Abstract
Binding of the Escherichia coli global transcription factor FIS to the upstream activating sequence (UAS) of stable RNA promoters activates transcription on the outgrowth of cells from stationary phase. Paradoxically, while these promoters require negative supercoiling of DNA for optimal activity, FIS counteracts the increase of negative superhelical density by DNA gyrase. We demonstrate that binding of FIS at the UAS protects the rrnA P1 promoter from inactivation at suboptimal superhelical densities. This effect is correlated with FIS-dependent constraint of writhe and facilitated untwisting of promoter DNA. We infer that FIS maintains stable RNA transcription by stabilizing local writhe in the UAS. These results suggest a novel mechanism of transcriptional regulation by a transcription factor acting as a local topological homeostat.
INTRODUCTION
The Escherichia coli global transcriptional regulator FIS activates the promoters of stable RNA (rRNA and tRNA) operons during the adjustment of cellular translation machinery to rapid growth conditions (for recent review, see Travers et al., 2001). These promoters contain two cis-acting positive regulatory elements: the UP element involved in binding the α-CTD of RNA polymerase (RNAP) (Ross et al., 1993); and the upstream activating sequence (UAS) containing multiple FIS-binding sites, which are usually arranged in helical register. The deletion of UAS strongly reduces promoter activity, whereas inactivation of the fis gene did not diminish transcription, suggesting a compensatory mechanism derepressing stable RNA promoters in cells lacking FIS (Lamond and Travers, 1983; Ross et al., 1990; Lazarus and Travers, 1993). The activation of transcription by FIS requires the bending of UAS DNA (Nilsson et al., 1990; Newlands et al., 1991; Zacharias et al., 1992; Lazarus and Travers, 1993). Notably, the UAS itself is anisotropically flexible and presumably forms a DNA microloop delimited by RNAP (Nachaliel et al., 1989; Muskhelishvili et al., 1997; I.K. Pemberton, G. Muskhelishvili, A.A. Travers and M. Buckle, submitted for publication). FIS is thought to stabilize this DNA microloop on binding at multiple phased sites in the UAS (Travers and Muskhelishvili, 1998). However, a recent study using the seven E. coli rrn P1 promoters demonstrated differences in the contribution of multiple FIS-binding sites to overall promoter activity (Hirvonen et al., 2001). The input of distal FIS sites upstream of promoter-proximal site I is small at the rrnB P1 and rrnG P1 promoters but substantial at the rrnA P1 and rrnE P1 promoters, accounting at the former for ∼75% of total activation.
Furthermore, the activity of stable RNA promoters strongly depends on negative superhelical density of DNA both in vitro and in vivo (Glaser et al., 1983; Lamond, 1985; Ohlsen and Gralla, 1992a; Bowater et al., 1994; Free and Dorman, 1994). In this class of promoter, the ‘sensing’ of superlelical density involves the GC-rich ‘discriminator’ sequence between the –10 hexamer and the startpoint of transcription (Travers, 1980). This structural element determines the unusually short lifetime of initial transcription complexes by acting as a barrier to promoter opening, which can be partially overcome by negative supercoiling of DNA (Figueroa-Bossi et al., 1998; Pemberton et al., 2000).
Whereas the activity of stable RNA promoters strongly depends on DNA superhelical density, FIS counteracts the elevation of negative superhelicity by DNA gyrase on the outgrowth of cells from stationary phase (Schneider et al., 1999). This general effect of FIS is at variance with the maximal activity of stable RNA promoters observed at the same growth stage. We resolve this apparent paradox by providing evidence that stabilization of local writhe in UAS by FIS protects the rrnA P1 promoter from inactivation at suboptimal superhelical densities. This novel mechanism of transcriptional regulation reveals the property of a global transcription factor to act as a local topological homeostat maintaining selective gene expression.
