CBR)
The CBR (Biosurface Technologies, Bozeman, MT, USA) was run by following published protocols (Donlan et al. 2004; Goeres et al. 2005). The CBR contains eight coupon holders with space for three disk coupons per holder. These were filled with PVC coupons (13 mm diameter, 4 mm thickness; Biosurface Technologies, Bozeman, MT, USA) that had been washed in laboratory detergent (Versaclean, Fisher Scientific, Pittsburgh, PA), rinsed 5x in deionized water, rinsed a final time in 70% ethanol, and air-dried. The assembled CBR was sterilized with ethylene oxide, the outflow line was clamped, and the reactor was filled with 350 ml of autoclaved drinking water. Suspensions of the four model bacteria were prepared as described above and inoculated into the CBR at a final concentration of ~105 CFU ml−1 per species. The CBR was run in batch mode for 24 h, with the inner paddle rotating at 100 rpm, in an environmental chamber at 25°C. After 24 h, the reactor was continuously supplied with autoclaved drinking water contained in a 20-l carboy, at 2.5 ml min−1 (140 min residence time), for at least 13 additional days.
Biofilm sample processing
PVC coupons were harvested by aseptically removing a sampling rod from the reactor and transferring them to conical 50 ml tubes containing 10 ml of PBST. CBR water was sampled by inserting a pipet through the empty sample rod slot before closing the hole with a sterile replacement rod. Coupons were subjected to three alternating 30 s cycles of sonication (frequency of 42 kHz [±6%], Branson Ultrasonics Corp., Danbury, CT) and vortexing. Previously determined recovery efficiency of NTM biofilm from plastic and stainless steel coupon surfaces ranged between 90 and 99% (Williams et al. 2009). Biofilm suspensions were serially diluted and spread-plated on R2A agar. Plates were incubated at room temperature for up to 2 weeks. Colonies of each organism were differentiated and enumerated based on colony morphology and time of appearance on R2A (Table 1).
Table 1.
Characteristics of each model PWDS organism colony formation on R2A medium.
| Organism | Colony morphology | Days until first appearance |
|---|---|---|
| Delftia acidovorans | Translucent, undulate | 2–3 |
| Sphingomonas paucimobilis | Yellow, round | 2–3 |
| Mycobacterium mucogenicum | White, round | 3–5 |
| Methylobacterium sp. | Pink, round | 3–5 |
NH2Cl disinfection assays
NH2Cl was generated by adding ammonium chloride (NH4Cl) and sodium hypochlorite (NaOCl) in a 3:1 molar ratio to 0.0035M phosphate buffer at pH ≥ 8.5. The NH2Cl concentration was determined with a Monochlor F kit (Hach Company, Loveland, CO), and read in a Hach DR 4000 spectrophotometer at 655 nm. The batch disinfection assays were performed on 14-day old biofilm grown on PVC coupons in the CBR, placed in chlorine demand-free conical glass tubes containing 45 ml of NH2Cl solution at the target concentration for disinfection (1 or 2 mg l−1). At each sampling time point, coupons were removed from the disinfectant solution and placed in 10 ml of 0.01% sodium thiosulfate to inactivate the NH2Cl before processing. The NH2Cl residual concentration and plate count following neutralization with sodium thiosulfate were measured on the remaining 45 ml of disinfectant solution. The log CFU cm−2 of each species for each time point or NH2Cl concentration, triplicate coupons from at least two replicate batch disinfection experiments, were used to calculate the log10 reduction for biofilm and water samples. Two direct viability measurements were performed on a subset of biofilm samples exposed to 0 or 2 mg l−1 of NH2Cl for 5 or 24 h (see Methods subsection ‘Direct viability detection of suspended PWDS biofilm’).
Continuous flow disinfection assays were performed by filling the CBR and supply carboy with autoclaved drinking water containing the target NH2Cl concentration of 1 mg l−1. To achieve the target concentration of NH2Cl in the reactor immediately at the beginning of the continuous flow disinfection, the entire top of the reactor (including the sampling rods) was removed aseptically and the volume of NH2Cl stock needed to achieve the target concentration of NH2Cl was added directly into the reactor. The solution in the CBR was mixed for 5 s before the top was replaced and flow of treated drinking water was initiated at a rate of 2.5 ml min−1. At each time point, a rod was removed as previously described (Donlan et al. 2004; Goeres et al. 2005), neutralized in 30 ml of 0.01% sodium thiosulfate, and then processed to recover the biofilms, as described above. Water samples were also neutralized by adding 20 μl of 0.1 N sodium thiosulfate into 8 ml of reactor water, before diluting and plating. The concentration of NH2Cl in the reactor was monitored over time, and kept at 1.0 ± 0.2 mg l−1 by adding a calculated amount of the stock solution directly into the reactor, when needed. The log10 CFU cm−2 of each species was calculated as an average of between 9 and 12 biofilm samples, for each time point, from at least three replicate continuous flow disinfection experiments. The log10 reduction was calculated for biofilm and water samples using pre-disinfection Day 14 samples as the control.
