Abstract
Neurotrophic receptor kinase (NTRK) fusions are actionable oncogenic drivers of multiple pediatric and adult solid tumors, and tropomyosin receptor kinase (TRK) has been considered as an attractive therapeutic target for “pan-cancer” harboring these fusions. Currently, two generations TRK inhibitors have been developed. The representative second-generation inhibitors selitrectinib and repotrectinib were designed to overcome clinic acquired resistance of the first-generation inhibitors larotrectinib or entrectinib resulted from solvent-front and gatekeeper on-target mutations. However, xDFG (TRKAG667C/A/S, homologous TRKCG696C/A/S) and some double mutations still confer resistance to selitrectinib and repotrectinib, and overcoming these resistances represents a major unmet clinical need. In this review, we summarize the acquired resistance mechanism of the first- and second-generation TRK inhibitors, and firstly put forward the emerging selective type II TRK inhibitors to overcome xDFG mutations mediated resistance. Additionally, we concluded our perspectives on new challenges and future directions in this field.
Key words: NTRK fusions, TRK kinase, Clinical resistance, xDFG mutations, Selective type II inhibitors
Graphical abstract
This review mainly summarizes the acquired resistance mechanisms of the first- and second-generation TRK inhibitors and firstly update the emerging selective type II TRK inhibitors to overcome the xDFG mutations mediated resistance.
1. Introduction
Tropomyosin receptor kinase (TRK) family including TRKA, TRKB and TRKC are well-known cell surface transmembrane receptor tyrosine kinases, and encoded by the neurotrophic receptor kinase 1, 2 and 3 genes (NTRK1, NTRK2 and NTRK3), respectively1,2. Under normal physiological conditions, the binding of TRKs to neurotrophin ligands induces TRKs dimerization, phosphorylation and thereby activation of the downstream signaling pathways, including RAS/MAPKs, PI3K/AKT and PLC-γ1 pathways, which is essential for maintaining cell proliferation, differentiation and even apoptosis3,4. However, TRKs can be constitutively activated via NTRK gene fusions in the pathogenesis of human malignant cancers, in which the 3′ region of the NTRK gene, including the kinase domain, achieves an in-frame fusion to the 5′ sequence of fusion partner gene5,6. The first NTRK fusion gene TPM3-NTRK1 was discovered in human colorectal carcinoma in 19827,8, and to date, over 100 different NTRK fusion partners have been identified in various cancers9,10. TRK proteins have been considered as attractive “pan-cancer” targets for the treatment of various cancers harboring NTRK fusions11. Because most oncogenic forms of NTRK fusions alter or eliminate the extracellular domain, traditional monoclonal antibody therapy is not effective. Thus, the main approach to target oncogenic NTRK fusions is focused on TRKs small molecule inhibitors12, 13, 14.
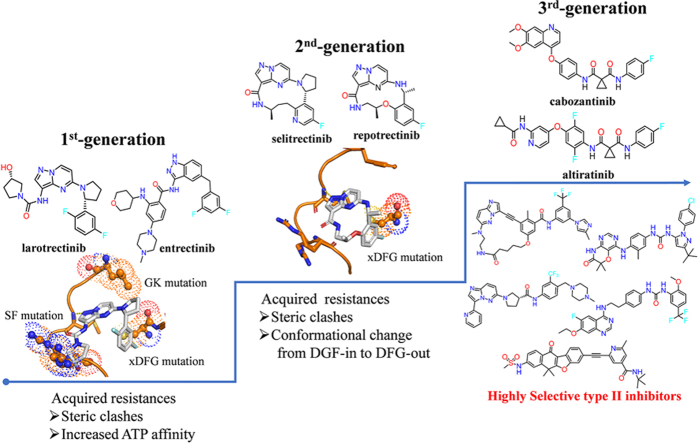
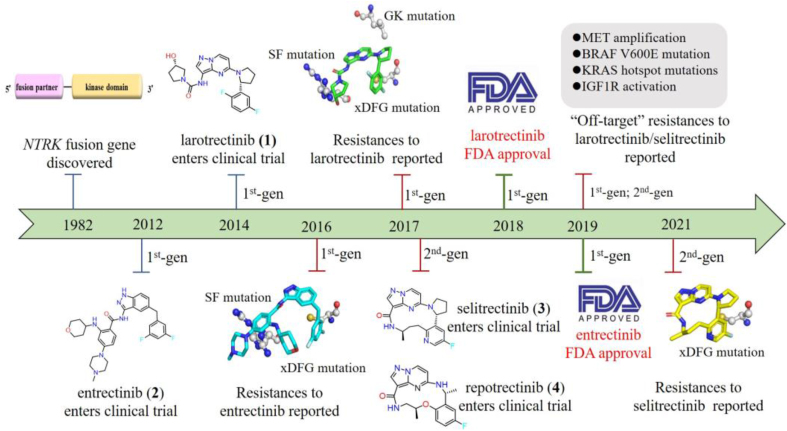
Larotrectinib (1, Vitrakvi®) and entrectinib (2, Rozlytrek®) are two first-generation TRK inhibitors approved by the U.S. Food and Drug Administration (FDA) in 2018 and 2019, respectively (Fig. 1)15, 16, 17, 18. Both inhibitors achieved rapid and massive clinical responses in adult and pediatric patients with advanced solid tumors harboring NTRK fusions19, 20, 21. However, drug resistances against larotrectinib and entrectinib have emerged in clinic. Point mutations in the kinase domain (KD) of TRK, including solvent-front (SF) mutations, gatekeeper (GK) mutations and xDFG motif mutations, are the main resistance mechanisms to these first-generation TRK inhibitors (Fig. 1). SF mutations located on the solvent front region of TRKs are commonly a smaller glycine substituted by arginine or glutamic acid with a larger side chain, e.g., TRKAG595R, TRKBG639R, TRKCG623R/E mutations12. GK mutations anchor on the gatekeeper site of TRKs, which is an aromatic phenylalanine mutated as an alkyl leucine, e.g., TRKAF589L, TRKBF633L, TRKCF617L 12. The xDFG mutations map to the ‘x’ position of the xDFG motif with the ‘x’ indicating the position preceding the activation loop DFG motif. And xDFG mutations containing a small glycine are commonly replaced by cysteine, serine or alanine with large volume, e.g., TRKAG667C/A/S, TRKBG709C/A/S and TRKCG696C/A/S 12. These point alterations in the kinase domain of TRKs cause steric hindrances and/or increase the affinity of ATP to the mutant kinases, thus compromising the binding of the first-generation TRK inhibitors21, 22, 23, 24, 25.
Figure 1.
Development pipeline of TRK inhibitors and associated drug resistances.
Subsequently, second-generation TRK inhibitors were developed to overcome these resistances. The representatives, macrocycle-based selitrectinib (3, Loxo-195) and repotrectinib (4, TPX-0005), are currently undergoing phase II and phase III clinical trials, respectively (Fig. 1). Although the two drugs displayed significant clinical effects against SF and GK mutations mediated resistances24,26,27, they showed limited efficacy against tumors with acquired xDFG mutations and compound mutations28, 29, 30. Moreover, several second-generation TRK inhibitors, such as taletrectinib (DS-6051b, phase II, NCT04395677)31, PBI-200 (phase I, NCT04901806)32, SIM1803-1A (phase I, NCT04671849)33 etc., are undergoing early clinical trials and the related drug resistances are not presented yet. Recently, several reviews described the resistance mechanisms of first-generation TRK inhibitors and the clinical activities of developed second-generation TRK inhibitors12,27,34, 35, 36. However, no review includes the detailed analysis of the resistance mechanisms of both first- and second-generation inhibitors and the potential strategies that may overcome the resistances of second-generation inhibitors. In this review, we focus on summarizing reported resistance mechanisms of the first- and second-generation TRK inhibitors and giving the insight of emerging type II TRK inhibitors that effectively address the xDFG alterations.
2. Structure and downstream signaling pathways of TRK
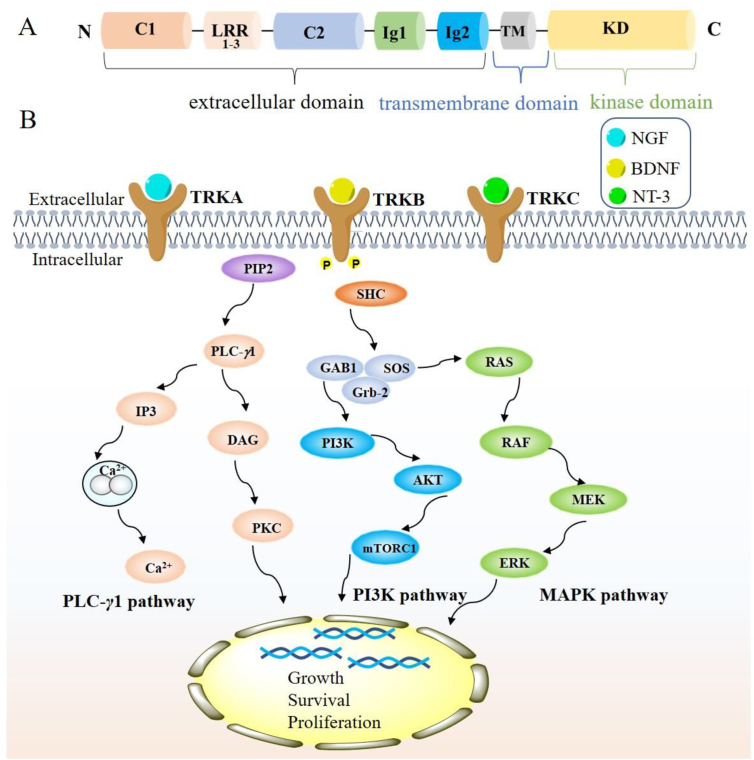
The full-length structures of TRKA, TRKB and TRKC are highly homologous37,38, which are mainly composed of three parts: an extracellular domain, a transmembrane domain (TM) and an intracellular kinase domain (KD) (Fig. 2A). The extracellular domain of TRK consists of two cysteine-rich clusters (C1‒2), three leucine-rich regions (LRR1‒3) and two immunoglobulin-like domains (Ig1, Ig2), which are responsible for ligand recognition and binding39, 40, 41. The KD of TRK possesses the similar architectural features to other protein kinases, which contains one N-terminal lobe mainly composed of β-strands, one C-terminal lobe constituted of α-helices and a hinge linker bridging these two lobes42. Although TRKA, TRKB, TRKC share similar tertiary protein structure, their extracellular neurotrophin binding ligands are different. TRKA binds preferentially to nerve growth factor (NGF)2,42, TRKB specifically binds to brain-derived neurotrophic factor (BDNF)43,44, while TRKC selectively binds to neurotrophin-3 (NT-3)45,46. Extracellular neurotrophin ligands binding to TRK receptors results in receptor dimerization, autophosphorylation and eventually activates the kinase activity of TRK (Fig. 2B)3,4. The NGF binding to TRKA triggers the activating of downstream rat sarcoma oncogene (RAS)-mitogen-activated protein kinase (MAPK) signaling pathways, controlling cell growth and proliferation2. The NGF/TRKA pathway also plays an important role in mediating chronic pain, inflammation, itch and the survival and differentiation of sympathetic and sensory neurons47. The BDNF-TRKB signaling causes the activation of RAS-extracellular-signal-regulated kinase (ERK), phospholipase C-γ1 (PLC-γ1), phosphatidylinositol-3-kinase (PI3K) pathways, regulating neural cell plasticity, survival and metabolism43. The NT-3 binding to TRKC stimulates PI3K-v-AKT murine thymoma viral oncogene homolog (AKT) pathway, which is critical for sustaining cell survival and preventing apoptosis45. However, TRK aberrations, including NTRK fusion, mutation and TRK overexpression, can abnormally activate TRK in a neurotrophin ligands-independent manner, leading continuously activation of downstream PI3K/AKT, RAS/MAPK, and PLC-γ1 pathways and high risks of carcinogenesis14,48.
