Abstract
The mRNA encoding Escherichia coli polypeptide chain release factor 2 (RF2) has two partially overlapping reading frames. Synthesis of RF2 involves ribosomes shifting to the +1 reading frame at the end of the first open reading frame (ORF). Frameshifting serves an autoregulatory function. The RF2 gene sequences from the 86 additional bacterial species now available have been analyzed. Thirty percent of them have a single ORF and their expression does not require frameshifting. In the ∼70% that utilize frameshifting, the sequence cassette responsible for frameshifting is highly conserved. In the E. coli RF2 gene, an internal Shine–Dalgarno (SD) sequence just before the shift site was shown earlier to be important for frameshifting. Mutagenic data presented here show that the spacer region between the SD sequence and the shift site influences frameshifting, and possible mechanisms are discussed. Internal translation initiation occurs at the shift site, but any functional role is obscure.
INTRODUCTION
The majority of eubacteria use two peptide release factors, release factor 1 (RF1) and release factor 2 (RF2), for recognition of translation termination codons. RF1 mediates termination at UAG and UAA, and RF2 mediates termination at UAA and UGA. In a few circumstances, only one factor is sufficient: in the minimal genome eubacteria, Mycoplasma and Ureplasma and some organelles of bacterial origin, one of the release factors has been lost; certain mutants of Escherichia coli release factors acquire the ability to mediate termination at all three termination codons (Ito et al., 1998). However, in bacteria it seems advantageous to have two distinct stop codon discriminating release factors (Tate et al., 1999), since both are found in nearly all bacteria. In contrast, archaea and eukaryotes have only one release factor that recognizes all three stop codons.
Translation of E. coli RF2 mRNA requires ribosomes to switch to the +1 reading frame to synthesize RF2 (Craigen et al., 1985; Craigen and Caskey, 1986; Weiss et al., 1987). The first- or zero-frame is short and ends with CUU UGA. When RF2 is plentiful, a high proportion of ribosomes terminate at UGA to synthesize a short peptide that is rapidly degraded. However, codon CUU can detach from the anticodon of peptidyl-tRNALeu to effect a shift to the +1 frame that encodes the bulk of RF2. There is competition between frameshifting and termination that is affected by the level of RF2. Termination greatly predominates at high RF2 concentration, whereas termination efficiency is decreased at low concentration, allowing increased frameshifting. Thus, the frameshifting required for RF2 synthesis serves an autoregulatory function.
Advances in bacterial genome sequencing permit the generality and significant features of this mechanism to be assessed. The frameshifting mechanism is present in 70% of the 87 bacteria whose gene for RF2 has been sequenced (Baranov et al., 2002). All those that utilize frameshifting have the shift site/stop codon CUU UGA—except for Chlorobium tepidum, where it is CUU UAA. Whether there is some alternative reguloratory mechanism in the 30% that do not utilize frameshifting is unknown. Probably, the frameshifting mechanism was not utilized in a common bacterial ancestor but arose subsequent to divergence (Baranov et al., 2002). This study exploits the sequencing bonanza for insights into RF2 frameshifting.
Part of this work focuses on signals present in RF2 mRNA that were previously shown to be important for the frameshifting required for synthesis of E. coli RF2. An internal Shine–Dalgarno (SD) sequence 5′ of the CUU U shift site facilitates frameshifting. The exact position of the SD sequence with respect to the shift site is critical for its stimulatory effect, which requires pairing between the SD sequence and the complementary sequence near the 3′ end of 16S rRNA in translating ribosomes (Weiss et al., 1988; Atkins et al., 2002). Also facilitating the frameshifting in E. coli is the identity of the base 3′ of the UGA. The C at this position in E. coli makes UGA a relatively inefficient terminator (Tate et al., 1995).