RESULTS
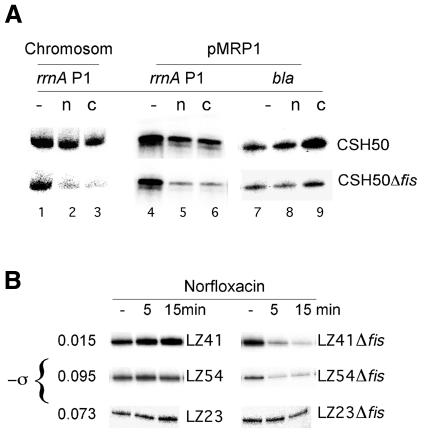
To monitor stable RNA transcription under conditions of DNA relaxation, two different coumarin inhibitors of DNA gyrase—novobiocin and coumermycin (Maxwell, 1997)—were added to exponentially growing CSH50 wild-type and CSH50Δfis cells at OD600 = 0.1. The cells were harvested after 5 min, total RNA isolated and the amount of chromosomal rrnA P1 transcripts measured by primer extension. Since the 5′ end of rRNA has been found to turn over rapidly, the relative amount of detected transcript essentially reflects the efficiency of transcription initiation (Sarmientos et al., 1983; Aviv et al., 1996). On addition of DNA relaxing agents, we observed a significant reduction in the amount of chromosomal rrnA P1 transcripts in fis mutant cells in comparison to wild-type cells (Figure 1A, lanes 1–3). Likewise, these agents reduced transcription of plasmid-borne rrnA P1 (pMRP1) on average to 40 and 80% in the wild-type and fis cells, respectively, whereas transcription of reference bla promoter was not reduced (Figure 1A, lanes 4–9). To exclude any role of toxic effects of drugs, we next used a set of isogenic strains containing different combinations of drug-sensitive and drug-resistant alleles of gyrase (gyrAL83) and topoisomerase IV (parCK84) genes. In these strains, the addition of a quinolone (norfloxacin) allows the variation of the overall superhelical density from almost fully relaxed (σ = –0.015, strain LZ41) to near physiological (σ = –0.073, strain LZ23) to hypernegatively supercoiled (σ = –0.095, strain LZ54) state. Without norfloxacin, these strains have a similar overall superhelical density of around –0.07 (Khodursky et al., 1995; Zechiedrich et al., 1997; Schneider et al., 2000). We compared chromosomal rrnA P1 transcription in these strains and their fis derivatives grown to OD600 = 0.1. A significant reduction of transcripts on relaxation of DNA was observed only in fis mutant cells (Figure 1B). Hypernegative supercoiling of DNA also led to a reduction of rrnA P1 transcription in the fis mutant cells but not in the wild-type cells, whereas no significant differences were observed at near physiological levels of overall negative superhelicity (σ = –0.073).
Fig. 1. FIS protects rrnA P1 transcription at suboptimal superhelical densities in vivo. (A) Relaxation of DNA by novobiocin (n) and coumermycin (c). Chromosomal rrnA P1 (lanes 1–3), pMRP1-borne rrnA P1 (lanes 4–6) and bla (lanes 7–9) transcripts detected in CSH50 and CSH50Δfis cells. (B) Transcription of chromosomal rrnA P1 in LZ41, LZ54 and LZ23 strains and their fis derivatives 5 and 15 min after norfloxacin treatment. The overall negative superhelical densities (–σ) estimated in these strains after treatment with norfoxacin are indicated on the left.
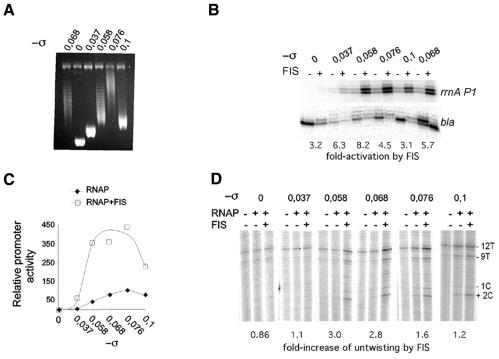
Taken together, these results strongly suggest that fis counteracts either directly or indirectly the reduction of rrnA P1 transcription by any deviations from optimal superhelical densities. To distinguish between these possibilities, we measured in vitro transcription of the rrnA P1 promoter construct pMRP1 with and without FIS using topoisomers of different negative superhelical densities (Figure 2A). In the absence of FIS, the transcription of rrnA P1 demonstrated a clear optimum at a negative superhelical density of –0.076 (Figure 2B and C). This value is significantly higher than the expected unconstrained superhelicity in vivo, although similarly high in vitro optimum levels have been obtained for the supercoiling-dependent fis and tyrT promoters (Schneider et al., 2000; H. Auner, M. Buckle, A. Deufel, T. Kutateladze, L. Lazarus, G. Muskhelishvili, I. Pemberton, R. Schneider and A. Travers, submitted for publication). Addition of FIS supported high levels of rrnA P1 transcription over a range of superhelical densities, including those at which the transcription by RNAP alone was suboptimal (Figure 2C). This result is consistent with the in vivo data and suggests that FIS compensates for the suboptimal template topology in vitro.