Figure 2.
Overview of the structure and major signal transduction pathways of TRK. (A) A cartoon shows three domains of TRK: an extracellular domain containing two cysteine-rich clusters (C1, C2), three leucine-rich regions (LRR1‒3), two immunoglobulin-like domains (Ig1, Ig2); a transmembrane domain (TM) and an intracellular kinase domain (KD). (B) TRK mediates the activation of RAS–RAF–MEK–ERK, PI3K–AKT, and PLC-γ1–DAG–PKC signaling pathways.
3. NTRK fusions
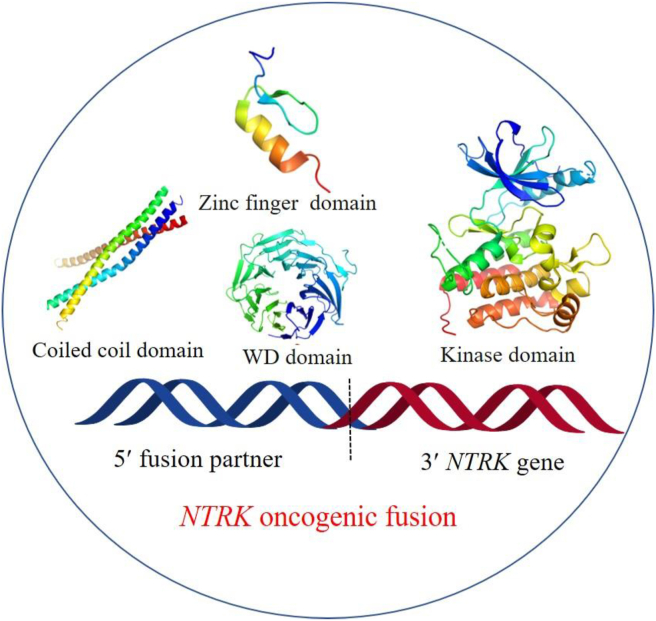
NTRK fusions are the most common mechanisms of oncogenic TRK activation. Fusions involving all three NTRK genes have been identified as oncogenic drivers in a broad range of pediatric and adult tumors5,6,12. These fusions arise from inter- or intrachromosomal rearrangements, in which the 3′ region of NTRK gene containing kinase domain is fused with the 5′ fusion partner gene5,49. The expressed NTRK fusion protein commonly lacks an extracellular ligand binding domain, but contains a 5′ gene partner encoding an oligomerization domain, such as coiled-coil domains, zinc finder domains or WD repeats, which are required for full activation of downstream kinases (Fig. 3). Driving by the 5′ upstream partner, the NTRK chimeric oncoprotein can be constitutively activated through a ligand-independent manner, leading persistent activation of TRK downstream signaling pathways and high risks of oncogenesis50, 51, 52. Since the first NTRK fusion protein TPM3-NTRK1 was detected in colon cancer in 1982, over 100 different 5′ fusion partners have been identified in more than 20 types of human tumors8,10,12,53,54. Among three NTRK isoforms, NTRK1 and NTRK3 fusions are widely distributed in a series of different cancer types, while NTRK2 fusions are mainly detected in central nervous system (CNS) tumors54, 55, 56, 57. NTRK fusions are the major oncogenic driving events in some rare tumors, such as infantile fibrosarcoma (IFS)58, secretory breast cancer (SBC)59, mammary analog secretory carcinoma (MASC)60, and congenital mesoderm nephroma (CMN)61. The prevalence of NTRK fusions can reach 90% or higher in these rare tumors, with the ETV6-NTRK3 fusion diagnosed most6,62, 63, 64, 65, while NTRK fusions are found at low incidence (<5%) in a series of common cancer types, such as lung cancer, breast cancer, colorectal cancer, pancreatic cancer etc.21,12,66,67. In addition to NTRK fusions, other TRK aberrations have also been found in some tumors, including TRK point mutations68, 69, 70, 71, 72, splice variants48,68,73, 74, 75, and TRK overexpression76, 77, 78.
Figure 3.
The structure of NTRK fusion gene.
4. Resistances to the first-generation TRK inhibitors
DFG motif, a highly conserved three amino acids (Asp-Phe-Gly) in the activation loop, directly affects the conformation of kinases. In an active or DFG-in conformation, the Asp of DFG motif directs the ATP binding site. In an inactive or DFG-out conformation, the Asp points outward the ATP binding site and the Phe of DFG motif flips into the ATP active site to form an accessible hydrophobic back pocket79. TRK small molecule inhibitors can be classified into three different types, namely type I, type II and type III, according to their different binding modes with TRK kinases. Inhibitors that bind to the ATP-binding site of kinase with an active conformation (DFG-in) are defined as type I inhibitors80, such as the first- and second-generation TRK inhibitors larotrectinib, entrectinib, selitrectinib, repotrectinib, taletrectinib etc.14. Inhibitors that bind to the ATP-binding site and an adjacent hydrophobic back pocket of kinase with an inactive conformation (DFG-out) are classified as type II inhibitors, e.g., multi-targeted kinase inhibitors cabozantinib, foretinib, ponatinib80. Inhibitors that bind to a hydrophobic pocket distant from the ATP-binding site of kinase are defined as type III inhibitors, which can induce conformational changes to regulate the kinase activity80.
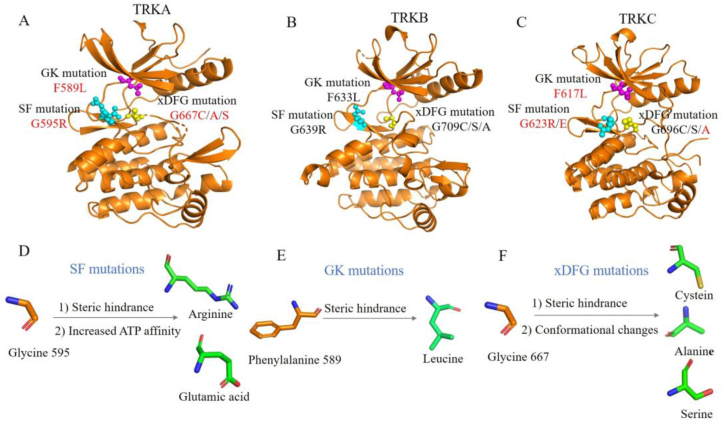
Despite the initial marked tumor shrinkage achieved by the treatment with larotrectinib and entrectinib19, 20, 21, unfortunately, duration of response was eventually limited by acquired resistances. Tumor sequencing or sequential circulating tumor DNA (ctDNA) obtained from patient's tumor specimen or plasma at progression revealed that point mutations in the KD of TRKs are the main mechanisms of acquired resistances21, 22, 23, 24, 25. There are three major point mutations in the KD of TRKs: solvent-front (SF) mutations, gatekeeper (GK) mutations as well as xDFG motif mutations (Fig. 4A‒C)19,22. SF mutations located on the solvent front region of TRKs are commonly a smaller glycine substituted by arginine or glutamic acid with a larger side chain, e.g., TRKAG595R, TRKBG639R, TRKCG623R/E mutations (Fig. 4D), which are paralogous to ALKG1202R and ROS1G2032R mutations81. Structural modeling indicates that the large side chain of mutant arginine or glutamic acid introduce steric hindrance with the hydroxypyrrolidine group of larotrectinib or methylpiperazine motif of entrectinib (Fig. 5B‒C)22,24,26. Besides, biochemical analyses demonstrate that the TRKAG595R mutation leads the increased binding affinity of ATP to the mutant kinase (Km 6 μmol/L for TRKAG595R vs. 51 μmol/L for TRKA), thus improving the intrinsic kinase activity24. Both steric hindrances and increased intrinsic kinase activity impair the binding of larotrectinib and entrectinib.
Figure 4.
Representative point mutations in the KD of TRKA (PDB code 7VKN) (A); TRKB (PDB code 4ASZ) (B); TRKC (PDB code 6KZD) (C). The solvent-front (SF), gatekeeper (GK), xDFG substitutions are shown in cyan, magenta, yellow spheres, respectively. Mutations detected in patients are shown in red font. Resistance mechanisms caused by SF mutations (D); GK mutations (E); xDFG mutations (F).
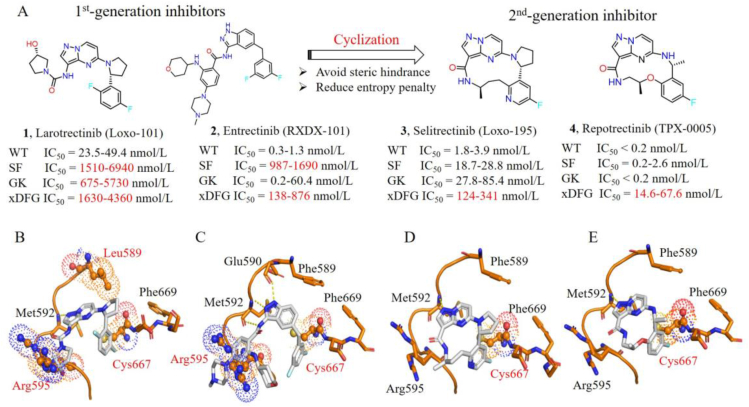
Figure 5.
Chemical structures, cell-proliferation inhibitory activities and binding modes of the first- and second-generations TRKs inhibitors. (A) Chemical structures of larotrectinib, entrectinib, selitrectinib, repotrectinib and their cell-proliferation inhibitory activities against Ba/F3 cells engineered with wild-type (WT), solvent-front (SF) mutant, gatekeeper (GK) mutant and xDFG mutant TRKs. Structural models showing steric clashes between solvent-front (G595R), gatekeeper (F589L) or xDFG (G667C) mutations and larotrectinib (B); entrectinib (C); selitrectinib (D); repotrectinib (E) (generated from PDB code 7VKN). Steric clashes are highlighted in spheres.
The GK mutations anchor on the gatekeeper site of TRKs, which is an aromatic phenylalanine mutated as a alkyl leucine, e.g., TRKAF589L, TRKBF633L, TRKCF617L (Fig. 4E)12. Structural modeling indicates that the mutant leucine may generate steric clash with pyrrole fragment in larotrectinib and hinder its binding (Fig. 5B)12,82. Whereas entrectinib is a methylene group at this position, which effectively avoids steric hindrance with the mutated Leu589 (Fig. 5C)83. The xDFG mutations locate on the activate loop of kinase, which contains a small glycine replaced by cysteine, serine or alanine with large volume, e.g., TRKAG667C/A/S, TRKBG709C/A/S and TRKCG696C/A/S (Fig. 4F)12. These mutations cause potential steric clashes between the difluorophenyl groups of larotrectinib and entrectinib and mutated residues (Fig. 5B‒C)22,24,36.