Direct observation of drinking water biofilm
Biofilm on PVC coupon surfaces was directly observed using epifluorescence microscopy after staining with Sybr Green I (Invitrogen, Carlsbad, CA), as previously described (Williams et al. 2009). Briefly, coupons containing 14-day biofilm were fixed in 3.7% formaldehyde and stored at 4°C until needed. Coupons were rinsed by dipping into PBS (phosphate buffered saline, Fisher Scientific, Pittsburgh, PA) before immersing in 2 ml of a 5x solution of Sybr Green I (Invitrogen) in room temperature TE buffer at pH 8 (10 mM Tris, 1 mM EDTA; Mediatech, Inc., Manassus, VA). Coupons were stained in the dark at room temperature for 10 min, rinsed in sterile deionized water for 1 min, and air-dried in the dark. Coupons were attached to microscope slides and coverslips were mounted with ProLong Gold antifade solution (Invitrogen). Biofilm was viewed through a Zeiss Axioplan epifluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with a fluorescein filter set (480/40 nm excitation, 505 nm long-pass dichroic mirror, 535/50 nm emission) and an ApoTome. Image z-stacks were obtained through a 63x oil immersion objective.
Direct viability detection of suspended PWDS biofilm
Biofilm viability after batch disinfection with 2 mg l−1 of NH2Cl was determined in treated and control suspended biofilm samples by two direct methods, viz. detection of esterase activity, measured with ChemChrome V6 and a solid-phase cytometer (ScanRDI; Chemunex, Ivy-Sur-Seine, France), and membrane permeability, measured by BacLight™ viability staining (Molecular Probes, Life Technologies, Grand Island, NY). Aliquots of suspended biofilm from triplicate coupons were processed for esterase activity as previously described (Baudart et al. 2002; Shams et al. 2007). Second aliquots were processed to detect membrane permeability with BacLight by following the manufacturer’s protocol. Stained suspensions were filtered through black polycarbonate membrane filters, 0.2 μm pore size (Whatman, Inc. Piscataway, NJ), and air-dried on microscope slides in the dark. Coverslips were secured on filters with BacLight™ mounting fluid. Samples were viewed through a Zeiss Axioplan epifluorescence microscope described above. Green and red bacteria were enumerated in 10 microscope fields per coupon, totaling at least 500 bacteria.
Statistics
All statistical analyses were performed on log10 transformed colony enumeration data using SPSS PASW Statistics 18, Release Version 18.0.0 (SPSS, Inc., 2009, Chicago, IL). One-way ANOVA was performed on Day 14 base model data from multiple experiments to determine the source of variation among mean total log10 cell densities. The repeatability SD of the base model was calculated as previously described for the CBR (Goeres et al. 2005), comparing the mean log10 total CFU cm−2 for all four species combined, per coupon harvested on Day 14, between three replicate experiments. Log10 reductions of viable cells from batch and continuous flow disinfection experiments were calculated using different controls. For the batch disinfections, the control coupons were manipulated in the same way as the experimental coupons, however they were not exposed to NH2Cl. For the continuous flow disinfections, control coupons were harvested on Day 14, immediately prior to the addition of NH2Cl to the CBR. Significant differences in log10 values between disinfected and untreated samples were determined by t-test (p < 0.05).
Results
Potable water distribution system (PWDS) biofilm model
All four species of the base model grew in the CBR and remained in the biofilm during the entire 2 week period, over three experiments (Figure 1a). Of the four species, α-Proteobacteria, typically Methylobacterium sp., were the dominant bacteria in the biofilm. Total culturable biofilm (colony enumeration of all four species combined) on Day 14 for replicate experiments ranged from 6.16 to 6.65 log10 CFU cm−2. The repeatability SD was 0.24, indicating that the model system provides repeatable total viable biofilm quantities on PVC surfaces. The estimated within-experiment variance was 0.0724 (6 df) and the estimated between-experiment variance was 0.0316 (2 df). There was no significant difference in the mean total log10 biofilm cell densities for the three experiments (ANOVA, p = 0.34), with 70% of the variance coming from within the experiments. Direct observation of biofilm collected on Day 14 demonstrated that the four species formed a patchy biofilm on the PVC surface, the majority of which occurred in a scattered monolayer, with some microcolonies up to 11 mm thick (Figure 2).