Fig. 2. FIS compensates for suboptimal superhelical densities in vitro. (A) The pMRP1 preparations used for in vitro experiments and the superhelical densities of each. (B) In vitro transcription from pMRP1 preparations shown in (A). Concentrations of DNA, RNAP and FIS were 10, 100 and 40 nM, respectively. Fold-activation by FIS for each σ is indicated below. The repression of the divergently oriented bla promoter on pMRP1 by FIS is presumably due to bending the UAS DNA towards rrnA P1. (C) Graphical representation of the experiment shown in (B). The relative promoter activities were normalized to the maximum rrnA P1 activity (100%) in the absence of FIS. (D) Permanganate footprinting of initiation complexes assembled with pMRP1. Concentration of DNA and proteins was as in (B). The reactive bases around the start and within the –10 region are indicated. The fold-increase of untwisting represents the ratio of summarized intensities of reactive bases in lanes with FIS divided by those without FIS.
To gain insight into the mechanism of this compensatory effect of FIS, we monitored open complex formation by permanganate reactivity assay using RNAP in excess of pMRP1 DNA and high salt conditions (230 mM NaCl) unfavourable for promoter opening (Ohlsen and Gralla, 1992a). Under these conditions, the permanganate reactivity signals in the –10 element and around the transcription startpoint indicative of RNAP open (initiation) complexes were detected only with templates of high negative σ levels (–0.076 and –0.1). Again, addition of FIS increased the range of superhelical densities at which promoter opening was detectable (Figure 2D). From these data, we infer that at suboptimal superhelical densities FIS maintains high levels of rrnA P1 transcription by facilitating initiation complex formation.
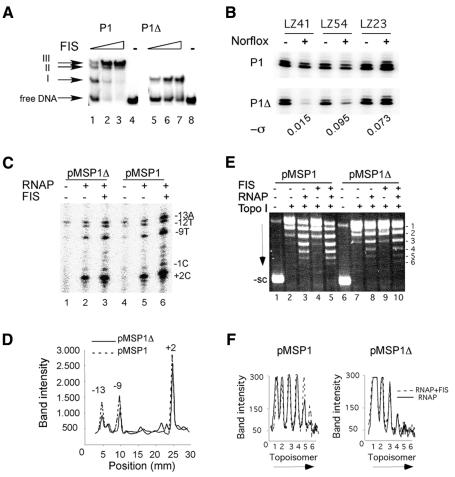
To prove the role of the UAS in the FIS-dependent protection of promoter activity, we compared the wild-type rrnA P1 with the deletion mutant rrnA P1Δ containing only the promoter-proximal FIS-binding site I (Figure 3A). The constructs pMSP1 and pMSP1Δ were transformed in the LZ41, LZ54 and LZ23 strains and the transcription of the wild-type and mutant promoters measured after norfloxacin treatment. As expected, FIS prevented the reduction of the plasmid-borne rrnA P1 activity under the conditions of both DNA relaxation and hypernegative supercoiling. In contrast, the transcription of the P1Δ mutant promoter could not be protected by FIS, indicating that an intact UAS is required (Figure 3B).