In order to overcome the acquired point mutations of larotrectinib and entrectinib, great efforts have been made in exploiting the second-generation TRK inhibitors12,14. The rigid macrocyclic selitrectinib (3) and repotrectinib (4) are two representative second-generation TRK inhibitors, which are undergoing phase II and phase III clinical studies, respectively. Selitrectinib is a selective pan-TRKs inhibitor24, while repotrectinib is a multi-targeted TRK/anaplastic lymphoma kinase (ALK)/c-ros oncogene 1 (ROS1) inhibitor26. Selitrectinib and repotrectinib are designed by a cyclized conformationally restricted strategy based on the scaffold of larotrectinib, thus avoiding steric hindrances with the mutated residues and reducing the binding entropy penalty (Fig. 5A)24,26. Selitrectinib and repotrectinib effectively induce visible tumor regression in patients with the SF acquired resistance mutations (e.g., TRKAG595R, TRKCG623R and TRKCG623E) relapsed from the treatment of larotrectinib (1) or entrectinib (2)23,24,26,84,85. An X-ray cocrystal structure of repotrectinib with TRKAG595R demonstrates that it precisely anchors in the ATP-binding pocket without any extra motifs extending to the solvent area to clash with the bulky Arg595 (Fig. 5E)24,83. The docking model of selitrectinib with TRKAG595R indicates that it displays similar binding mode to that of repotrectinib (Fig. 5D). Besides, selitrectinib and repotrectinib with a compact macrocyclic structure can better accommodate GK mutations and show improved in vitro kinase inhibitory activity against TRKAF589L mutation, compared with larotrectinib (Fig. 5A)24,83. However, structural modeling indicates that the mutated Cys667 still generates steric hindrances with the fluoroaromatic group in selitrectinib and repotrectinib and hinders their binding (Fig. 5D‒E)28,29.
5. Emerging resistances to selitrectinib and repotrectinib
Despite the superior activities of selitrectinib and repotrectinib against a variety of SF and GK alterations in vitro and in vivo, acquired drug resistances still emerged. Clinical trials and in vitro experimental models indicate multiple resistance mechanisms of selitrectinib and repotrectinib, which can be classified into two categories: TRK-dependent and TRK-independent resistance mechanisms. The former involves acquired xDFG mutations and compound mutations (such as SF/xDFG, SF/GK compound mutations)28,29,36,86, while the latter is associated to abnormal activation of TRK bypass signaling, such as MAPK pathway30,85.
5.1. TRK-dependent resistance mechanisms
5.1.1. xDFG mutations
It was reported that the xDFG mutations in the KD of TRKs, e.g., TRKAG667C/A/S alterations, result in acquired resistances to selitrectinib in breast and colorectal cancer patients28. One patient harboring a TPM3-NTRK1-fused TRKAG595R-mutant sarcoma progressed on selitrectinib therapy. Target next-generation sequencing (NGS) assays based on the tumor samples revealed the acquisition of TRKAG667C mutation and loss of TRKAG595R mutation28. Another patient with LMNA-NTRK1-fused, TRKAG595R-mutant breast cancer developed resistance to selitrectinib after 2 months of treatment. Cell-free DNA (cfDNA) collected from plasma samples and molecular profiling using NGS platform demonstrated a novel TRKAG667C alteration28. Besides, a TRKAG595R mutated primary colorectal cancer cell line developed resistance to repotrectinib following chronic drug exposure due to an emerged TRKAG667C mutation28. CellTiter-Glo-based proliferation and Western blot assays confirmed that repotrectinib fails to suppress cell growth and TRK-activated downstream signaling, suggesting that the xDFG mutation TRKAG667C may also confer acquired resistance to it28. The decreased potencies of selitrectinib and repotrectinib against xDFG alterations have also confirmed by in vivo cell proliferation inhibitory activities with IC50 values of 124–341 nmol/L and 14.6–67.6 nmol/L against Ba/F3 cells engineered with xDFG mutant TRKs, respectively, which are more than 68-fold less potent than their wide-type counterpart (Fig. 5A)28,83. Besides, taletrectinib, another second-generation TRK inhibitor, also shows poor inhibitory efficacy towards TRKAG667C mutation with an IC50 value of 304.1 nmol/L against Ba/F3 TPM3-NTRK1G667C cells31.
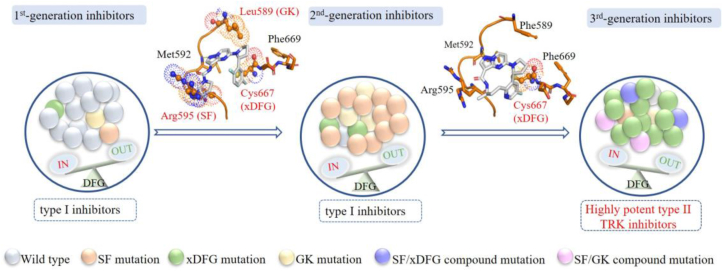
Although the cyclization strategy effectively avoids steric clashes in the solvent front region, the macrocycle inhibitors selitrectinib and repotrectinib do not essentially change the binding conformation of the fluoroaromatic group in the original first generations. Similar to the resistance mechanisms of larotrectinib and entrectinib14,16, the mutated residues (e.g., alanine, cysteine or serine) at Gly667 in TRKA still cause steric impedances with the fluoroaromatic group and disturb the binding of selitrectinib and repotrectinib (Fig. 5D‒E)28,29. Cocco E et al. reported that the xDFG mutations (TRKAG667C or TRKCG696C) induced conformational changes of TRK to preferentially adopt a DFG-out state28,29, thus favorable for binding with type II inhibitors (Fig. 6). Interestingly, several multi-targeted type II inhibitors, e.g., cabozantinib, foretinib, ponatinib, indeed display high potencies against TRK xDFG alterations in vitro and in vivo28,29. In a word, the steric clashes and conformational change resulted from the xDFG mutations will abrogate the binding of most type I-based 1st- and 2nd-generation TRK inhibitors and sensitize to type II inhibitors, providing insights into rational type II drug design to address recalcitrant resistant alterations.
Figure 6.
Proposed therapeutical pipeline for NTRK fusion cancers. Upon diagnosed, tumors mainly harbor wild-type NTRK fusion cells. These wide-type kinases adopt a DFG-in active conformation and type I based 1st-generation TRK inhibitors larotrectinib or entrectinib are approved for use. Subsequently developed point mutations resistances and SF alterations dominate. The kinases remain in DFG-in active conformation and type I based 2nd-generation inhibitors are applied. At progression, the emerged xDFG mutations and some compound mutations cause steric clashes, impeding the binding of type I based 2nd-generation inhibitors, and induce the kinase preferring a DFG-out inactive conformation, favoring the binding of type II inhibitors. Each colored ball represents a distinct clone. Steric clashes are highlighted in spheres.
5.1.2. Compound mutations
In addition to the xDFG mutations, some compound mutations were also detected in patients after the treatment of selitrectinib28,86. A patient with a LMNA-NTRK1G595R-mutant colorectal cancer developed resistance to selitrectinib after 11 months of treatment. Targeted next-generation sequencing of the resistant tumor revealed a novel TRKAG667A mutation and retained TRKAG595R mutation. RNA sequencing indicated that TRKAG595R and TRKAG667A mutations are located on the same allele in cis with similar allele frequencies (33% and 27%, respectively)28. Selitrectinib and repotrectinib display decreased inhibitory activities against TRKAG595R/G667C (SF/xDFG) compound mutation with IC50 values of 596 and 205 nmol/L, respectively28. Besides, a patient with ETV6-NTRK3 fused infantile fibrosarcoma underwent resistance to selitrectinib due to a new TRKCF617L alteration in cis-form with TRKCG623R, which was definitized as SF/GK compound mutation86. Selitrectinib displays weak antiproliferative activity against Ba/F3 cells expressed TRKAG595R/F589L alteration with an IC50 value of 468 nmol/L, which is 100-fold less potent than that of wild-type83. Although the resistance mechanisms of compound mutations have not been illustrated in detail, there is an urgent need to exploit novel TRK inhibitors to address recalcitrant xDFG or compound mutations.
5.2. TRK-independent resistance mechanisms
Similar to other receptor tyrosine kinase inhibitors, off-target alterations also mediated acquired resistances to TRK inhibitors30,87,88. Previous studies have demonstrated acquired BRAFV600E, KRASG12D mutations as well as MET amplification render resistance to the 1st-generation inhibitors larotrectinib and entrectinib in clinic30. These off-target alterations are predicated to TRK-independently activate downstream MAPK signaling pathways and cannot be adequately addressed by the 2nd-generation TRK inhibitor selitrectinib alone. Furthermore, the emerged KRASG12A/V/D substitutions can also cause resistance to patients who receive selitrectinib30,85. Combination therapy of selitrectinib and a MET inhibitor crizotinib can achieve marked tumor shrinkage in a cholangiocarcinoma patient harboring NTRK-fusion and MET amplification30.
6. Type II TRK inhibitors overcome xDFG mutations
6.1. Multi-targeted type II inhibitors
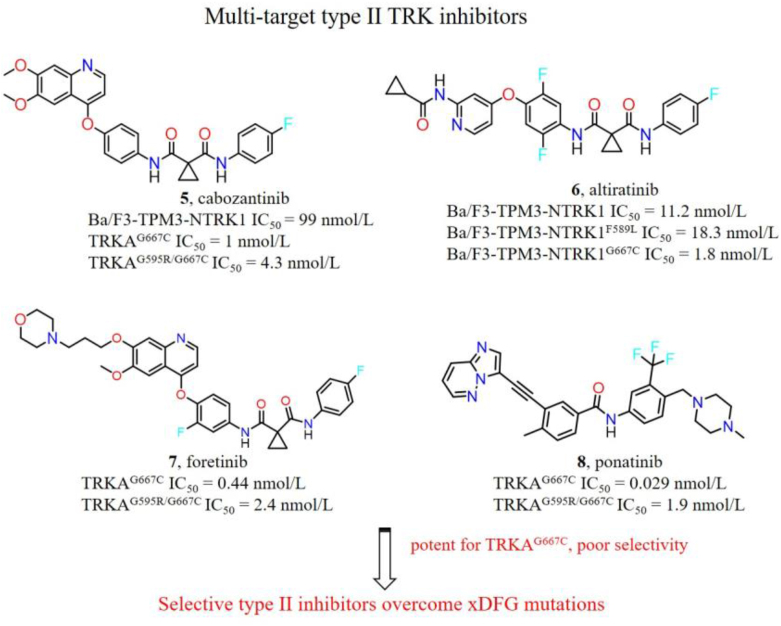
The majority of the 1st- and 2nd-generation TRK inhibitors are type I inhibitors and bind in the DFG-in active conformation of TRK13,14. However, the xDFG mutations stabilize the kinases in DFG-out inactive conformation, thus sensitizing the binding of type II TRK inhibitors28,29. Some multi-targeted type II kinase inhibitors, e.g., cabozantinib (5), altiratinib (6), foretinib (7) and ponatinib (8), preferentially bind to TRK xDFG mutations and exhibit strong in vitro and in vivo inhibitory activities (Fig. 7). Cabozantinib, a multi-kinase inhibitor for hepatocyte growth factor receptor (c-Met), vascular endothelial growth factor receptor 2 (VEGFR2), and rearranged during transfection (RET)89, displays strong biochemical and cellular inhibitory activities against TRKAG667C and Ba/F3-TPM3-NTRK1 cells with IC50 values of 1 and 99 nmol/L, respectively. Furthermore, cabozantinib exhibits potent and durable tumor progression inhibition in selitrectinib resistant (TRKAG667C) patient derived xenografts (PDX) model28. Currently, cabozantinib is undergoing multiple phase II trials in patients with RET fusion-positive advanced non-small cell lung cancer (NSCLC) and those with other genotypes: ROS1 or NTRK fusions or increased MET or AXL activity (NCT01639508). Altiratinib, regarded as a c-Met, tunica intima endothelial kinase 2 (TIE2) and VEGFR2 kinase inhibitor90, suppresses the growth of Ba/F3-TPM3-NTRK1F589L and Ba/F3-TPM3-NTRK1G667C cells with IC50 values of 18.3 and 1.8 nmol/L, respectively. Besides, foretinib and ponatinib also exhibit potent kinase inhibitory efficacies against TRKAG667C mutation (IC50 < 1 nmol/L)28,91. More importantly, cabozantinib, foretinib and ponatinib can strongly inhibit TRKAG595R/G667C compound mutation with IC50 values below 5 nmol/L, while corresponding IC50 values of selitrectinib and repotrectinib are 596 and 205 nmol/L, respectively28. Although the aforementioned type II inhibitors can potently inhibit xDFG alterations in vitro and in vivo, substantial off-target inhibition of these agents may cause poor plasma exposures and high frequency of side effects. It is desired that the focus is put on exploitation of highly selective type II TRK inhibitors.