RESULTS AND DISCUSSION
Statistical analysis of the RF2 frameshifting site
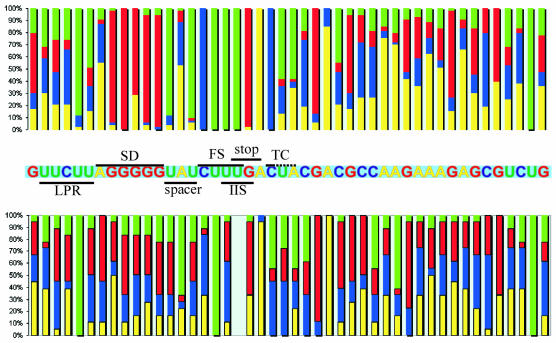
A BLAST search (Altschul et al., 1990) in sequenced and partially sequenced bacterial genomes has identified the RF2 genes in 87 bacteria. Some of these sequences can be found in the database of recoding events ‘Recode’ (Baranov et al., 2001). The distribution of nucleotides around the frameshift site, and the corresponding location in RF2 mRNA, translated without frameshifting, are shown in Figure 1. Conserved sequences that are present in frameshift-dependent RF2 (FS RF2) mRNAs but absent in frameshift-independent RF2 (non-FS RF2) mRNAs may reflect some sequence features important for the frameshifting. However, some differences may instead reflect differences in codon bias. This comparison is illustrated in Figure 1. The first highly conservative element is the frameshifting site itself, which is always CUU U in FS RF2 mRNAs. In non-FS RF2 mRNAs, only one U is conserved at the corresponding position: this is at the second codon position and it may reflect conservation at the amino acid level. Other elements relevant for the frameshifting can be found both upstream and downstream of the shift site. The stop codon that is part of the frameshifting site is UGA with one exception. In non-FS RF2 mRNAs, the predominant codon corresponding to the terminator for zero-frame translation of FS-RF2 mRNAs is of the form NRA, where N is any nucleotide. In these non-FS RF2 mRNAs, N is at the third codon position. R and A, the first and second nucleotides of the next codon, respectively, may be conserved because of the functional importance of the corresponding amino acid. Further downstream there is a consensus sequence of CUU in FS RF2 mRNAs, but YYY in non-FS RF2 mRNAs. The first C is 100% conserved and it is crucially important for efficient frameshifting. UGA, with a 3′ flanking C, is the least efficient termination signal (Major et al., 1996; Pavlov et al., 1998; Poole et al., 1998). By reducing the efficiency of termination, this C shifts the competition between termination and frameshifting in favor of frameshifting. The following 2 nts are also known to influence termination efficiency and it was shown that they interact with RF2 during termination (Poole et al., 1998). However, they are less conserved, in accordance with their weaker effect on termination.
Fig. 1. Statistical comparative analysis of the RF2 frameshifting cassette (top panel) with the corresponding regions of RF2 mRNAs that do not utilize frameshifting for their expression (lower panel). Each column represents the frequency within a subset of bacterial RF2 genes of a particular nucleotide at the position corresponding to the given E. coli RF2 mRNA sequence. Yellow represents A; light blue, C; red, G; green, U. Particular elements within mRNA are indicated with a black line as follows: LPR, low purine region; SD, Shine–Dalgarno sequence; FS, frameshifting site; IIS, internal initiation site; TC, termination context.