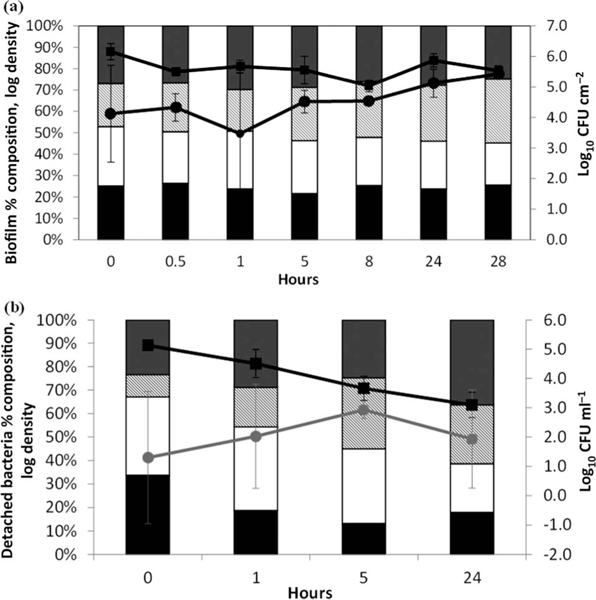
Figure 1.
(a) PWDS biofilm model growth during a 2-week incubation. Total log10 CFU cm−2 (all four species) is displayed on the line plot. The percentage composition of the four species in biofilms from each sample day is represented by the bar chart (log density of each species divided by the log density of all four species, multiplied by 100). (b) Log10 CFU ml−1 of unattached bacteria in CBR liquid during incubation for 2 weeks; the three Gram-negative species are shown by the black line. Unattached M. mucogenicum was present in CBR erratically, as demonstrated by the grey line. The percentage composition of each species within water samples on each sample day are presented in the bar chart.
Figure 2.
Image of 14-day old multi-species base model PWDS biofilm formed in drinking water on PVC substratum. The image is a compiled z-stack obtained through a 63x objective. Biofilm was visualized after staining with Sybr Green I. Micrometers are labeled along the X- and Y-axis.
At the end of the 24 h batch mode the total viable unattached or suspended cell density had increased slightly in the CBR liquid (Figure 1b). The unattached cell density decreased when the reactor was switched from batch mode to continuous flow mode at 24 h, and stabilized at 5.1 log10 CFU ml−1 (SD = 0.3) from Day 4 through the end of the reactor run on Day 14. Unattached M. mucogenicum was not detected in CBR water samples after 24 h (nexp = 5), but was present in the biofilm (3.9 log10 CFU cm−2; SD = 0.4, n = 3). Low log density of unattached M. mucogenicum was detected sporadically in water samples collected over the remaining incubation days, with maximum log density observed on Day 11, 1.3 log10 CFU ml−1, SD = 1.7.
Paired-growth biofilm assays
As preliminary work to confirm compatibility of the four species in a multi-species biofilm, each of the model system organisms was tested for its ability to enhance or inhibit biofilm formation of the other species using a paired-growth biofilm assay. M. mucogenicum produced significantly more biofilm when grown in the presence of D. acidovorans (p = 0.043), S. paucimobilis (p = 0.0052), and Methylobacterium sp. (p = 0.0042) than when grown alone. Biofilm growth of D. acidovorans, S. paucimobilis, or Methylobacterium sp. was neither enhanced nor inhibited by the presence of the other model organisms.
Batch biofilm disinfection with NH2Cl
Achieving concentrations of 1 or 2 mg l−1 of NH2Cl initially during batch disinfection resulted in actual concentrations of 0.8 mg l−1 (SD = 0.1, nexp = 3) or 1.6 mg l−1 (SD = 0.3, nexp = 5) of NH2Cl, respectively, over 24 h, calculated as weighted averages. Total culturable biofilm experienced one log reduction (LR) after exposure to 2 mg l−1 of NH2Cl (Figure 3, p < 0.01). Exposure for 5 or 24 h resulted in essentially the same LR, 1.05 (SD = 0.51, nexp = 5) and 1.03 (SD 0.6, nexp = 5), respectively. D. acidovorans and S. paucimobilis were the most susceptible to disinfection. However, the log density of M. mucogenicum remained the same or increased slightly during disinfection while the log density of the three Proteobacteria species decreased, resulting in a population shift to favor M. mucogenicum as the largest component of the biofilm (Figure 3a, b). One mg l−1 of NH2Cl had little effect on the biofilm in 5 h, with 0.37 LR (SD = 0.32, nexp = 4, p = 0.06). After 24 h, however, LR was slightly higher: 0.6 (SD = 0.15, nexp = 2, p < 0.05).
Figure 3.
(a) Culturable biofilm after batch NH2Cl disinfection for 5 h, and (b) 24 h, (c) detached bacteria after batch NH2Cl disinfection for 5 h, and (d) 24 h. Bars are percentage composition: solid grey (top), Methylobacterium sp.; diagonal stripes, M. mucogenicum; white bar, S. paucimobilis; and solid black bar, D. acidovorans. Line plots, solid square is total log density of culturable biofilm bacteria, and solid circle represents the log density of M. mucogenicum.