Figure 7.
Chemical structures of multi-targeted type II TRK inhibitors and their kinase or cellular inhibitory activities towards wild-type (WT) or mutant TRK.
6.2. Selective type II TRK inhibitors
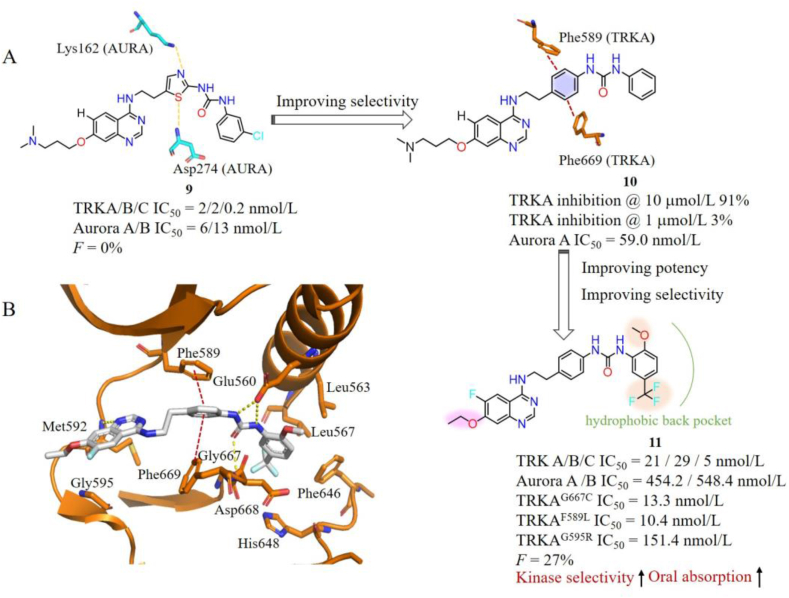
More recently, diverse selective TRK type II inhibitors that can overcome acquired resistance mutations were developed. Li et al.92 reported a series of selective type II TRK inhibitors with a quinazoline scaffold based on compound 9, which is potent against TRKs (IC50 = 0.2–2 nmol/L) and Aurora A/B (IC50 = 6/13 nmol/L) with poor oral bioavailability93. Structure analysis indicated that the thiazole ring of 9 forms crucial hydrogen bond interactions with Lys162 and Asp274 on Aurora A, respectively, while corresponding residues on TRKA are Phe589 and Phe669 (Fig. 8A). In order to improve its TRK selectivity, the thiazole ring of 9 was replaced by a phenyl group to reduce the hydrogen interactions with Aurora A and increase a π‒π interaction with TRKs. The resulting compound 10 indeed shows decreased activity against Aurora A but greatly lost potency towards TRKA92. Considering the back hydrophobic pocket of TRKA is more spacious than that of Aurora A, diverse substituents were introduced to the terminal phenyl ring of 10 to occupy the hydrophobic pocket of TRKA. The final compound 11 with 3-CF3/6-OCH3 phenyl group exhibits improved kinase inhibitory activity for TRKs (IC50 ranging from 5 to 29 nmol/L) and high selectivity over Aurora A/B with IC50 values of 454.2 and 548.4 nmol/L, respectively. Moreover, 11 potently inhibits TRKAG667C, TRKAF589L, TRKCG696A alterations as well as TRKAG595R/G667C with IC50 values of 13.3, 10.4, 22.5 and 19.6 nmol/L, respectively, while exhibiting moderate activities against TRKAG595R and TRKCG623R with IC50 values of 151.4 and 75.4 nmol/L, respectively92. The docking mode of 11 with TRKA illustrates that it anchors the ATP-binding pocket in a type II binding mode with the quinazoline core and urea moiety forming tight hydrogen bonds with Met592, Glu560 and Asp668. The middle phenyl group forms additional π‒π interactions with Phe589 and Phe669. The terminal substituted phenyl group occupies the back pocket of TRKA and makes favorable hydrophobic interactions with surrounding residues (Fig. 8B). Most importantly, 11 exhibits acceptable in vivo pharmacokinetics (PK) profiles in rats with good oral absorption (AUC0‒inf = 3012 ng/mL × h), moderate half-life (t1/2 = 5.1 h) and reasonable oral bioavailability (F = 27.2%). 11 shows good in vivo antitumor efficacy in KM12 xenograft mouse model with TGI value of 64% at a dosage of 75 mg/kg three times per week by orally administered92.
Figure 8.
(A) Optimization process of compound 11. (B) The binding mode of 11 with TRKA (docking based on PDB code 6PL1). 11 and key residues are showed in sticks, hydrogen bonds are presented as yellow dashed lines, and π‒π interactions are presented as red dashed lines.
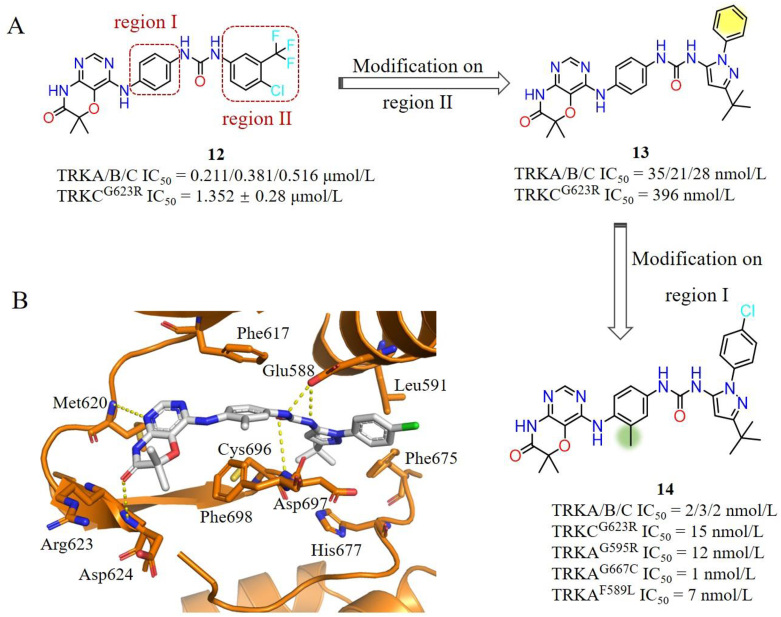
Pan et al.94 screened their in-house kinase inhibitors-like compound library and identified a lead compound 12, which has moderate inhibitory activities for TRKs (IC50 ranging from 0.211 to 0.516 μmol/L) and weak potency for TRKCG623R (IC50 = 1.352 μmol/L) (Fig. 9A). In order to improve its potency against TRKs, the researchers have carried out abundant structural optimizations to increase additional interactions with two spacious pockets in TRKs. One pocket is surrounding residues Phe589 and Phe669 (region I), and another is a big hydrophobic back pocket partly occupied by the “tail” 4-chloro-3-(trifluoromethyl) phenyl moiety of 12 (region II) (Fig. 9A). Firstly, they replaced the 4-chloro-3-(trifluoromethyl) phenyl group of 12 with diverse hydrophobic groups to fill the region II pocket. Compound 13, with a 3-(tert-butyl)-1-phenyl-1H-pyrazole hydrophobic “tail” interacting with Leu591, Phe675, His677 and Asp697, displays improved activities against TRKAWT and TRKCG623R with IC50 values of 35 and 396 nmol/L, respectively (Fig. 9B). Further modifications on the middle phenyl moiety (region I) yielded compound 14 with a meta-methyl, which displays low nanomolar inhibitory activities against both WT and various TRK mutants (TRKAG595R, TRKCG623R, TRKAG667C and TRKAF589L) (IC50 ranging from 0.5 to 26 nmol/L) in biochemical and cellular levels (Fig. 9A). The binding mode of compound 14 with mutant TRKCG623R/G696C shows that 14 occupies the ATP-binding pocket in a type II mode without any steric hindrances with mutant Arg623 and Cys696 (Fig. 9B). Compound 14 displays evident tumor growth inhibition in Ba/F3-TEL-TRKA and Ba/F3-TEL-TRKCG623R allograft mouse models without apparent toxicity94.
Figure 9.
(A) Optimization process of compound 14. (B) The binding mode of compound 14 with TRKCG623R/G696C kinase (generated based on PDB code 6KZD). 14 and key residues are showed in sticks and the hydrogen bonds are presented as yellow dashed lines.
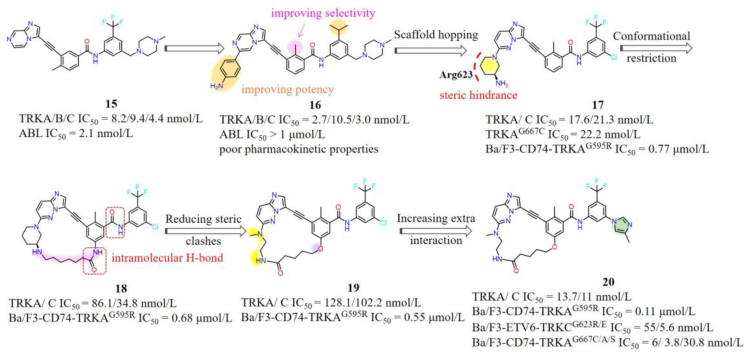
Our group has also developed several selective TRK inhibitors in recent years95, 96, 97, 98, 99. Compound 15 is a potent ABL (IC50 = 2.1 nmol/L) and TRKs (IC50 ranging from 4.4 to 9.4 nmol/L) inhibitor developed in our laboratory100. Switching the methyl group in the middle phenyl moiety from 4′-position to 2′-position effectively improves the TRK selectivity over ABL. The representative compound 16 shows digital nanomolar inhibitory activities against TRKs while sparing ABL (Fig. 10). However, the poor pharmacokinetic properties of 16 hinders its further development95,101. In order to exploit novel selective TRK inhibitors, scaffold hopping strategy was employed to obtain a series of imidazo[1,2-b]pyridazine derivatives based on 16. The resulting compound 17 displays nanomolar inhibitory activities against both WT and G667C mutant, but less potent against the G595R mutant (Ba/F3-CD74-TRKAG595R IC50 = 0.77 μmol/L) (Fig. 10). Structural modeling suggested that the aminopiperidine group of 17 causes steric hindrance with mutated Arg623 (Fig. 11A)98. Considering that cyclized conformationally restricted type I inhibitors (e.g., selitrectinib or repotrectinib) effectively overcome SF mutants, a series of macrocycle-based type II molecules with different sizes were designed. Compound 18 bearing an 8-atom linker displays a minor activity improvement against TRKAG595R with an IC50 value of 0.68 μmol/L for Ba/F3-CD74-TRKAG595R cells. In order to further reduce the steric hindrance with SF mutation, the aminopiperidine of 18 was replaced by a smaller size methylethanediamine yielding compound 19, which shows increased activity against TRKAG595R with an IC50 value of 0.55 μmol/L. 19 with a freely rotating oxygen atom at the linker position effectively prevents intramolecular hydrogen bond between the two amide groups in 18 and maintains key hydrogen bond interactions with Glu560 and Asp668 (Fig. 11B). Introducing extra hydrophilic groups pointing towards the solvent-accessible area generated the representative compound 20, which demonstrates high potencies towards WT and various TRKs variants with IC50 values of 110 and 6 nmol/L against Ba/F3-CD74-TRKAG595R and Ba/F3-CD74-TRKAG667C cells. The binding model of 20 with TRKAG595R indicated that it occupies the ATP binding pocket with a type II binding mode, the imidazo[1,2-b]pyridazine core and carbonyl of amide form key hydrogen bond interactions with Met592 and Asp668, respectively. Methylimidazole ring points to the solvent region and forms an additional π‒π interaction with the flipped Phe669. Methylethanediamine is far away from the mutant Arg595, which can explain the improved potency for the SF mutations (Fig. 11B)98. More importantly, such first macrocycle-based type II TRK inhibitor shows extraordinary kinome selectivity among 373 wild-type kinases. Further drug-like properties optimizations for this series compounds are still in progress in our lab98.