Fig. 3. The deletion mutant containing only proximal FIS-binding site I fails to protect rrnA P1 transcription. (A) Gel retardation analysis of the wild-type and mutant DNA fragments. The three complexes formed on binding FIS at the wild-type fragment are indicated. Note that the mutant forms only one complex, even at the highest FIS concentrations used. The concentration of FIS in lanes 1–3 and 5–7 was 4, 11 and 22 nM, respectively. (B) Transcription of rrnA P1 from pMRP1 and pMRP1Δ in LZ41, LZ54 and LZ23 strains after norfloxacin treatment. The overall negative superhelical densities estimated in these strains after norfoxacin treatment are indicated. (C) Permanganate footprinting of initiation complexes. The concentration of RNAP and FIS was 100 and 40 nM, respectively. The reactive bases are indicated. (D) Graphical representation of band intensities of lanes 3 and 6 shown in (C). Note the increased reactivity of bases –13 and –9 for wild-type rrnA P1. (E) Relaxation of pMSP1 DNA (10 nM) in the presence of RNAP (50 nM) with or without FIS. The concentration of FIS in lanes 4, 5, 9 and 10 was 33 nM. Untreated supercoiled plasmid (-sc). (F) Graphical representation of topoisomer distribution in lanes 3 and 5 (pMSP1, left panel) and 8 and 9 (pMSP1Δ, right panel) shown in (E). Note the increase in the abundance of pMSP1 topoisomers 5 and 6. Arrows indicate the direction of electrophoresis.
We next analysed promoter opening with supercoiled pMSP1 and pMSP1Δ in the presence of excess RNAP with or without FIS. We found that FIS facilitates open complex formation at both the rrnA P1 and rrnA P1Δ promoters. However, the effect of FIS on DNA untwisting was more pronounced with the wild-type promoter, especially in the –10 region (Figure 3C and D). We infer that, at suboptimal superhelical densities, the FIS-binding site I is insufficient to rescue rrnA P1 transcription and that this failure is correlated with impaired initiation complex formation.
Finally, we investigated whether the binding of FIS at rrnA P1 UAS involves any changes in DNA writhe. For this purpose, RNAP initiation complexes were assembled with supercoiled pMSP1 and pMSP1Δ in the presence and absence of FIS and treated with topoisomerase I. Addition of FIS to RNAP initiation complexes changed the equilibrium distribution of topoisomers with pMSP1 but not with pMSP1Δ (Figure 3E and F). We infer that the constraint of writhe by initiation complexes requires an intact UAS DNA.
DISCUSSION
In this study we have demonstrated that FIS protects the rrnA P1 promoter from inactivation at suboptimal DNA negative superhelicities in vivo. Several lines of evidence suggest that this effect is mediated by direct binding of FIS at UAS. First, an intact UAS containing multiple FIS-binding sites is required to observe this effect in vivo. Secondly, FIS activates the rrnA P1 promoter about 3- to 8-fold at the lower than optimal levels of superhelicity (σ of 0 to –0.058) and about 3-fold under conditions of hypernegative supercoiling in vitro. Thirdly, at suboptimal superhelical densities, FIS facilitates open complex formation, a step thought to be rate-limiting in this class of promoter (Ohlsen and Gralla, 1992b; Figueroa-Bossi et al., 1998). This latter result does not necessarily imply that FIS acts explicitly at the step of promoter opening. However, since we used a large excess of RNAP over DNA in all in vitro experiments, we infer that a step subsequent to initial complex formation is accelerated by FIS. This notion is supported by studies on rrnD P1, rrnB P1 and tyrT promoters (Sander et al., 1993; Muskhelishvili et al., 1997; Bartlett et al., 2000; H. Auner, M. Buckle, A. Deufel, T. Kutateladze, L. Lazarus, G. Muskhelishvili, I. Pemberton, R. Schneider and A. Travers, submitted for publication).
We have not addressed the specific roles of the multiple FIS-binding sites of rrnA P1 UAS in the promoter protection mechanism described here. However, the proximal site I is not sufficient either to maintain transcription at suboptimal superhelical densities in vivo or for the constraint of writhe by initiation complexes and efficient open complex formation in vitro. These findings are consistent with the reported requirement of all FIS-binding sites for maximum rrnA P1 activation in vivo (Hirvonen et al., 2001). We cannot exclude that other factors, in addition to FIS, may be involved in modulating the response of rrnA P1 to changes of supercoiling in vivo, but we consider it unlikely for two reasons: first, the putative factor should be able to activate the promoter under conditions of opposite changes in superhelical density, as we have demonstrated here for FIS; and, secondly, since the deletion of distal FIS-binding sites in UAS abolishes the protection effect, the putative factor would also require this same UAS region and, in addition, FIS bound at site I (see also Hirvonen et al., 2001).