Two direct viability measurements of biofilm bacteria after batch disinfection with 2 mg l−1 of NH2Cl provided similar results that contrasted with culturable bacterial enumeration. Esterase activity and membrane integrity were not reduced significantly in biofilm exposed to 2 mg l−1 of NH2Cl in comparison to untreated control biofilm (Table 2, n = 3 coupons). Similar biofilm log densities were measured in untreated control biofilm by culture, esterase activity, and viable bacteria with intact membranes. The log density of total biofilm bacteria detected with the BacLight™ kit was only slightly higher than the log density of viable bacteria, since the majority of bacteria were viable (green).
Table 2.
Comparison of culturable and viable biofilm bacteria after batch disinfection with 2 mg l−1 NH2Cl for 5 or 24 h.
| NH2Cl (mg l−1), | Culturable bacteria | Esterase activity | Intact membranes | BacLight™ Total |
|---|---|---|---|---|
| Time (h) | (Log10 CFU cm−2) | (Log10 cells cm−2) | (Log10 cells cm−2) | direct count (Log10 cells cm−2) |
| 0 mg l−1, 5 h | 6.26 (0.00) | 6.06 (0.13) | 6.07 (0.14) | 6.16 (0.15) |
| 2 mg l−1, 5 h | 5.56 (0.23)* | 5.87 (0.14) | 5.88 (0.07) | 6.08 (0.04) |
| LR | 0.7 | 0.19 | 0.19 | 0.08 |
| 0 mg l−1, 24h | 6.00 (0.09) | 5.89 (0.15) | 5.89 (0.09) | 5.96 (0.07) |
| 2 mg l−1, 24 h | 5.30 (0.13)* | 5.73 (0.09) | 5.76 (0.14) | 6.05 (0.12) |
| LR | 0.7 | 0.16 | 0.13 | −0.09 |
Note: Direct viability was quantified by detection of esterase activity and cell membrane integrity (n = 3 coupons; SD in parentheses; LR = log reduction). Total direct count of bacteria was calculated as the sum of viable and nonviable cells after BacLight™ staining (green and red cells).
= a statistically significant reduction from the untreated control (p < 0.01).
During the batch disinfection experiments, cells of each PWDS species detached from biofilm on PVC coupons and remained in suspension in vessels containing NH2Cl and control vessels with no NH2Cl. Control vessels contained 5.48 (SD 0.48) log density of detached culturable bacteria (range, 4.45–6.71 log10 CFU ml−1). Counting all four species together, detached bacteria experienced a 2.01 (SD = 0.66, nexp = 5) or 2.41 (SD = 0.88, nexp = 5) LR after exposure for 5 or 24 h to 2 mg l−1 NH2Cl, respectively (Figure 3c, d, p < 0.01), demonstrating greater susceptibility to disinfection than biofilm bacteria. Exposure to 1 mg l−1 of NH2Cl resulted in 1.38 (SD = 0.8, nexp = 4) or 1.41 (SD = 0.33, nexp = 2) LR of detached bacteria after 5 or 24 h, respectively. Within the overall enumeration, however, the quantity of detached M. mucogenicum increased significantly to 3 log10 CFU ml−1 after exposure for 5 h to both a target of 1 and 2 mg l−1 NH2Cl (Figure 3c). Also, although detached M. mucogenicum was only recovered erratically from water with no NH2Cl, it was consistently cultured from water at both time points and NH2Cl levels during batch disinfection.
Continuous flow NH2Cl disinfection in CBR
Studies were performed to assess the repeatability of the response of the PWDS biofilms to disinfection, with 1 mg l−1 of NH2Cl under continuous flow. Targeting 1 mg l−1 of NH2Cl in the CBR for the continuous flow disinfection experiments resulted in a mean concentration of 0.9 mg l−1 NH2Cl over 28 h (SD = 0.2, nexp = 3). The response of each species to continuous flow and batch disinfection with 1 mg l−1 of NH2Cl were similar. There was a statistically significant decrease in the total amount of bacteria recovered from the biofilm at all time points sampled during the continuous flow disinfection (range 0.06–1.30 log10 CFU cm−2, p < 0.05), although the overall decrease was less than that which occurred in the batch disinfection. The numbers of D. acidovorans and S. paucimobilis cultured from the biofilm in the continuous flow disinfection was significantly reduced at all time points (p < 0.01, data not shown). The quantity of M. mucogenicum recovered from the biofilm increased during exposure for 28 h to 1 mg l−1 of NH2Cl. However, this increase was not statistically significant, probably due to the large variability of M. mucogenicum log density found in 0 h samples (Figure 4a).
Figure 4.
(a) Culturable biofilm during continuous flow of 1 mg l−1 NH2Cl disinfection for 28 h and (b) culturable detached bacteria from the same experiments. Bars represent the percentage composition of each species, calculated from log densities: solid grey (top), Methylobacterium sp., diagonal stripes, M. mucogenicum; white bar, S. paucimobilis, and solid black bar, D. acidovorans. Line plots, solid square is total log density of culturable biofilm bacteria, and solid circle represents the log density of M. mucogenicum.