Figure 10.
Optimization process of compound 20 and the optimization strategy employed to address the SF mutation.
Figure 11.
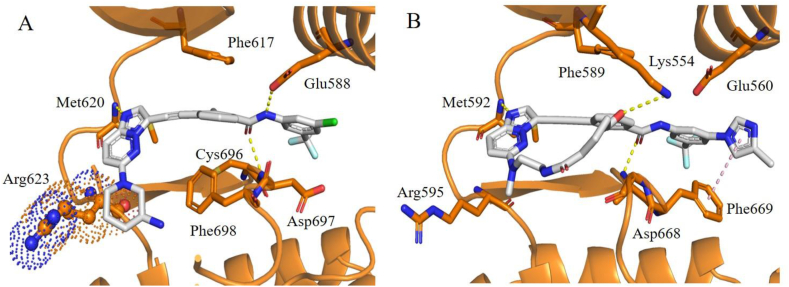
(A) The predicted binding mode of 17 with TRKCG623R (generated from PDB code 6KZD). (B) The predicated binding mode of 20 with TRKAG595R (generated from PDB code 7XAF). Steric clashes are highlighted in spheres. Compounds and key residues are showed in sticks, the hydrogen bonds are presented as yellow dashed lines, and π‒π interactions are presented as pink dashed lines.
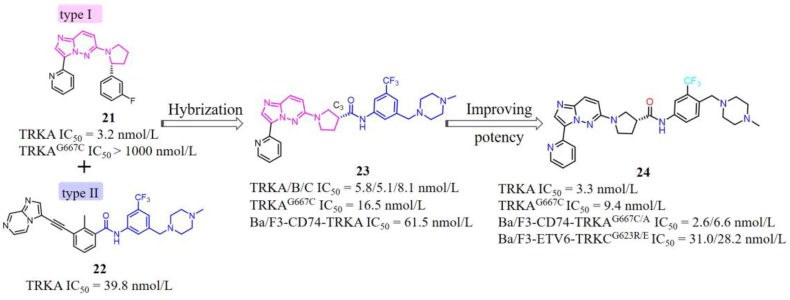
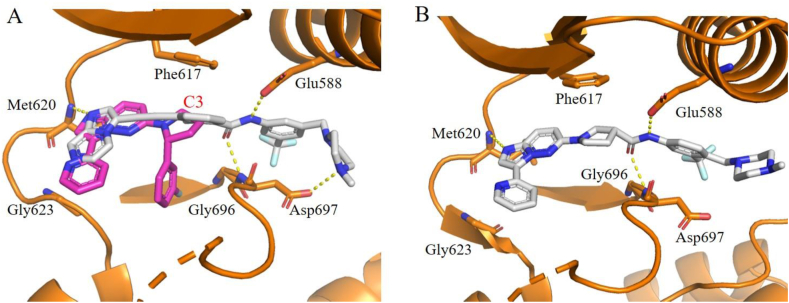
Our group also employed hybridization strategy to design a series of 6-(pyrrolidin-1-yl)imidazo[1,2-b]pyridazine-based type II TRK inhibitors99. Compound 21 is a potent type I TRK inhibitor developed by Novartis, which displays high potency towards WT TRKs but weak inhibitory activity against TRKAG667C mutation (Fig. 12)102. Based on the recent studies that TRK xDFG mutations stabilize the DFG-out inactive conformation and sensitize these mutant kinases to type II inhibitors28,29, we intended to switch this type I inhibitor 21 to a type II inhibitor to overcome xDFG resistances. Superimposition of the X-ray cocrystal structures of 21 and our reported type II inhibitor 2295 indicates that the C3 position of pyrrole ring in 21 is an optimal site to introduce a type II “tail” (Fig. 13A). Subsequently, the hybrid compound 23 was yielded by attaching the N-(3-((4-methyl-piperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)formamide “tail” of 22 and removing the fluorophenyl of 21 that is closed to the xDFG site to avoid steric hindrance99. Compound 23 exhibits potent kinase inhibitory activities against WT TRKs (IC50 ranging from 5.1 to 8.1 nmol/L) as well as TRKAG667C mutation (IC50 = 16.5 nmol/L), but moderate cellular inhibitory activity against Ba/F3-CD74-TRKA cells (IC50 = 61.5 nmol/L). In order to enhance the cellular activities of 23, different types of classical type II “tails” were employed to replace the initial “tail” of 23. And the most potent compound 24 displays superior cellular inhibitory activities against xDFG mutations with IC50 values of 2.6 and 6.1 nmol/L against Ba/F3-CD74-TRKAG667C and Ba/F3-ETV6-TRKCG696C cells, respectively, compared to type I-based larotrectinib and selitrectinib. Encouragingly, 24 also shows potent antiproliferation inhibitory activities against SF alterations with IC50 values of 31.0 and 28.2 nmol/L towards Ba/F3-ETV6-TRKCG623R and Ba/F3-ETV6-TRKCG623E cells (Fig. 12). The docking model of 24 with TRKC suggests that it binds to TRKC in a type II mode with the imidazo[1,2-b]pyridazine core forming key hydrogen bond with Met620 in the hinge region, the amide group forming two hydrogen bonds with Glu588 in the α-C helix and Asp697 in the DFG motif, respectively (Fig. 13B)99. Compound 24 provides a potential novel scaffold for the discovery of potent and selective type II TRK inhibitors to overcome TRK clinical resistances.
Figure 12.
Hybridization of type I TRK inhibitor 21 and type II TRK inhibitor 22 resulted in 24.
Figure 13.
(A) Superimposition of the X-ray cocrystal of compound 21 with TRKA (PDB code 4YNE) and 22 with TRKC (PDB code 6KZC). Compounds 21 and 22 are shown in magenta and gray sticks, respectively. (B) The predicated binding mode of compound 24 complexed with TRKC (docking based on PDB code 6KZC). Compounds and key residues are showed in sticks and the hydrogen bonds are presented as yellow dashed lines.
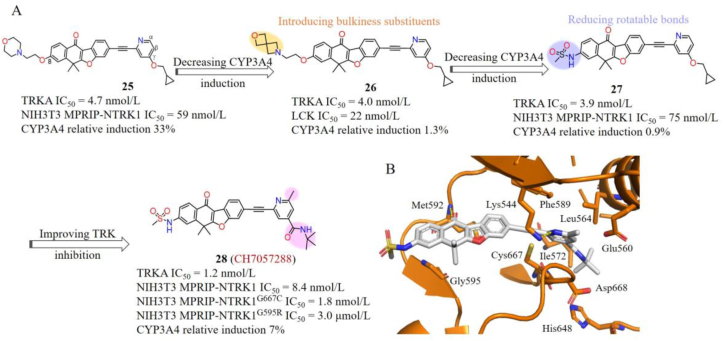
Chugai Pharmaceutical Co., Ltd.103 reported a series of potent type II TRK inhibitors with a unique tetracyclic scaffold. Hit compound 25 was obtained by screening a kinase-inhibitors library, which displayed single digital nanomolar inhibition against TRKs, but moderate antiproliferative activities towards NIH3T3 MPRIP-NTRK1 cells (IC50 = 59 nmol/L) as well as a potential risk of CYP3A4 induction (Fig. 14A). Several reports demonstrated that the CYP3A4 induction is caused by the activation and increased expression of nuclear hormone receptors as well as stabilization interactions of mRNAs or proteins104, 105, 106. Therefore, they intended to conduct some structural modifications on compound 25 to destabilize interactions with mRNAs and/or proteins to attenuate the CYP3A4 induction. The 8-position morpholinoethoxy group extending to the solvent-accessible region was firstly replaced by different bulkier substituents to generate steric repulsion with mRNAs and/or proteins without losing interactions with TRK. Compound 26 with a morpholine substituent at the 8-position significantly reduces CYP3A4 induction. However, 26 also shows strong inhibitory activity against lymphocyte-specific protein kinase (LCK) (IC50 = 22 nmol/L) (Fig. 14A). Subsequently, they aimed to weaken favorable interactions with proteins to decrease the CYP3A4 induction by increasing the rigidification of 25. Compound 27 with a methanesulfonamide group effectively avoids the CYP3A4 induction and maintains TRK inhibitory activity (IC50 = 3.9 nmol/L), selectivity as well as antiproliferation activity towards NIH3T3 MPRIP-NTRK1 cells (IC50 = 75 nmol/L). In order to further improve the potency of 27, the γ-position cyclopropylmethoxy substituent of pyridine was replaced by tert-butyl amide and a methyl was introduced to the α-position of pyridine. The target compound 28 (CH7057288) displays high TRKA inhibitory activity and antiproliferative activity with IC50 values of 1.2 nmol/L against TRKA and 8.4 nmol/L against NIH3T3 MPRIP-NTRK1 cells. The oral administered CH7057288 once a day shows strong antitumor activity in the NTH3T3 MPRIP-NTRK1-fusion NSCLC xenograft model with the TGI value of 171%. More importantly, CH7057288 displays comparable antiproliferative inhibitory activity against NIH3T3 MPRIP-NTRK1G667C cells (IC50 = 1.8 nmol/L) and less potent towards NIH3T3 MPRIP-NTRK1G595R cells (IC50 = 3.0 μmol/L)103,107. The structural modeling of CH7057288 with TRKAG667C indicated that CH7057288 binds to a DFG-out conformation of TRKA with the carbonyl oxygen and pyridine nitrogen forming hydrogen bonds with Met592 and Lys544, respectively. The mutant Cys667 forms favorable sulfur‒π interaction with the pyridine moiety, while the tetracyclic core is very closed to the Gly595, which explains the high potency of CH7057288 against TRKAG667C mutation and weak activity to TRKG595R mutation (Fig. 14B)103,107.
Figure 14.
(A) The discovery of CH7057288 and optimization strategy employed to address the CYP3A4 induction. (B) The predicated binding mode of CH7057288 with TRKAG667C (generated from PDB code 5WR7). CH7057288 and key residues are showed in sticks and the hydrogen bonds are presented as yellow dashed lines.
7. Conclusions and perspectives
Development of new generation TRK inhibitors that can overcome acquired resistance of larotrectinib and entrectinib becomes a research hotspot and unmet clinical need. Large numerous of second-generation macrocyclic inhibitors have been developed and entered in clinical trials. Among them, the representative selitrectinib and repotrectinib successfully combat SF mutations and GK mutations relapsed from the treatment of larotrectinib or entrectinib in patients, but TRK-dependent and TRK-independent resistance still emerge. As TRK-dependent resistance, acquired xDFG mutations are a factor in significant resistance to the second-generation TRK inhibitors. The xDFG mutation can not only cause steric hindrances with the fluoropyrimidine of selitrectinib and the fluorophenyl groups of repotrectinib, but also stable the mutant TRK in a DFG-out inactive conformation, thus limiting the sensitivity of current type I second-generation inhibitors and preferring to type II TRK inhibitors.