The position of FIS-binding site I (centred around –71) is highly conserved in all stable RNA promoters, whereas that of distal sites is more variable, although they are usually arranged in helical register (Verbeek et al., 1990; Condon et al., 1992; Lazarus and Travers, 1993; Hirvonen et al., 2001). FIS bound at site I may interact with the α-CTD of RNAP (Bokal et al., 1997), and this site has been shown to suffice for maximum activation of rrnB P1 in vitro (Gosink et al., 1993). It is thus possible that, at least at some stable RNA promoters, the proximal site I is sufficient for high activity under optimal growth conditions, whereas the upstream sites are required to maintain promoter activity under conditions of suboptimal superhelicitiy caused by environmental stresses (Dorman, 1995; Tse-Dinh et al., 1997). In UAS regions of stable RNA promoters so far characterized in detail, the local helical repeat separating the FIS-binding sites is lower than the intrinsic helical repeat of negatively supercoiled DNA (Herzel et al., 1999), implying that bending of DNA by FIS would stabilize left-handed writhe (Travers and Muskhelishvili, 1998). Indeed, we observe FIS-dependent constraint of writhe in the ternary complexes assembled with wild-type promoter but not the UAS mutant rrnA P1Δ promoter. We estimate that the ternary complex assembled on rrnA P1 changes the linking number of pMSP1 DNA by 1. This correlates with facile initiation complex formation and argues for coupling between local writhe stabilized in rrnA P1 UAS and promoter untwisting. Such repartition of writhe and twist mediated by torsional transmission mechanism has been inferred for the closely related tyrT promoter, which also requires phased FIS-binding sites in UAS for maximal activity (Muskhelishvili et al., 1997).
We thus infer that UAS is an important element involved in the physiological response of the rrnA P1 promoter to alterations in negative superhelicity. Notably, in contrast to the GC-rich discriminator and other core promoter elements implicated in sensing the supercoiling level (H. Auner, M. Buckle, A. Deufel, T. Kutateladze, L. Lazarus, G. Muskhelishvili, I. Pemberton, R. Schneider and A. Travers, submitted for publication), the UAS appears primarily involved in the adaptation to deviations of DNA superhelicity. This ‘buffering’ function of UAS requires binding of FIS at multiple phased sites and indicates, as proposed earlier (Travers and Muskhelishvili, 1998), that FIS acts as a local topological homeostat.
Expression of fis is activated by high levels of negative supercoiling (Schneider et al., 2000). FIS is produced in large amounts on the outgrowth of cells from stationary phase and counteracts the elevation of negative superhelicity by gyrase (Schneider et al., 1999). This FIS-dependent reduction of overall superhelicity is thought to prevent the deleterious effects of excessive supercoiling, but it would also limit the activity of the stable RNA as well as many other promoters that strongly depend on DNA superhelicity (Dorman, 1995; Tse-Dinh et al., 1997). Our finding that FIS stabilizes a local DNA writhe (a microloop) in the upstream regions of stable RNA promoters resolves this apparent paradox and reveals a mechanism enabling a selective response of rrn operons, and hence of the translational machinery, to increased demands of protein synthesis on the commitment of cells to rapid growth. In fis cells, the overall level of negative superhelicity is elevated (Schneider et al., 1997, 1999) and would thus derepress stable RNA promoters and compensate for the absence of FIS during the outgrowth from stationary phase.