The recovery of detached D. acidovorans and S. paucimobilis in the CBR decreased significantly during continuous flow disinfection (Figure 4b). Detached M. mucogenicum was recovered from the majority of water samples containing NH2Cl from batch and continuous flow disinfection experiments, with higher log density than found in untreated water (Table 3).
Table 3.
Percentage of water samples that contained detached M. mucogenicum from batch and continuous flow disinfection at all disinfectant levels and time points, compared to untreated samples without NH2Cl.
| Percentage of water samples containing M. mucogenicum |
||
|---|---|---|
| Disinfection type | Treated (NH2Cl) | Untreated |
| Batch | 92% | 31% |
| Continuous flow | 85% | 21% |
Discussion
A stable, repeatable, PWDS multispecies biofilm model system was developed to aid investigation into the interaction of opportunistic pathogenic bacteria with biofilms and to determine the efficacy of potable water disinfectants. This model is relevant to in situ studies of PWDS bacteria, in which Proteobacteria dominate the known species of the community analyses (Williams et al. 2004; Revetta et al. 2010, 2011). The log density of culturable biofilm formed in this model, 6 log10 CFU cm−2 within 7 days, was similar to the density of biofilm formed by undefined heterotrophic plate count (HPC) bacteria in simulated drinking water produced with defined nutrients in a separate experimental design (Camper et al. 1996). Nutrient limitation may be the overriding factor in determining the plateau of biofilm formation by mixed HPC bacteria in continuous flow drinking water model systems, regardless of the particular mix of species or biofilm sampling device (LeChevallier et al. 1991; Van der Kooij 1992). Direct observation through epifluorescence microscopy of the total, intact biofilm (Sybr Green I staining), and the viable suspended biofilm (ChemChrome V6 and BacLight™ kits) confirmed that the architecture and quantity of PWDS biofilm was limited, and matched the CFU enumeration results. Other researchers also have observed the formation of ‘patchy’ multi-species PWDS biofilm with limited architecture (Paris et al. 2007).
Several biofilm sampling devices (Deines et al. 2010) have been used to sample or model drinking water biofilms, including the CBR (Goeres et al. 2005). The CBR is a practical device for conducting disinfection experiments because replicate biofilm samples can be exposed to disinfectant in a continuous flow system or after removal from the reactor (batch disinfection). The CBR and other devices have been used to study the tolerance to disinfectant exposure of water biofilms with various combinations of defined, undefined or partially defined inocula in defined, undefined, or partially defined media (Stewart et al. 1994; Camper et al. 1996; Butterfield et al. 2002; Norton et al. 2004; Donlan et al. 2005). The defined inoculum in the current study allowed observation of the interactions between the four species in the PWDS biofilm model system. Differing colony morphologies and growth rates of these species made it practical to rely on CFU enumeration to track changes in the composition of biofilm and detached cells. No competition or inhibition was observed between species on R2A plates that contained 300 or fewer colonies. Paired growth studies demonstrated that biofilm growth of M. mucogenicum was enhanced by the presence of the Proteobacteria and that no species was inhibited by the presence of another species. In contrast, a previous study found that isolates of Methylobacterium sp. and M. mucogenicum produced similar amounts of biofilm when grown as dual and single species (Simões et al. 2007). The different response of M. mucogenicum to biofilm formation in the presence of Methylobacterium sp. may be strain dependent, or due to incubation in higher nutrient medium in the previous work. This preliminary work confirmed that differences in the log density of the biofilm of the four species were caused by differences in growth rate, rather than an antagonistic effect of one species toward another in the biofilm. Recovery efficiency of NTM biofilm from plastic and stainless steel coupon surfaces by the current method was determined previously to range between 90 and 99% (Williams et al. 2009), indicating consistency in the processing protocol. The residence time of water in the CBR, 140 min, was adequate to stabilize bacteria in the reactor water without growth in the unattached mode. This was confirmed by determining that these organisms did not exhibit measurable growth as unattached bacteria in unamended potable water within 7 days (unpublished data).
While this model used an undefined medium (autoclaved drinking water), water quality parameters, monitored by DeKalb County Watershed Management, indicated that no major changes to the quality of water had occurred over the duration of the experiments, such as a large difference in TOC or turbidity due to seasonal changes or an extreme weather event. Goeres et al. (2005) demonstrated that the CBR can be a rugged model system in which small variations to the procedures, including minor changes in nutrient levels in the medium, may be made without impacting the overall predicted outcome of the experiment.