Type II TRK inhibitors can be divided into two categories: multi-targeted inhibitors and selective inhibitors. Although several multi-target type II TRK inhibitors can potently combat xDFG variants, side effects caused by substantial off-target inhibition may impede their future development. Currently, a series of selective type II TRK inhibitors with different scaffolds present potent inhibitory activities towards the xDFG mutations in vitro and in vivo. Among them, compound 14 and the first macrocycle-based type II TRK inhibitor 20 display significant inhibitory activities against both xDFG and SF mutations, presenting promising agents for drug discovery and deserving further investigation.
Despite these significant progresses, several limitations and challenges remain to be addressed. First, there are no second generation TRK inhibitor approved for the treatment of acquired resistance of larotrectinib and entrectinib. Although several type II selective TRK inhibitors have been reported, they are all in early biological investigation for cancer therapy. Further exploitation of potent type II TRK inhibitors with druggability becomes a major research focus. Second, most selective type II TRK inhibitors only overcame xDFG mutations and still showed weak efficacy against SF mutations, such as compounds 11 and CH7057288. In addition, compound mutations also occur in cis-form in patients who relapse after the treatment with selitrectinib, but there are no reported inhibitors designed to overcome these mutations. Therefore, there is an urgent need to develop novel TRK inhibitors that can simultaneously overcome major mutations in SF, xDFG and GK. Given that these point mutations are located around the ATP binding pocket, it is suggested that TRK allosteric inhibitors may have the potential to combat existing mutations due to no direct interactions with ATP-binding pocket108,109. On the other hand, it is revealed that proteolysis targeting chimeras (PROTAC) is iterative and less susceptible to target overexpression or mutations, which have been widely used in Bruton's tyrosine kinase (BTK) and Bcr-Abl, etc.110, 111, 112. Currently, a series of TRK allosteric inhibitors have emerged in attempt to develop TRKA selective inhibitors for the treatment of chronic pain based on the NGF-TRKA in pain signaling113, 114, 115, 116, 117. Several TRK degraders were also disclosed with potent degradation efficacy118, 119, 120. However, resistant associated activities of these TRK allosteric inhibitors and degraders were not presented currently. It is necessary to launch further studies to evaluate their potency in overcoming TRK clinical acquisition resistant mutations. Lastly, besides on-target resistances, bypass signaling resistances also cannot be addressed by new generation TRK inhibitors and the strategies to solve these off-target resistances also present a greater unmet need.
Currently, the general strategies for overcoming the TRK point mutations include avoiding steric hindrance or increasing additional interactions, e.g., hydrogen bonds or hydrophobic interactions. Forming a covalent bond with a conserved cysteine or arginine is also an effective way to overcome point mutations in EGFRT790M resistance121, KRASG12C122 and KRASG12R123 mutations. Thus, covalent targeting the mutant residues Cys667 or Arg595 in TRKA may be a novel approach to increase the binding affinity of compounds with mutant kinase. Moreover, it is suggested that the use of computer-aided drug design and the emerging artificial intelligence (AI) generative models may help accelerate the discovery of TRK inhibitors to combat drug resistance. For example, Wang et al.124,125 utilized a KinaFrag web platform to conduct fragment virtual screening and fragment growing and developed some highly potent TRK inhibitors that effectively overcome resistance mediated by point mutations. To date, the cocrystal structures of TRK xDFG mutations with TRK type II inhibitors have not been resolved, therefore more researches are still needed to disclose the detailed mechanisms of the type II inhibitors to overcome drug resistance.
Author contributions
Xiaoyun Lu conceived the project and supervised the project. Shuang Xiang summed up the literature and drafted the manuscript. All authors approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
We gratefully acknowledge the financial support from the National Natural Science Foundation of China (82273763), the Natural Science Foundation of Guangdong Province (2022A1515011939, China), the Opening Project of State Key Laboratory of Respiratory Disease (SKLRD-OP-202313, China), the Opening Project of Guangdong Provincial Key Laboratory of New Drug Design and Evaluation (2020B1212060034, China) and Wang Kuancheng Young Scholar of Jinan University.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
References
- 1.Huang E.J., Reichardt L.F. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107–114. doi: 10.1016/s0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham M.E., Stephens R.M., Kaplan D.R., Greene L.A. Autophosphorylation of activation loop tyrosines regulates signaling by the TRK nerve growth factor receptor. J Biol Chem. 1997;272:10957–10967. doi: 10.1074/jbc.272.16.10957. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham M.E., Greene L.A. A function-structure model for NGF-activated TRK. Embo J. 1998;17:7282–7293. doi: 10.1093/emboj/17.24.7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaishnavi A., Le A.T., Doebele R.C. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5:25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amatu A., Sartore-Bianchi A., Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1 doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pulciani S., Santos E., Lauver A.V., Long L.K., Aaronson S.A., Barbacid M. Oncogenes in solid human tumours. Nature. 1982;300:539–542. doi: 10.1038/300539a0. [DOI] [PubMed] [Google Scholar]
- 8.Martin-Zanca D., Hughes S.H., Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–748. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 9.Amatu A., Sartore-Bianchi A., Bencardino K., Pizzutilo E.G., Tosi F., Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol. 2019;30:viii5–viii15. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aepala M.R., Peiris M.N., Jiang Z., Yang W., Meyer A.N., Donoghue D.J. Nefarious NTRK oncogenic fusions in pediatric sarcomas:too many to Trk. Cytokine Growth Factor Rev. 2022;68:93–106. doi: 10.1016/j.cytogfr.2022.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Pestana R.C., Sen S., Hobbs B.P., Hong D.S. Histology-agnostic drug development―considering issues beyond the tissue. Nat Rev Clin Oncol. 2020;17:555–568. doi: 10.1038/s41571-020-0384-0. [DOI] [PubMed] [Google Scholar]
- 12.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan W., Lakkaniga N.R., Carlomagno F., Santoro M., McDonald N.Q., Lv F., et al. Insights into current tropomyosin receptor kinase (TRK) inhibitors: development and clinical application. J Med Chem. 2018;62:1731–1760. doi: 10.1021/acs.jmedchem.8b01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang T., Wang G., Liu Y., Feng L., Wang M., Liu J., et al. Development of small-molecule tropomyosin receptor kinase (TRK) inhibitors for NTRK fusion cancers. Acta Pharm Sin B. 2021;11:355–372. doi: 10.1016/j.apsb.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott L.J. Larotrectinib: first global approval. Drugs. 2019;79:201–206. doi: 10.1007/s40265-018-1044-x. [DOI] [PubMed] [Google Scholar]
- 16.Doebele R.C., Davis L.E., Vaishnavi A., Le A.T., Estrada-Bernal A., Keysar S., et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5:1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menichincheri M., Ardini E., Magnaghi P., Avanzi N., Banfi P., Bossi R., et al. Discovery of entrectinib: a new 3-aminoindazole as a potent anaplastic lymphoma kinase (ALK), c-ros oncogene 1 kinase (ROS1), and pan-tropomyosin receptor kinases (Pan-TRKs) inhibitor. J Med Chem. 2016;59:3392–3408. doi: 10.1021/acs.jmedchem.6b00064. [DOI] [PubMed] [Google Scholar]
- 18.Al-Salama Z.T., Keam S.J. Entrectinib: first global approval. Drugs. 2019;79:1477–1483. doi: 10.1007/s40265-019-01177-y. [DOI] [PubMed] [Google Scholar]
- 19.Hong D.S., DuBois S.G., Kummar S., Farago A.F., Albert C.M., Rohrberg K.S., et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–540. doi: 10.1016/S1470-2045(19)30856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doebele R.C., Drilon A., Paz-Ares L., Siena S., Shaw A.T., Farago A.F., et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:271–282. doi: 10.1016/S1470-2045(19)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drilon A., Laetsch T.W., Kummar S., DuBois S.G., Lassen U.N., Demetri G.D., et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russo M., Misale S., Wei G., Siravegna G., Crisafulli G., Lazzari L., et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6:36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 23.Goh X.N., Seng M.S., Loh A.H.P., Gupta A., Chang K.T.E., Iyer P. Larotrectinib followed by selitrectinib in a novel DCTN1-NTRK1 fusion undifferentiated pleomorphic sarcoma. J Oncol Pharm Pract. 2021;27:485–489. doi: 10.1177/1078155220938849. [DOI] [PubMed] [Google Scholar]
- 24.Drilon A., Nagasubramanian R., Blake J.F., Ku N., Tuch B.B., Ebata K., et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 2017;7:963–972. doi: 10.1158/2159-8290.CD-17-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers C., Morrissette J.J.D., Sussman R.T. NTRK point mutations and their functional consequences. Cancer Genet. 2022;262–263:5–15. doi: 10.1016/j.cancergen.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Drilon A., Ou S.I., Cho B.C., Kim D.W., Lee J., Lin J.J., et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent- front mutations. Cancer Discov. 2018;8:1227–1236. doi: 10.1158/2159-8290.CD-18-0484. [DOI] [PubMed] [Google Scholar]
- 27.Drilon A. TRK inhibitors in TRK fusion-positive cancers. Ann Oncol. 2019;30(Suppl 8):viii23–viii30. doi: 10.1093/annonc/mdz282. [DOI] [PubMed] [Google Scholar]
- 28.Cocco E., Lee J.E., Kannan S., Schram A.M., Won H.H., Shifman S., et al. TRK xDFG mutations trigger a sensitivity switch from type I to II kinase inhibitors. Cancer Discov. 2021;11:126–141. doi: 10.1158/2159-8290.CD-20-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somwar R., Hofmann N.E., Smith B., Odintsov I., Vojnic M., Linkov I., et al. NTRK kinase domain mutations in cancer variably impact sensitivity to type I and type II inhibitors. Commun Biol. 2020;3:1–13. doi: 10.1038/s42003-020-01508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cocco E., Schram A.M., Kulick A., Misale S., Won H.H., Yaeger R., et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med. 2019;25:1422–1427. doi: 10.1038/s41591-019-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katayama R., Gong B., Togashi N., Miyamoto M., Kiga M., Iwasaki S., et al. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kollol P., Anthony R., Aram E., Vincent A., Johnathan B., Marc O., et al. EXTH-80. PBI-200: in vivo efficacy of a novel, highly CNS-penetrant next generation TRK inhibitor. Neuro Oncol. 2021;23:vi181–vi182. [Google Scholar]