METHODS
Bacterial strains and plasmids. Bacterial strains used in this study were E. coli K12 derivatives. The genotype of LZ41 is parCK84gyrA+topA10, that of LZ54 is parC+gyrAL83topA10 and that of LZ23 is parC+gyrAL83topA+ (Khodursky et al., 1995; Zechiedrich et al., 1997; Schneider et al., 2000). The fis derivatives of strains LZ41, LZ54 and LZ23 were obtained by phage P1 transduction from strain CSH50Δfis described by González-Gil et al. (1998). All strains were grown in rich 2× YT medium (Schneider et al., 2000).
pMRP1 contains the region ∼300 bp upstream of the transcription startpoint of rrnA P1 and strong rrnB operon terminators ∼250 bp downstream (Gafny et al., 1994). pMRP1Δ was generated by PCR-aided internal deletion of the region between –94 and –147, removing all FIS-binding sites except the promoter-proximal site I. pMSP1 and pMSP1Δ were generated by internal deletion of a 3-kb fragment containing the lacZ gene from pMRP1 and pMRP1Δ, respectively.
Proteins. FIS was purified as described previously (Koch and Kahmann, 1986). Escherichia coli RNAP came from Boehringer-Mannheim, and Vaccinia topoisomerase was a kind gift of Karin Schnetz (Institute for Genetics, Cologne, Germany).
RNA analysis. Primer extension using total E. coli RNA was performed as described by Nasser et al. (2001). Extension of RNA templates transcribed from the chromosomal rrnA P1 promoter with the primer G11 (5′-GAGCAGTGCCGCTTCGC-3′) yields a 156-bp fragment. The extension products were resolved on a 6% sequencing gel and visualized by phosphoimaging. The length of the transcripts was identified by using corresponding dideoxy sequencing reactions as a reference.
High-resolution agarose gel electrophoresis. High-resolution agarose gel electrophoresis was carried out in 1× TBE buffer as described previously (Schneider et al., 1997).
Preparation of topoisomer distibutions and determination of σ. Topoisomer distributions were prepared using Vaccinia topoisomerase as described by Schneider et al. (2000). σ was calculated according to the formula: –σ = ΔLK/LK0, where LK0 for pMRP1 ∼6.500-bp/10.5-bp/turn = 619. The error for all σ is 10–15%.
In vitro transcription from supercoiled templates. In vitro transcription/coupled primer extension reactions using plasmids with distinct σ were carried out with reverse transcriptase (SuperScript II) as described by Lazarus and Travers (1993). Extension of RNA templates transcribed from the plasmid-borne rrnA P1 promoter with the primer G11 yields a 65-bp fragment. The mRNA transcribed from the bla promoter located on the same plasmid yields a 95-bp fragment on extension with the primer bla3B4 (5′-CAGGAAGGCAAAATGCCGC-3′). The samples were analysed on 6% denaturing polyacrylamide gels, visualized by phosphorimaging and quantitated using ImageQuant software.
Potassium permanganate reactivity assay. The reactions for potassium permanganate reactivity assays were assembled as for in vitro transcription reactions, but only ATP and CTP (100 µM) were present in the incubation mixture to allow formation of the dinucleotide pppApC. Footprinting was performed as described by Nasser et al. (2001).
Topological analysis of transcription complexes. The pMSP1 DNA was relaxed in the presence of proteins as indicated for 1 h by calf thymus topoisomerase I (Amersham) according to the manufacturer’s recommendations. The complexes were assembled as for potassium permanganate reactivity assays, and the samples were processed as described by Schneider et al. (1997) and run on 1% high-resolution agarose gels.
Gel-shift analysis. The ∼130-bp fragments comprising the UAS regions of pMRP1 and pMRP1Δ were obtained by PCR amplification, radioactively end-labelled by [γ−32P]ATP using T4 Polynucleotide kinase (Biolabs) and incubated at 37°C for 15 min in a buffer containing 10 mM Tris–HCl pH 7.9 and 100 mM NaCl with FIS as indicated. The samples were loaded on a 5% non-denaturing polyacrylamide gel and visualized on an X-ray film.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andrew Travers for critical comments. M.R. was supported by short-term fellowship grant ASTF Nr 9610 from EMBO and a Boehringer Ingelheim Fonds travel allowance grant. This work was supported in part by Deutsche Forschungsgemeinschaft through SFB 397.