The batch and continuous flow NH2Cl disinfection methods used on the base biofilm at Day 14 yielded similar results. In both cases, statistically significant biofilm LR was too little to be of practical value. Typical demonstrations of disinfectant efficacy target 2 or 3 LR (Rose et al. 2007), while the highest LR of biofilm observed under these conditions was 1.05 (2 mg l−1 of NH2Cl, 5 h batch disinfection). The low LR was caused by the inclusion of M. mucogenicum in the biofilm, and limiting the NH2Cl concentration to levels expected in PWDS (Seidel et al. 2005). It is well established that NTM are highly tolerant to exposure to chlorine disinfectant compared to other bacteria (Taylor et al. 2000; Le Dantec et al. 2002), and that multi-species biofilm is more tolerant than single-species biofilm (Simões et al. 2010). The population shift toward M. mucogenicum in biofilm and water observed in this study is comparable to a previous study of the survival of M. avium in a model PWDS biofilm, in which the biofilm consisted of nearly 100% M. avium after competitive heterotrophic bacteria had been eliminated by disinfection with free chlorine (Norton et al. 2004).
A previous examination of monochloramine inactivation of a dual-species biofilm demonstrated greater LR (Stewart et al. 1994), probably due to the higher nutrient level in the reactor medium, a higher NH2Cl concentration, and the greater sensitivity of the two species of Proteobacteria comprising the biofilm. Bacteria tend to be more sensitive to inactivation by chlorine disinfectants when grown in high nutrient medium than when grown in drinking water (Taylor et al. 2000).
The mode of action of NH2Cl is not well characterized (USEPA 1999). Continued esterase activity and the maintenance of cell membrane potential after batch disinfection indicated that the same log density of biofilm bacteria was still metabolically active, despite decreased culturability. The contradictory results may indicate that a portion of bacteria were injured, rather than truly inactivated, as has been demonstrated to occur with other water organisms (McFeters et al. 1986). Viability indicators that depend on different aspects of cell function may give variable results (Stewart et al. 1994). Regrowth experiments to determine if the PWDS biofilm could recover culturability after NH2Cl disinfection would be a useful next step.
The batch disinfection method effectively demonstrated that bacteria were released from the biofilm. Because non-adherent cells were rinsed from the biofilm before the coupons were transferred into vessels for batch disinfection, any bacteria cultured from the liquid phase during batch disinfection experiments had been released from the biofilm. While detached bacteria cultured during the continuous flow disinfection experiments cannot be conclusively attributed to cells releasing or sloughing from the biofilm, the commonality in quantity and species of detached bacteria in the bulk fluid of the CBR and in the batch disinfection liquid confirms the findings. The greater sensitivity of detached bacteria to monochloramine or free chlorine disinfection than biofilm bacteria has been demonstrated previously (Cochran et al. 2000; Steed and Falkinham 2006; Behnke et al. 2011).
Detached M. mucogenicum was consistently cultured during disinfection experiments, but not from CBR bulk water obtained during the 14 days leading up to the disinfection, suggesting that M. mucogenicum preferentially inhabits biofilm in PWDS in the absence of monochoramine. Addition of NH2Cl increased the frequency of occurrence, as well as the log density, of M. mucogenicum in the water. This work demonstrates that concentrations of NH2Cl that are typically found in PWDS encourage release of viable M. mucogenicum cells from model multi-species biofilm into the bulk water. The public health implications of this finding should be explored further by determining if NH2Cl causes M. mucogenicum cells to detach from biofilm in actual PWDS, if hypochlorite (free chlorine) has a similar effect on M. mucogenicum, and if NH2Cl increases release of other viable, clinically relevant NTM from biofilm into potable water.
Conclusions
A repeatable potable water biofilm model has been developed comprising four drinking water bacteria. All four species persisted in the biofilm for 14 days. This model was designed to determine the fate of specific pathogenic bacteria in biofilm as a function of time and disinfectant concentration. Biofilms persisted in 1 mg l−1 monochloramine over 24 h but detached bacteria suspended in water were reduced. PWDS biofilms were more tolerant to continuous flow disinfection, which mimicked conditions in distribution systems more closely than batch disinfection. Disinfection with monochloramine selected for M. mucogenicum in the biofilm and increased detachment of culturable M. mucogenicum into the water.
Acknowledgements
The authors thank Jody Shoemaker of DeKalb County Watershed Management for providing water quality data. Sarah Gilbert and Elizabeth Perez are also thanked for technical assistance. Funding for this work was provided by the National Center for Environmental Health in the Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US CDC.