- 33.Wang J., Yu X., Zhu S., Chen Q., Sun J., Xia Y., et al. Preclinical evaluation of SIM1803-1A, a small molecule Trk/ROS1 dual inhibitor for wild and mutate NTRK/ROS1 fusion solid malignancies. J Clin Oncol. 2020;38 [Google Scholar]
- 34.Liu F., Wei Y., Zhang H., Jiang J., Zhang P., Chu Q. NTRK fusion in non-small cell lung cancer: diagnosis, therapy, and TRK inhibitor resistance. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.864666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada G., Drilon A. TRK inhibitor activity and resistance in TRK fusion-positive cancers in adults. Cancer Genet. 2022;264–265:33–39. doi: 10.1016/j.cancergen.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blauel E.R., Laetsch T.W. The promise of TRK inhibitors in pediatric cancers with NTRK fusions. Cancer Genet. 2022;262–263:71–79. doi: 10.1016/j.cancergen.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertrand T., Kothe M., Liu J., Dupuy A., Rak A., Berne P.F., et al. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol. 2012;423:439–453. doi: 10.1016/j.jmb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 39.Reichardt L.F. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ultsch M.H., Wiesmann C., Simmons L.C., Henrich J., Yang M., Reilly D., et al. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J Mol Biol. 1999;290:149–159. doi: 10.1006/jmbi.1999.2816. [DOI] [PubMed] [Google Scholar]
- 41.Wiesmann C., Ultsch M.H., Bass S.H., de Vos A.M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 42.Klein R., Jing S.Q., Nanduri V., O'Rourke E., Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 43.Julius D., Basbaum A.I. Molecular mechanisms of nociception. Nature. 2001;413:203–210. doi: 10.1038/35093019. [DOI] [PubMed] [Google Scholar]
- 44.Squinto S.P., Stitt T.N., Aldrich T.H., Davis S., Bianco S.M., Radziejewski C., et al. TrkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell. 1991;65:885–893. doi: 10.1016/0092-8674(91)90395-F. [DOI] [PubMed] [Google Scholar]
- 45.Li M., Dai F.R., Du X.P., Yang Q.D., Zhang X., Chen Y. Infusion of BDNF into the nucleus accumbens of aged rats improves cognition and structural synaptic plasticity through PI3K-ILK-Akt signaling. Behav Brain Res. 2012;231:146–153. doi: 10.1016/j.bbr.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Lamballe F., Klein R., Barbacid M. TrkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell. 1991;66:967–979. doi: 10.1016/0092-8674(91)90442-2. [DOI] [PubMed] [Google Scholar]
- 47.Singer H.S., Hansen B., Martinie D., Karp C.L. Mitogenesis in glioblastoma multiforme cell lines: a role for NGF and its TrkA receptors. J Neuro Oncol. 1999;45:1–8. doi: 10.1023/a:1006323523437. [DOI] [PubMed] [Google Scholar]
- 48.Tacconelli A., Farina A.R., Cappabianca L., Desantis G., Tessitore A., Vetuschi A., et al. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer Cell. 2004;6:347–360. doi: 10.1016/j.ccr.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Kheder E.S., Hong D.S. Emerging targeted therapy for tumors with NTRK fusion proteins. Clin Cancer Res. 2018;24:5807–5814. doi: 10.1158/1078-0432.CCR-18-1156. [DOI] [PubMed] [Google Scholar]
- 50.Schram A.M., Chang M.T., Jonsson P., Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14:735–748. doi: 10.1038/nrclinonc.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coulier F., Martin-Zanca D., Ernst M., Barbacid M. Mechanism of activation of the human trk oncogene. Mol Cell Biol. 1989;9:15–23. doi: 10.1128/mcb.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knezevich S.R., McFadden D.E., Tao W., Lim J.F., Sorensen P.H. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 53.Hechtman J.F. NTRK insights: best practices for pathologists. Mod Pathol. 2022;35:298–305. doi: 10.1038/s41379-021-00913-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laetsch T.W., Hong D.S. Tropomyosin receptor kinase inhibitors for the treatment of TRK fusion cancer. Clin Cancer Res. 2021;27:4974–4982. doi: 10.1158/1078-0432.CCR-21-0465. [DOI] [PubMed] [Google Scholar]
- 55.Zhao X., Kotch C., Fox E., Surrey L.F., Wertheim G.B., Baloch Z.W., et al. NTRK fusions identified in pediatric tumors: the frequency, fusion partners, and clinical outcome. JCO Precis Oncol. 2021;5:204–214. doi: 10.1200/PO.20.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roviello G., D'Angelo A., Sciortino M., Mini E., Nobili S., De Logu F., et al. TRK fusion positive cancers: from first clinical data of a TRK inhibitor to future directions. Crit Rev Oncol Hematol. 2020;152 doi: 10.1016/j.critrevonc.2020.103011. [DOI] [PubMed] [Google Scholar]
- 57.Gambella A., Senetta R., Collemi G., Vallero S.G., Monticelli M., Cofano F., et al. NTRK fusions in central nervous system tumors: a rare, but worthy target. Int J Mol Sci. 2020;21:753. doi: 10.3390/ijms21030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgeois J.M., Knezevich S.R., Mathers J.A., Sorensen P.H. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol. 2000;24:937–946. doi: 10.1097/00000478-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 59.Tognon C., Knezevich S.R., Huntsman D., Roskelley C.D., Melnyk N., Mathers J.A., et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 60.Skálová A., Vanecek T., Sima R., Laco J., Weinreb I., Perez-Ordonez B., et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 61.El Demellawy D., Cundiff C.A., Nasr A., Ozolek J.A., Elawabdeh N., Caltharp S.A., et al. Congenital mesoblastic nephroma: a study of 19 cases using immunohistochemistry and ETV6-NTRK3 fusion gene rearrangement. Pathology. 2016;48:47–50. doi: 10.1016/j.pathol.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 62.Klein R., Nanduri V., Jing S.A., Lamballe F., Tapley P., Bryant S., et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66:395–403. doi: 10.1016/0092-8674(91)90628-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Halalsheh H., McCarville M.B., Neel M., Reynolds M., Cox M.C., Pappo A.S. Dramatic bone remodeling following larotrectinib administration for bone metastasis in a patient with TRK fusion congenital mesoblastic nephroma. Pediatr Blood Cancer. 2018;65 doi: 10.1002/pbc.27271. [DOI] [PubMed] [Google Scholar]
- 64.Davis J.L., Lockwood C.M., Albert C.M., Tsuchiya K., Hawkins D.S., Rudzinski E.R. Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol. 2018;21:68–78. doi: 10.1177/1093526617712639. [DOI] [PubMed] [Google Scholar]
- 65.Laetsch T.W., DuBois S.G., Mascarenhas L., Turpin B., Federman N., Albert C.M., et al. Larotrectinib for paediatric solid tumours harbouring NTRK gene fusions: phase 1 results from a multicentre, open-label, phase 1/2 study. Lancet Oncol. 2018;19:705–714. doi: 10.1016/S1470-2045(18)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsiao S.J., Zehir A., Sireci A.N., Aisner D.L. Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn. 2019;21:553–571. doi: 10.1016/j.jmoldx.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu C., Si L., Wang W., Li Z., Song Z., Wang Q., et al. Expert consensus on the diagnosis and treatment of NTRK gene fusion solid tumors in China. Thorac Cancer. 2022;13:3084–3097. doi: 10.1111/1759-7714.14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teng H.K., Teng K.K., Lee R., Wright S., Tevar S., Almeida R.D., et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda C., Mazzoni M., Sensi M., Pierotti M.A., Greco A. Functional characterization of NTRK1 mutations identified in melanoma. Genes Chromosomes Cancer. 2014;53:875–880. doi: 10.1002/gcc.22200. [DOI] [PubMed] [Google Scholar]
- 70.Geiger T.R., Song J.Y., Rosado A., Peeper D.S. Functional characterization of human cancer-derived TRKB mutations. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harada T., Yatabe Y., Takeshita M., Koga T., Yano T., Wang Y., et al. Role and relevance of TrkB mutations and expression in non-small cell lung cancer. Clin Cancer Res. 2011;17:2638–2645. doi: 10.1158/1078-0432.CCR-10-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchetti A., Felicioni L., Pelosi G., Del Grammastro M., Fumagalli C., Sciarrotta M., et al. Frequent mutations in the neurotrophic tyrosine receptor kinase gene family in large cell neuroendocrine carcinoma of the lung. Hum Mutat. 2008;29:609–616. doi: 10.1002/humu.20707. [DOI] [PubMed] [Google Scholar]
- 73.Tomasson M.H., Xiang Z., Walgren R., Zhao Y., Kasai Y., Miner T., et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reuther G.W., Lambert Q.T., Caligiuri M.A., Der C.J. Identification and characterization of an activating TrkA deletion mutation in acute myeloid leukemia. Mol Cell Biol. 2000;20:8655–8666. doi: 10.1128/mcb.20.23.8655-8666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tacconelli A., Farina A.R., Cappabianca L., Gulino A., Mackay A.R. Alternative TrkAIII splicing: a potential regulated tumor-promoting switch and therapeutic target in neuroblastoma. Future Oncol. 2005;1:689–698. doi: 10.2217/14796694.1.5.689. [DOI] [PubMed] [Google Scholar]
- 76.Eggert A., Grotzer M.A., Ikegaki N., Liu X.G., Evans A.E., Brodeur G.M. Expression of the neurotrophin receptor TrkA down-regulates expression and function of angiogenic stimulators in SH-SY5Y neuroblastoma cells. Cancer Res. 2002;62:1802–1808. [PubMed] [Google Scholar]
- 77.Lagadec C., Meignan S., Adriaenssens E., Foveau B., Vanhecke E., Romon R., et al. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene. 2009;28:1960–1970. doi: 10.1038/onc.2009.61. [DOI] [PubMed] [Google Scholar]
- 78.Lange A.M., Lo H.W. Inhibiting TRK proteins in clinical cancer therapy. Cancers. 2018;10:105–119. doi: 10.3390/cancers10040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Modi V., Dunbrack R.L., Jr. Defining a new nomenclature for the structures of active and inactive kinases. Proc Natl Acad Sci U S A. 2019;116:6818–6827. doi: 10.1073/pnas.1814279116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roskoski R., Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug‒enzyme complexes. Pharmacol Res. 2016;103:26–48. doi: 10.1016/j.phrs.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 81.Lu X., Smaill J.B., Ding K. Medicinal chemistry strategies for the development of kinase inhibitors targeting point mutations. J Med Chem. 2020;63:10726–10741. doi: 10.1021/acs.jmedchem.0c00507. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Y., Xiang S., Yang F., Lu X. Targeting gatekeeper mutations for kinase drug discovery. J Med Chem. 2022;65:15540–15558. doi: 10.1021/acs.jmedchem.2c01361. [DOI] [PubMed] [Google Scholar]
- 83.Murray B.W., Rogers E., Zhai D., Deng W., Chen X., Sprengeler P.A., et al. Molecular characteristics of repotrectinib that enable potent inhibition of TRK fusion proteins and resistant mutations. Mol Cancer Ther. 2021;20:2446–2456. doi: 10.1158/1535-7163.MCT-21-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Florou V., Nevala-Plagemann C., Whisenant J., Maeda P., Gilcrease G.W., Garrido-Laguna I. Clinical activity of selitrectinib in a patient with mammary analogue secretory carcinoma of the parotid gland with secondary resistance to entrectinib. J Natl Compr Cancer Netw. 2021;19:478–482. doi: 10.6004/jnccn.2021.7022. [DOI] [PubMed] [Google Scholar]