There are at least three mRNA elements specific for FS RF2 mRNAs 5′ of the frameshift site. First, there is an SD sequence 3 nt upstream of the frameshift site (Curran and Yarus, 1988). It interacts with the 3′ end of 16S rRNA (anti-SD sequence) of ribosomes translating RF2 mRNA (Weiss et al., 1988). The exact position of the SD sequence is important (Weiss et al., 1987). The very short distance between the SD sequence and the frameshift site is inferred to create tension between the anti-SD sequence and the decoding center. This tension destabilizes the initial P-site codon–anticodon interactions, whereas optimization of the distance between mRNA sites that interact with the P-site tRNA and the anti-SD sequence may stabilize P-site codon–anticodon interactions in the +1 frame. Thus, the rRNA between the P-site and the anti-SD sequence acts like a ‘compressed spring’. In accordance with this, the SD sequence can enhance –1 frameshifting in several bacterial genes if placed 10–15 nt 5′ of the frameshifting site (Larsen et al., 1994), probably by a similar mechanism whereby rRNA between the anti-SD sequence and the P-site acts as a ‘stretched spring’ (reviewed in Atkins et al., 2002). Introduction of an additional SD sequence upstream of the stimulatory SD sequence in RF2 mRNA causes a reduction in frameshifting (Weiss et al., 1987, 1990). This artificial SD sequence interacts with the 3′ end of 16S rRNA, preventing its binding to the stimulatory SD sequence. For this reason, a low purine region (LPR) probably exists just upstream of the SD sequence in FS RF2 mRNAs (Figure 1). Another upstream feature is the spacer region between the SD sequence and the frameshift site. Figure 1 clearly indicates that conservation of this region may be associated with frameshifting efficiency, and the most abundant spacer is U(A/C)U. The importance of this feature is investigated in the next section.
Mutagenic analysis of the spacer region between the stimulatory SD sequence and the frameshifting site
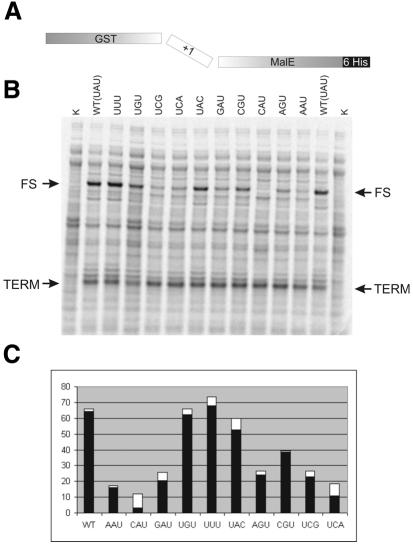
The genetic constructs employed a feature of a GST reporter for monitoring zero-phase translation and the 3′ malE gene in the +1 frame for monitoring transframe fusion products. The test frameshift cassette is inserted between GST and malE genes and fused to both (Figure 2A). Every nucleotide of the spacer region between the stimulatory SD sequence and the shift site has been mutated to all others. The exception is third position U, where substitution of U to A or G would create a stop codon. For this reason the spacers UCA and UCG were used instead. The frameshifting efficiencies were analyzed by pulse–chase labeling (Figure 2B and C). The highest effect has been observed for the 5′ U: substitution of this nucleotide to any other drops the level of frameshifting almost 4-fold (AAU, CAU and GAU). The second and third positions of the spacer do not seem to be so important if changed alone. UGU, UUU and UAC show the same frameshifting efficiency as E. coli wild type (WT) (UAU). However, substitution of both the second and third nucleotides have a dramatic effect on frameshifting (UCA and UCG). Few bacterial RF2 mRNAs have spacers without U at the first position (AAG in Borrelia burgdorferi, CGA in Dehalococcoides ethenogenes, AGU in Treponema pallidum and CGU in Caulobacter crescentus and Treponema denticola). Analysis of these spacers (AAG, AGU and CGU) in E. coli has shown reduction in frameshifting, although in the case of CGU the reduction was only 2-fold. Of course, such analysis may not reflect the level of frameshifting in other bacteria with these spacers, but it does indicate the importance of this region.
Fig. 2. Analysis of the effect of nucleotide identity in the spacer region between the stimulatory SD sequence and the frameshift site. (A) Outline of genetic constructs. (B) PAGE separation of the pulse–chase-labeled lysates of E. coli expressing genetic constructs containing the RF2 frameshifting cassette with different spacers. K, non-induced cells; WT, frameshift cassette with wild-type spacer (UAU); sequences of other spacers are written above each lane. TERM, product of termination; FS, product of frameshifting. (C) Plot representing the efficiency of termination as calculated from several independent experiments. White boxes are error bars.