REFERENCES
- Aviv M., Giladi, H., Oppenheim, A. and Glaser, G. (1996) Analysis of the shut-off of ribosomal RNA promoters in E. coli upon entering the stationary phase of growth. FEMS Microbiol. Lett., 140, 71–76. [DOI] [PubMed] [Google Scholar]
- Bartlett M.S., Gaal, T., Ross, W. and Gourse, R.L. (2000) Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters. J. Bacteriol., 182, 1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokal A., Ross, W., Gaal, T., Johnson, R.C. and Gourse, R.L. (1997) Molecular anatomy of a transcription activation patch: FIS–RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J., 16, 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R.P., Chen, D. and Lilley, D.M.J. (1994) Modulation of tyrT promoter activity by template supercoiling in vivo. EMBO J., 13, 5647–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C., Philips, J., Fu, Z.-Y., Squires, C. and Squires, C.L. (1992) Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J., 11, 4175–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C.J. (1995) DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology, 141, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Guerin, M., Rahmouni, R., Leng, M. and Bossi, L. (1998) The supercoiling sensitivity of a bacterial tRNA promoter parallels its responsiveness to stringent control. EMBO J., 17, 2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free A. and Dorman, C.J. (1994) Escherichia coli tyrT gene transcription is sensitive to DNA supercoiling in its native chromosomal context: effect of DNA topoisomerase IV overexpression on tyrT promoter function. Mol. Microbiol., 14, 151–161. [DOI] [PubMed] [Google Scholar]
- Gafny R., Cohen, S., Nachaliel, N. and Glaser, G. (1994) Isolated P2 rRNA promoters of Escherichia coli are strong promoters that are subject to stringent control. J. Mol. Biol., 243, 152–156. [DOI] [PubMed] [Google Scholar]
- Glaser G., Sarmientos, P. and Cashel, M. (1983) Functional interrelationship between two tandem E. coli ribosomal RNA promoters. Nature, 302, 74–76. [DOI] [PubMed] [Google Scholar]
- González-Gil G., Kahmann, R. and Muskhelishvili, G. (1998) Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. EMBO J., 17, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink K.K., Ross, W., Leirmo, S., Osuna R., Finkel, S.E., Johnson, R.C. and Gourse, R.L. (1993) DNA binding and bending are necessary but not sufficient for Fis-dependent activation of rrnB P1. J. Bacteriol., 175, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel H., Weiss, O. and Trifonov, E.N. (1999) 10–11 bp periodicities in complete genomes reflect protein structure and DNA folding. Bioinformatics, 15, 187–193. [DOI] [PubMed] [Google Scholar]
- Hirvonen C.A., Ross, W., Wozniak, C.E., Marasco, E., Anthony, J.R., Aiar, S.E., Newburn, V.H. and Gourse, R.L. (2001) Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol., 183, 6305–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky A.B., Zechiedrich, E.L. and Cozzarelli, N.R. (1995) Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 11801–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. and Kahmann, R. (1986) Purification and properties of the Escherichia coli host factor required for the inversion of the G segment in bacteriophage Mu. J. Biol. Chem., 261, 15673–15678. [PubMed] [Google Scholar]
- Lamond A.I. (1985) Supercoiling response of a bacterial tRNA gene. EMBO J., 4, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I. and Travers, A.A. (1983) Requirement for an upstream element for optimal transcription of a bacterial tRNA gene. Nature, 305, 248–250. [DOI] [PubMed] [Google Scholar]
- Lazarus L.R. and Travers, A.A. (1993) The E. coli FIS protein is not required for the activation of tyrT transcription on simple nutritional upshift. EMBO J., 12, 2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A. (1997) DNA gyrase as a drug target. Trends Microbiol., 5, 102–108. [DOI] [PubMed] [Google Scholar]