Footnotes
Publisher's Disclaimer: The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
References
- Anaissie EJ, Penzak SR, Dignani MC. 2002. The hospital water supply as a source of nosocomial infections: a plea for action. Arch Int Med 162:1483–1492. [DOI] [PubMed] [Google Scholar]
- APHA AWWA, WEF. 2005. Standard methods for the examination of water and wastewater. 21st ed. Washington (DC: ): American Public Health Association. p. 4–56 to 4–76. [Google Scholar]
- Baudart J, Coallier J, Laurent P, Prévost M. 2002. Rapid and sensitive enumeration of viable diluted cells of members of the family Enterobacteriaceae in freshwater and drinking water. Appl Environ Microbiol 68:5057–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke S, Parker AE, Woodall D, Camper AK. 2011. Comparing the chlorine disinfection of detached biofilm clusters with those of sessile biofilms and planktonic cells in single- and dual-species cultures. Appl Environ Microbiol 77:7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Xi C, Raskin L. 2006. Microbial ecology of drinking water distribution systems. Curr Opin Biotech 17:297–302. [DOI] [PubMed] [Google Scholar]
- Butterfield PW, Camper AK, Ellis BD, Jones WL. 2002. Chlorination of model drinking water biofilm: implications for growth and organic carbon removal. Water Res 36:4391–4405. [DOI] [PubMed] [Google Scholar]
- Camper AK, Jones WL, Hayes JT. 1996. Effect of growth conditions and substratum composition on the persistence of coliforms in mixed-population biofilms. Appl Environ Microbiol 62:4014–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camper AK, Burr M, Ellis B, Butterfield P, Abernathy C. 1999. Development and structure of drinking water biofilms and techniques for their study. J Appl Microbiol Symp Suppl 85:1S–12S. [DOI] [PubMed] [Google Scholar]
- Carson LA, Petersen NJ, Favero MS, Aguero SM. 1978. Growth characteristics of atypical mycobacteria in water and their comparative resistance to disinfectants. Appl Environ Microbiol 36:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotikanatis K, Bäcker M, Rosas-Garcia G, Hammerschlag MR. 2011. Recurrent intravascular-catheter-related bacteremia caused by Delftia acidovorans in a hemodialysis patient. J Clin Microbiol 49:3418–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WL, McFeters GA, Stewart PS. 2000. Reduced susceptibility of thin Pseudomonas aeruginosa biofilms to hydrogen peroxide and monochloramine. J Appl Microbiol 88:22–30. [DOI] [PubMed] [Google Scholar]
- Cooksey RC, Jhung MA, Yakrus MA, Butler WR, Adekambi T, Morlock GP, Williams MM, Shams AM, Jensen BJ, Morey RE, et al. 2008. Multiphasic approach reveals genetic diversity of environmental and patient isolates of Mycobacterium mucogenicum and Mycobacterium phocaicum associated with an outbreak of Bacteremias at a Texas hospital. Appl Environ Microbiol 74:2480–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert TC, Rodgers MR, Reyes AL, Stelma GN Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol 65:2492–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote MA, Huitt G. 2006. Infections due to rapidly growing mycobacteria. Clin Inf Dis 42:1756–1763. [DOI] [PubMed] [Google Scholar]
- Deines P, Sekar R, Husband P, Boxall J, Osborn A, Biggs C. 2010. A new coupon design for simultaneous analysis of in situ microbial biofilm formation and community structure in drinking water distribution systems. Appl Microbiol Biotech 87:749–756. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Forster TS, Murga R, Brown E, Lucas C, Carpenter J, Fields BS. 2005. Legionella pneumophila associated with the protozoan Hartmannella vermiformis in a model multi-species biofilm has reduced susceptibility to disinfectants. Biofouling 21:1–7. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, El-Sayed I, El-Sayed MA. 2004. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol 70:4980–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler S, Christen R, Holtje C, Westphal P, Botel J, Brettar I, Mehling A, Hofle MG. 2006. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol 72:1858–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming GA, Frangoul H, Dermody TS, Halasa N. 2006. A cord blood transplant recipient with Mycobacterium mucogenicum central venous catheter infection after infusion of tap water. Pediatr Infect Dis J 25:567–569. [DOI] [PubMed] [Google Scholar]
- Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, Donlan RM. 2005. Statistical assessment of a laboratory method for growing biofilms. Microbiology 151:757–762. [DOI] [PubMed] [Google Scholar]
- Hong P-Y, Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu W-T. 2010. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Appl Environ Microbiol 76:5631–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S, Cameron S, Streifel A, Yakrus MA, Kairis F, Peacock K, Besser J, Cooksey RC. 2004. An outbreak of bacteremias associated with Mycobacterium mucogenicum in a hospital water supply. Inf Control Hosp Epi 25:1042–1049. [DOI] [PubMed] [Google Scholar]
- Kressel AB, Kidd F. 2001. Pseudo-outbreak of Mycobacterium chelonae and Methylobacterium mesophilicum caused by contamination of an automated endoscopy washer. Infect Control Hosp Epidem 22:414–418. [DOI] [PubMed] [Google Scholar]
- Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V. 2002. Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Appl Environ Microbiol 68:1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeChevallier MW, Schulz W, Lee RG. 1991. Bacterial nutrients in drinking water. Appl Environ Microbiol 57:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livni G, Yaniv I, Samra Z, Kaufman L, Solter E, Ashkenazi S, Levy I. 2008. Outbreak of Mycobacterium mucogenicum bacteraemia due to contaminated water supply in a paediatric haematology-oncology department. J Hosp Inf 70:253–258. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Albrechtsen HJ, Arvin E, Molin S. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl Environ Microbiol − 1:8611–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeters GA, Kippin JS, LeChevallier MW. 1986. Injured coliforms in drinking water. Appl Environ Microbiol 51:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga R, Forster TS, Brown E, Pruckley JM, Fields BS, Donlan RM. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121–3126. [DOI] [PubMed] [Google Scholar]
- Norton CD, LeChevallier M, Falkinham III JO. 2004. Survival of Mycobacterium avium in a model distribution system. Water Res 38:1457–1466. [DOI] [PubMed] [Google Scholar]
- Paris T, Skali-Lami S, Block JC. 2007. Effect of wall shear rate on biofilm deposition and grazing in drinking water flow chambers. Biotech Bioeng 97:1550–1561. [DOI] [PubMed] [Google Scholar]
- Poitelon JB, Joyeux M, Welte B, Duguet JP, Prestel E, Lespinet O, DuBow MS. 2009. Assessment of phylogenetic diversity of bacterial microflora in drinking water using serial analysis of ribosomal sequence tags. Water Res 43:4197–4206. [DOI] [PubMed] [Google Scholar]
- Revetta RP, Matlib R, Santo Domingo JW. 2011. 16S rRNA gene sequence analysis of drinking water using RNA and DNA extracts as targets for clone library development. Curr Microbiol 63:50–59. [DOI] [PubMed] [Google Scholar]
- Revetta RP, Pemberton A, Lamendella R, Iker B, Santo Domingo JW. 2010. Identification of bacterial populations in drinking water using 16S rRNA-based sequence analyses. Water Res 44:1353–1360. [DOI] [PubMed] [Google Scholar]
- Rose LJ, Rice EW, Hodges L, Peterson A, Arduino MJ. 2007. Monochloramine inactivation of bacterial select agents. Appl Environ Microbiol 73:3437–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, Adley CC. 2010. Sphingomonas paucimobilis: a persistent gram-negative nosocomial infectious organism. J Hosp Inf 75:153–157. [DOI] [PubMed] [Google Scholar]
- Schulze-Röbbecke R, Janning B, Fischeder R. 1992. Occurrence of mycobacteria in biofilm samples. Tuberc Lung Dis 73:141–144. [DOI] [PubMed] [Google Scholar]
- Seidel CJ, McGuire MJ, Summers RS, Via S. 2005. Have utilities switched to chloramines? J Am Water Works Assoc 97:87–97. [Google Scholar]
- September SM, Brozel VS, Venter SN. 2004. Diversity of nontuberculoid Mycobacterium species in biofilms of urban and semiurban drinking water distribution systems. Appl Environ Microbiol 70:75−1–7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams AM, Rose LJ, Hodges L, Arduino MJ. 2007. Survival of Burkholderia pseudomallei on environmental surfaces. Appl Environ Microbiol 73:8001–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões LC, Simões M, Vieira MJ. 2007. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl Environ Microbiol 73:6192–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões LC, Simões M, Vieira MJ. 2010. Influence of the diversity of bacterial isolates from drinking water on resistance of biofilms to disinfection. Appl Environ Microbiol 76:6673–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed KA, Falkinham III JO. 2006. Effect of growth in biofilms on chlorine susceptibility of Mycobacterium avium and Mycobacterium intracellulare. Appl Environ Microbiol 72:4007–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Griebe T, Srinivasan R, Chen CI, Yu FP, deBeer D, McFeters GA. 1994. Comparison of respiratory activity and culturability during monochloramine disinfection of binary population biofilms. Appl Environ Microbiol 60:1690–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RH, Falkinham JO III, Norton CD, LeChevallier MW. 2000. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl Environ Microbiol 66:1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency. 1999. Alternative disinfectants and oxidants guidance manual. EPA 815-R-99–014. Chapter 6: Chloramines. Washington (DC): Government Printing Office. p. 1–29. [Google Scholar]
- United States Environmental Protection Agency. 2006. 2006 Edition of the Drinking Water Standards and Health Advisories. EPA 822-R-06–013. Washington (DC): Government Printing Office. p. 11. [cited 2012 Feb 9]. Available from: http://water.epa.gov/lasregs/rulesregs/sdwa/ [Google Scholar]
- Van der Kooij D. 1992. Assimilable organic carbon as an indicator of bacterial regrowth. J Am Water Works Assoc 84:57–65. [Google Scholar]
- Williams MM, Santo Domingo JW, Meckes MC, Kelty CA, Rochon HS. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J Appl Microbiol 96:954–964. [DOI] [PubMed] [Google Scholar]
- Williams MM, Yakrus MA, Arduino MJ, Cooksey RC, Crane CB, Banerjee SN, Hilborn ED, Donlan RM. 2009. Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl Environ Microbiol 75:2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]