- 85.Hemming M.L., Nathenson M.J., Lin J.R., Mei S., Du Z., Malik K., et al. Response and mechanisms of resistance to larotrectinib and selitrectinib in metastatic undifferentiated sarcoma harboring oncogenic fusion of NTRK1. JCO Precis Oncol. 2020;4:79–90. doi: 10.1200/PO.19.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oliver T.R.W., Jackson T.J., Ogunbiyi O., Sebire N., Slater O., Jorgensen M., et al. Comment on: “tumour-agnostic drugs in paediatric cancers”. Br J Cancer. 2021;124:524–526. doi: 10.1038/s41416-020-01103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fuse M.J., Okada K., Oh-Hara T., Ogura H., Fujita N., Katayama R. Mechanisms of resistance to NTRK inhibitors and therapeutic strategies in NTRK1-rearranged cancers. Mol Cancer Ther. 2017;16:2130–2143. doi: 10.1158/1535-7163.MCT-16-0909. [DOI] [PubMed] [Google Scholar]
- 88.El-Nassan H.B., Al-Qadhi M.A. Recent advances in the discovery of tropomyosin receptor kinases TRKs inhibitors: a mini review. Eur J Med Chem. 2023;258 doi: 10.1016/j.ejmech.2023.115618. [DOI] [PubMed] [Google Scholar]
- 89.Yakes F.M., Chen J., Tan J., Yamaguchi K., Shi Y., Yu P., et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 90.Smith B.D., Kaufman M.D., Leary C.B., Turner B.A., Wise S.C., Ahn Y.M., et al. Altiratinib inhibits tumor growth, invasion, angiogenesis, and microenvironment-mediated drug resistance via balanced inhibition of MET, TIE2, and VEGFR2. Mol Cancer Ther. 2015;14:2023–2034. doi: 10.1158/1535-7163.MCT-14-1105. [DOI] [PubMed] [Google Scholar]
- 91.Nishiyama A., Yamada T., Kita K., Wang R., Arai S., Fukuda K., et al. Foretinib overcomes entrectinib resistance associated with the NTRK1 G667C mutation in NTRK1 fusion-positive tumor cells in a brain metastasis model. Clin Cancer Res. 2018;24:2357–2369. doi: 10.1158/1078-0432.CCR-17-1623. [DOI] [PubMed] [Google Scholar]
- 92.Li M.C., Lin W.H., Wang P.C., Su Y.C., Chen P.Y., Fan C.M., et al. Design and synthesis of novel orally selective and type II pan-TRK inhibitors to overcome mutations by property-driven optimization. Eur J Med Chem. 2021;224 doi: 10.1016/j.ejmech.2021.113673. [DOI] [PubMed] [Google Scholar]
- 93.de Siqueira L.R.P., de Moraes Gomes P.A.T., de Lima Ferreira L.P., de Melo Rêgo M.J.B., Leite A.C.L. Multi-target compounds acting in cancer progression: focus on thiosemicarbazone, thiazole and thiazolidinone analogues. Eur J Med Chem. 2019;170:237–260. doi: 10.1016/j.ejmech.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 94.Pan S., Zhang L., Luo X., Nan J., Yang W., Bin H., et al. Structural optimization and structure–activity relationship studies of 6,6-dimethyl-4-(phenylamino)-6H-pyrimido[5,4-b][1,4]oxazin-7(8H)-one derivatives as a new class of potent inhibitors of pan-Trk and their drug-resistant mutants. J Med Chem. 2022;65:2035–2058. doi: 10.1021/acs.jmedchem.1c01597. [DOI] [PubMed] [Google Scholar]
- 95.Cui S., Wang Y., Wang Y., Tang X., Ren X., Zhang L., et al. Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors. Eur J Med Chem. 2019;179:470–482. doi: 10.1016/j.ejmech.2019.06.064. [DOI] [PubMed] [Google Scholar]
- 96.Duan Y., Wang J., Zhu S., Tu Z.C., Zhang Z., Chan S., et al. Design, synthesis, and structure‒activity relationships (SAR) of 3-vinylindazole derivatives as new selective tropomyosin receptor kinases (Trk) inhibitors. Eur J Med Chem. 2020;203 doi: 10.1016/j.ejmech.2020.112552. [DOI] [PubMed] [Google Scholar]
- 97.Guo J., Xiang S., Wang J., Zhou Y., Wang Z., Zhang Z., et al. Discovery of novel TrkA allosteric inhibitors: structure-based virtual screening, biological evaluation and preliminary SAR studies. Eur J Med Chem. 2022;228 doi: 10.1016/j.ejmech.2021.114022. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z., Wang J., Wang Y., Xiang S., Song X., Tu Z., et al. Discovery of the first highly selective and broadly effective macrocycle-based type II TRK inhibitors that overcome clinically acquired resistance. J Med Chem. 2022;65:6325–6337. doi: 10.1021/acs.jmedchem.2c00308. [DOI] [PubMed] [Google Scholar]
- 99.Xiang S., Wang J., Huang H., Wang Z., Song X., Zhou Y., et al. Switch type I to type II TRK inhibitors for combating clinical resistance induced by xDFG mutation for cancer therapy. Eur J Med Chem. 2023;245 doi: 10.1016/j.ejmech.2022.114899. [DOI] [PubMed] [Google Scholar]
- 100.Wang Z., Zhang Y., Pinkas D.M., Fox A.E., Luo J., Huang H., et al. Design, synthesis, and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-4-isopropyl-N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a dual inhibitor of discoidin domain receptors 1 and 2. J Med Chem. 2018;61:7977–7990. doi: 10.1021/acs.jmedchem.8b01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang J., Zhou Y., Tang X., Yu X., Wang Y., Chan S., et al. JND4135, a new type II TRK inhibitor, overcomes TRK xDFG and other mutation resistance in vitro and in vivo. Molecules. 2022;27:6500. doi: 10.3390/molecules27196500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi H.S., Rucker P.V., Wang Z., Fan Y., Albaugh P., Chopiuk G., et al. (R)-2-Phenylpyrrolidine substituted imidazopyridazines: a new class of potent and selective pan-TRK inhibitors. ACS Med Chem Lett. 2015;6:562–567. doi: 10.1021/acsmedchemlett.5b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito T., Kinoshita K., Tomizawa M., Shinohara S., Nishii H., Matsushita M., et al. Discovery of CH7057288 as an orally bioavailable, selective, and potent pan-TRK inhibitor. J Med Chem. 2022;65:12427–12444. doi: 10.1021/acs.jmedchem.2c01099. [DOI] [PubMed] [Google Scholar]
- 104.Gao Y.D., Olson S.H., Balkovec J.M., Zhu Y., Royo I., Yabut J., et al. Attenuating pregnane X receptor (PXR) activation: a molecular modelling approach. Xenobiotica. 2007;37:124–138. doi: 10.1080/00498250601050412. [DOI] [PubMed] [Google Scholar]
- 105.Hakkola J., Hukkanen J., Turpeinen M., Pelkonen O. Inhibition and induction of CYP enzymes in humans: an update. Arch Toxicol. 2020;94:3671–3722. doi: 10.1007/s00204-020-02936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burkhard J.A., Wagner B., Fischer H., Schuler F., Müller K., Carreira E.M. Synthesis of azaspirocycles and their evaluation in drug discovery. Angew Chem Int Ed Engl. 2010;49:3524–3527. doi: 10.1002/anie.200907108. [DOI] [PubMed] [Google Scholar]
- 107.Tanaka H., Sase H., Tsukaguchi T., Hasegawa M., Tanimura H., Yoshida M., et al. Selective TRK inhibitor CH7057288 against TRK fusion-driven cancer. Mol Cancer Ther. 2018;17:2519–2529. doi: 10.1158/1535-7163.MCT-17-1180. [DOI] [PubMed] [Google Scholar]
- 108.Lu X., Smaill J.B., Ding K. New promise and opportunities for allosteric kinase inhibitors. Angew Chem Int Ed Engl. 2020;59:13764–13776. doi: 10.1002/anie.201914525. [DOI] [PubMed] [Google Scholar]
- 109.Lu S., Qiu Y., Ni D., He X., Pu J., Zhang J. Emergence of allosteric drug-resistance mutations: new challenges for allosteric drug discovery. Drug Discov Today. 2020;25:177–184. doi: 10.1016/j.drudis.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 110.Burke M.R., Smith A.R., Zheng G. Overcoming cancer drug resistance utilizing PROTAC technology. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.872729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Békés M., Langley D.R., Crews C.M. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao C., He M., Wang L., He Y., Rao Y. Chemistries of bifunctional PROTAC degraders. Chem Soc Rev. 2022;51:7066–7114. doi: 10.1039/d2cs00220e. [DOI] [PubMed] [Google Scholar]
- 113.Subramanian G., Duclos B., Johnson P.D., Williams T., Ross J.T., Bowen S.J., et al. In pursuit of an allosteric human tropomyosin kinase A (hTrkA) inhibitor for chronic pain. ACS Med Chem Lett. 2021;12:1847–1852. doi: 10.1021/acsmedchemlett.1c00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee J.H., El-Damasy A.K., Seo S.H., Gadhe C.G., Pae A.N., Jeong N., et al. Novel 5,6-disubstituted pyrrolo[2,3-d]pyrimidine derivatives as broad spectrum antiproliferative agents: synthesis, cell based assays, kinase profile and molecular docking study. Bioorg Med Chem. 2018;26:5596–5611. doi: 10.1016/j.bmc.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 115.Furuya N., Momose T., Katsuno K., Fushimi N., Muranaka H., Handa C., et al. The juxtamembrane region of TrkA kinase is critical for inhibitor selectivity. Bioorg Med Chem Lett. 2017;27:1233–1236. doi: 10.1016/j.bmcl.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 116.Bagal S.K., Omoto K., Blakemore D.C., Bungay P.J., Bilsland J.G., Clarke P.J., et al. Discovery of allosteric, potent, subtype selective, and peripherally restricted TrkA kinase inhibitors. J Med Chem. 2019;62:247–265. doi: 10.1021/acs.jmedchem.8b00280. [DOI] [PubMed] [Google Scholar]
- 117.Su H.P., Rickert K., Burlein C., Narayan K., Bukhtiyarova M., Hurzy D.M., et al. Structural characterization of nonactive site, TrkA-selective kinase inhibitors. Proc Natl Acad Sci U S A. 2017;114:E297–e306. doi: 10.1073/pnas.1611577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen L., Chen Y., Zhang C., Jiao B., Liang S., Tan Q., et al. Discovery of first-in-class potent and selective tropomyosin receptor kinase degraders. J Med Chem. 2020;63:14562–14575. doi: 10.1021/acs.jmedchem.0c01342. [DOI] [PubMed] [Google Scholar]
- 119.Kargbo R.B. PROTAC compounds targeting TRK for use in cancer therapeutics. ACS Med Chem Lett. 2020;11:1090–1091. doi: 10.1021/acsmedchemlett.0c00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao B., Burgess K. TrkC-targeted kinase inhibitors and PROTACs. Mol Pharm. 2019;16:4313–4318. doi: 10.1021/acs.molpharmaceut.9b00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen L., Fu W., Zheng L., Liu Z., Liang G. Recent progress of small-molecule epidermal growth factor receptor (EGFR) inhibitors against C797S resistance in non-small-cell lung cancer. J Med Chem. 2018;61:4290–4300. doi: 10.1021/acs.jmedchem.7b01310. [DOI] [PubMed] [Google Scholar]
- 122.Chen H., Smaill J.B., Liu T., Ding K., Lu X. Small-molecule inhibitors directly targeting KRAS as anticancer therapeutics. J Med Chem. 2020;63:14404–14424. doi: 10.1021/acs.jmedchem.0c01312. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Z., Morstein J., Ecker A.K., Guiley K.Z., Shokat K.M. Chemoselective covalent modification of K-Ras(G12R) with a small molecule electrophile. J Am Chem Soc. 2022;144:15916–15921. doi: 10.1021/jacs.2c05377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Z.Z., Wang M.S., Wang F., Shi X.X., Huang W., Hao G.F., et al. Exploring the kinase-inhibitor fragment interaction space facilitates the discovery of kinase inhibitor overcoming resistance by mutations. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbac203. [DOI] [PubMed] [Google Scholar]
- 125.Zhuo L.S., Wang M.S., Wu F.X., Xu H.C., Gong Y., Yu Z.C., et al. Discovery of next-generation tropomyosin receptor kinase inhibitors for combating multiple resistance associated with protein mutation. J Med Chem. 2021;64:15503–15514. doi: 10.1021/acs.jmedchem.1c01539. [DOI] [PubMed] [Google Scholar]