- Muskhelishvili G., Buckle, M., Heumann, H., Kahmann, R. and Travers, A.A. (1997) FIS activates sequential steps during transcription initiation at a stable RNA promoter. EMBO J., 16, 3655–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachaliel M., Melnick, J., Gafny, R. and Glaser, G. (1989) Ribosome associated protein(s) specifically bind(s) to the upstream activator sequence of E. coli rrnA P1 promoter. Nucleic Acids Res., 17, 9811–9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser W., Schneider, R., Travers, A. and Muskhelishvili, G. (2001) CRP modulates fis transcription by alternate formation of activating and repressing nucleoprotein complexes. J. Biol. Chem., 276, 17878–17886. [DOI] [PubMed] [Google Scholar]
- Newlands J.T., Ross, W., Gosink, K. and Gourse, R. (1991) Factor-independent activation of Escherichia coli rRNA transcription. II. Characterisation of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J. Mol. Biol., 220, 569–583. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Vanet, A., Vijgenboom, E. and Bosch, L. (1990) The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J., 9, 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsen K.L. and Gralla, J.D. (1992a) Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli rrnB P1 promoter. Mol. Microbiol., 6, 2243–2251. [DOI] [PubMed] [Google Scholar]
- Ohlsen K.L. and Gralla, J.D. (1992b) Melting during steady-state transcription of the rrnB P1 promoter in vivo and in vitro. J. Bacteriol., 174, 6071–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton I.K., Muskhelishvili, G., Travers, A.A. and Buckle, M. (2000) The G+C rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. J. Mol. Biol., 299, 859–864. [DOI] [PubMed] [Google Scholar]
- Ross W., Thompson, J.F., Newlands, J.T. and Gourse, R. (1990) E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J., 9, 3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Gosink, K.K., Salomon, J., Igarishi, K., Zou, C., Ishihama, A., Severinov, K. and Gourse, R.L. (1993) A third recognition element in bacterial promoters: DNA binding by the a subunit of RNA polymerase. Science, 262, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Sander P., Langert, W. and Mueller, K. (1993) Mechanisms of upstream activation of the rrnD promoter P1 of Escherichia coli. J. Biol. Chem., 268, 16907–16916. [PubMed] [Google Scholar]
- Sarmientos P., Sylvester, J.E., Contente, S. and Cashel, M. (1983) Different stringent control of the tandem E. coli ribosomal RNA promoters from rrnA operon expressed in vivo in multicopy plasmids. Cell, 32, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Schneider R., Travers, A.A. and Muskhelishvili, G. (1997) FIS modulates the growth phase-dependent topological transitions of DNA in E. coli. Mol. Microbiol., 26, 519–530. [DOI] [PubMed] [Google Scholar]
- Schneider R., Travers, A., Kutateladse, T. and Muskhelishvili, G. (1999) A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol. Microbiol., 34, 953–964. [DOI] [PubMed] [Google Scholar]
- Schneider R., Travers, A. and Muskhelishvili, G. (2000) The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol. Microbiol., 38, 167–176. [DOI] [PubMed] [Google Scholar]
- Travers A.A. (1980) Promoter sequence for the stringent control of bacterial ribonucleic acid synthesis. J. Bacteriol., 141, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A.A. and Muskhelishvili, G. (1998) DNA microloops and microdomains—a general mechanism for transcription activation by torsional transmission. J. Mol. Biol., 279, 1027–1043. [DOI] [PubMed] [Google Scholar]
- Travers A., Schneider, R. and Muskhelishvili, G. (2001) DNA supercoiling and transcription in Escherichia coli—the FIS connection. Biochimie, 83, 213–217. [DOI] [PubMed] [Google Scholar]
- Tse-Dinh Y.-S., Qi, H. and Menzel, R. (1997) DNA supercoiling and bacterial adaptation: thermotolerance and thermoresistance. Trends Microbiol., 5, 323–326. [DOI] [PubMed] [Google Scholar]
- Verbeek H., Nilsson, L., Baliko, G. and Bosch, L. (1990) Potential binding sites of the trans-activator Fis are present upstream of all rRNA and of many but not all tRNA operons. Biochim. Biophys. Acta, 1050, 302–306. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Göringer, H.U. and Wagner, R. (1992) Analysis of the Fis-dependent and Fis-independent transcription activation mechanisms of the Escherichia coli ribosomal RNA P1 promoter. Biochemistry, 31, 2621–2628. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E.L., Khodursky, A.B. and Cozzarelli, N.R. (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev., 11, 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]