Why is the identity of the spacer between the SD sequence and frameshifting site important and why is the 5′ U the most important component? Crystallographic studies of the 70S ribosomal complex with mRNA shows that a U 3′ of an SD sequence (AGGAGG) can form a base pair with an A 5′ of an anti-SD sequence (CUUCUU) in 16S rRNA (Yusupova et al., 2001). If this happens during translation of RF2 mRNA, the real spacer between the SD sequence and P-site tRNA is even shorter than previously thought—just 2 nt. Although no codon–anticodon interactions between mRNA and tRNA in the E-site have yet been found in the crystal structure of a bacterial ribosome (Yusupov et al., 2001), there is a possibility of such interactions at certain stages of the elongation cycle (Nierhaus et al., 2000). Lill and Wintermeyer (1987) have shown that tRNA binding to the E-site is modulated by structural elements of the tRNA molecule, rather than by codon–anticodon interactions. However, if such interactions occur, they must be in competition with SD–anti-SD sequence base-pairing. But even without codon–anticodon interactions in the E-site, it is easy to imagine that the P-site tRNA and the structure, which is formed by the SD and anti-SD sequences, would surround the E-site tRNA very tightly. Thus, the E-site tRNA may play an important role in the destabilization of the initial P-site codon–anticodon interaction and the stabilization of codon–anticodon interactions in the +1 frame. This could also explain why the efficiency of frameshifting is different for different codons upstream of the frameshifting site. Even in those cases when U is the first nucleotide of the spacer (UCA and UCG), the frameshifting efficiency is reduced (Figure 2B and C). The effect of E-site tRNA may depend on its structural features.
Another well-studied example of +1 P-site frameshifting is in the decoding of eukaryotic antizyme 1, 2 and 3 mRNAs (Matsufuji et al., 1995; Ivanov et al., 2000). Antizyme frameshifting sites show some divergence between different organisms (Ivanov et al., 2000). However, there are two elements that are 100% conserved, the stimulatory UGA stop codon and the U at the first position of the codon corresponding to E-site tRNA. What is even more impressive is that this codon may be recognized by only three tRNAs (Cys, Trp and Tyr) out of the six possible with U at the corresponding position in the codon (Cys, Leu, Ser, Phe, Trp and Tyr). Is it possible that interactions between E-site and P-site tRNAs affect frameshifting also in decoding of antizyme mRNA?
In E. coli, the identities of the last two codons influence the efficiency of termination, and evidence has been presented that this is due to the character of the two last amino acids of the growing nascent peptide (Björnsson et al., 1996). Evidence has been obtained in yeast that characteristics of the tRNAs decoding the last two codons influence termination (Mottagui-Tabar et al., 1998).
It is likely that the effect of the spacer region is a combination of all the above-mentioned considerations (part of an anti-SD sequence, tRNA interactions in E- and P-sites, modulation of termination efficiency), but it is difficult to estimate their relative contribution.
Internal initiation at UUG
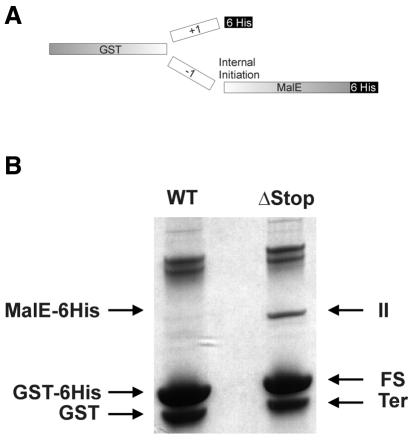
One of the codons used for initiation of translation in bacteria, in addition to AUG, is UUG. UUG is present in the frameshift site of RF2 mRNA, CUU UGA, in the –1 frame. Thus, the upstream stimulatory SD sequence can theoretically serve as a ribosomal binding site for an initiating ribosome. In contrast to the situation with non-FS genes, in all analyzed FS RF2 genes there is a stop codon in the corresponding frame either nine or 12 codons downstream of the UUG (or nearby), so that, if internal initiation occurs, the resulting product will be very short. In order to test whether ribosomes initiate at UUG in the RF2 frameshift site, the shift cassette of the E. coli RF2 gene (with the in-frame stop codon mutated to a sense codon) was placed between the coding sequence for GST and MalE proteins (Figure 3A), so that termination will result in production of GST protein. +1 frameshifting will result in GST protein with a small additional polypeptide containing a His6 tag; internal initiation will produce the MalE protein with a His6 tag; and –1 frameshifting will produce GST–MalE protein with a His6 tag. A frameshifting cassette with the WT stop codon for internal initiation was used as a control.
Fig. 3. Analysis of internal initiation in an RF2 frameshift cassette. (A) Outline of genetic constructs. (B) PAGE separation of expressed proteins after Ni–NTA isolation. II, product of internal initiation; FS, product of frameshifting; Ter, product of termination.
Proteins were purified using an Ni–NTA column and then analyzed by denaturing PAGE (Figure 3B). No significant products resulting from –1 frameshifting were detected. Double bands at the top of the gel are similar in size to GST–MalE–His6. However, if they were products of –1 frameshifting, they should be derived only from the construct with its in-frame stop deleted. These spots are related to proteins that were non-specifically co-isolated during the purification, as well as the termination product (GST without the His6 tag). The product of internal initiation is clearly seen in Figure 3B, but the efficiency of internal initiation is significantly lower than the efficiency of termination or +1 frameshifting. The intensities of termination and frameshifting products are the same for both constructs. This means that early termination of translation initiated at UUG does not affect the efficiency of frameshifting, and the high conservation of stop codons in the corresponding frame probably serves to prevent translation in the undesired open reading frame. It is unclear whether the presence of the initiator UUG is merely fortuitous because of the importance of its component nucleotides for frameshifting or whether there is functional significance in it acting as an initiator.
METHODS
Plasmids and bacterial strains. A GST–MalE fusion expression vector (GM1), containing BamHI and EcoRI restriction sites between the coding sequence of GST and MBP, has been described previously (Moore et al., 2000). Inserts made from complementary oligonucleotides (containing mutated RF2 frameshifting cassettes) were cloned between BamHI and EcoRI sites. Escherichia coli strain DH5α (Miller, 1992) was used in all studies.
Pulse–chase labeling. Measurements of frameshifting efficiency by [35S]methionine pulse–chase labeling were performed as described previously (Herr et al., 1999). Following pulse–chase labeling, total protein from each sample was separated on NuPAGE Bis–Tris SDS 4–12% (w/v) polyacrylamide gel (Invitrogen) and visualized with a Molecular Dynamics PhosphorImager.
Protein purification. Overnight cultures of strains expressing the appropriate plasmid were diluted 1:50 in Terrific Broth, grown for 2 h at 37°C, and then induced with 1 mM IPTG for an additional 4 h at 37°C. Harvested cells were lysed using Novagen’s BugBuster reagent. Recombinant proteins were purified by passage over Ni–NTA–agarose (Qiagen). Full-length TrxA–His6–gene 60–MBP fusion protein was purified by sequential passages over Ni–NTA–agarose (Qiagen). Purified protein was concentrated and washed extensively with Nanopure H2O using a Centricon 30 (Millipore).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Norma Wills for valuable technical suggestions. This work was supported by the NIH (grant R01-GM61200 to R.F.G. and R01-GM48152 to J.F.A.) and the DOE (grant DE-FG03-01ER63132 to R.F.G.).
REFERENCES
- Altschul S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Atkins J.F. et al. (2002) Over-riding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb. Symp. Quant. Biol., 66, 217–232. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Gurvich, O.L., Fayet, O., Prère, M.F., Miller, W.A., Gesteland, R.F., Atkins, J.F. and Giddings, M.C. (2001) RECODE: a database of frameshifting, bypassing and codon redefinition utilized for gene expression. Nucleic Acids Res., 29, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland, R.F. and Atkins, J.F. (2002) Recoding: translational bifurcations in gene expression. Gene, in press. [DOI] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar, S. and Isaksson, L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Craigen W.J. and Caskey, C.T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- Craigen W.J., Cook, R.G., Tate, W.P. and Caskey, C.T. (1985) Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl Acad. Sci. USA, 82, 3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J.F. and Yarus, M. (1988) Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J. Mol. Biol., 203, 75–83. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Atkins, J.F. and Gesteland, R.F. (1999) Mutations which alter the elbow region of tRNAGly2 reduce T4 gene 60 translational bypassing efficiency. EMBO J., 18, 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno, M. and Nakamura, Y. (1998) Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc. Natl Acad. Sci. USA, 95, 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Gesteland, R.F. and Atkins, J.F. (2000) Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res., 28, 3185–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Wills, N.M., Gesteland, R.F. and Atkins, J.F. (1994) rRNA and mRNA base pairing stimulates a programmed –1 ribosomal frameshift. J. Bacteriol., 176, 6842–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. and Wintermeyer, W. (1987) Destabilization of codon–anticodon interaction in the ribosomal exit site. J. Mol. Biol., 196, 137–148. [DOI] [PubMed] [Google Scholar]
- Major L.L., Poole, E.S., Dalphin, M.E., Mannering, S.A. and Tate, W.P. (1996) Is the in-frame termination signal of the Escherichia coli release factor-2 frameshift site weakened by a particularly poor context? Nucleic Acids Res., 24, 2673–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S., Matsufuji, T., Miyazaki, Y., Murakami, Y., Atkins, J.F., Gesteland, R.F. and Hayashi, S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell, 80, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Moore B., Persson, B.C., Nelson, C.C., Gesteland, R.F. and Atkins, J.F. (2000) Quadruplet codons: implications for code expansion and the specification of translation step size. J. Mol. Biol., 298, 195–209. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Tuite, M.F. and Isaksson, L.A. (1998) The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem., 257, 249–254. [DOI] [PubMed] [Google Scholar]
- Nierhaus K.H. et al. (2000) Ribosome elongation cycle. In Garrett, R.A., Douthwaite, S.R., Liljas, A., Matheson, A.T., Moore, P.B. and Noller, H.F. (eds), The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press, Washington, DC, pp. 319–335.
- Pavlov M.Y., Freistroffer, D.V., Dinçbas, V., MacDougall, J., Buckingham, R.H. and Ehrenberg, M. (1998) A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Major, L.L., Mannering, S.A. and Tate, W.P. (1998) Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res., 26, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W.P. et al. (1995) Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem. Cell Biol., 73, 1095–1103. [DOI] [PubMed] [Google Scholar]
- Tate W.P., Mansell, J.B., Mannering, S.A., Irvine, J.H., Major, L.L. and Wilson, D.N. (1999) UGA: a dual signal for ‘stop’ and for recoding in protein synthesis. Biochemistry (Moscow), 64, 1342–1353. [PubMed] [Google Scholar]
- Weiss R.B., Dunn, D.M., Atkins, J.F. and Gesteland, R.F. (1987) Slippery runs, shifty stops, backward steps, and forward hops: –2, –1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol., 52, 687–693. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn, D.M., Dahlberg, A.E., Atkins, J.F. and Gesteland, R.F. (1988) Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J., 7, 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.B., Dunn, D.M., Atkins, J.F. and Gesteland, R.F. (1990) Ribosomal frameshifting from –2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol., 39, 159–183. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H. and Noller, H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov, M.M., Cate, J.H. and Noller, H.F. (2001) The path of messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]