Abstract
pEPI-1, a vector in which a chromosomal scaffold/matrix-attached region (S/MAR) is linked to the simian virus 40 origin of replication, is propagated episomally in CHO cells in the absence of the virally encoded large T-antigen and is stably maintained in the absence of selection pressure. It has been suggested that mitotic stability is provided by a specific interaction of this vector with components of the nuclear matrix. We studied the interactions of pEPI-1 by crosslinking with cis-diamminedichloroplatinum II, after which it is found to copurify with the nuclear matrix. In a south-western analysis, the vector shows exclusive binding to hnRNP-U/SAF-A, a multifunctional scaffold/matrix specific factor. Immunoprecipitation of the crosslinked DNA–protein complex demonstrates that pEPI-1 is bound to this protein in vivo. These data provide the first experimental evidence for the binding of an artificial episome to a nuclear matrix protein in vivo and the basis for understanding the mitotic stability of this novel vector class.
INTRODUCTION
The eukaryotic genome is organized in a hierarchical fashion within the nucleus. On a first level, DNA interacts with the core histones to form the 10 nm fiber. This basic repetitive structure is then folded by the addition of chromatin proteins, such as histone H1, into a range of more highly condensed fibers and loops. Among these, the 30 nm fiber appears to be organized into looped domains by an interaction of specific sequences (scaffold/matrix-attached regions, S/MARs) with proteins of a subnuclear structure called nuclear scaffold or nuclear matrix. S/MARs of >1 kb have been considered as chromatin domain borders that play a critical role in nuclear architecture and function (Bode et al., 2000).
A number of proteins binding to S/MARs in vitro have been identified recently. These include ubiquitous nuclear proteins such as topoisomerase II (Adachi et al., 1989), lamin B1 (Luderus et al., 1992), SATB1 (Dickinson and Kohwi-Shigematsu, 1995), HMG I/Y (Zhao et al., 1993) and histone H1 (Izaurralde et al., 1989). Other highly conserved proteins that bind specifically to S/MARs via an evolutionary conserved protein domain include SAF-A (otherwise known as hnRNP-U) and SAF-B, which represent major components of the nuclear matrix and are therefore considered crucial for nuclear architecture in vivo (Renz and Fackelmayer, 1996; Kipp et al., 2000).
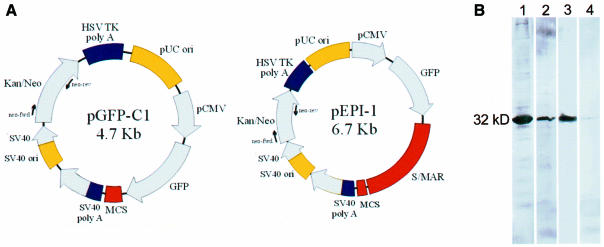
Organization of chromatin into topologically constrained loops by the interaction of S/MARs with nuclear matrix proteins not only has a major influence on transcriptional rates but, in addition, DNA replication seems to occur in tight association with the nuclear matrix, where the binding of the replication origin to this structure precedes the onset of S phase (Cook et al., 1999). We have shown recently that the viral simian virus 40 (SV40) origin linked to a human S/MAR allows sustained episomal replication of the vector that is independent of the expression of the virally encoded large T-antigen. This vector was stably maintained in CHO cells in the absence of selection pressure (Piechaczek et al., 1999). Further experiments suggested its specific interaction with the nuclear matrix and the chromosome scaffold. Following a nuclear fractionation protocol (Fey et al., 1986) and subsequent south-western analysis, its association with both a soluble nuclear protein and a protein copurifying with the nuclear matrix fraction was observed (Figure 1B). Both proteins had an approximate molecular mass of 32 kDa (Baiker et al., 2000) and were now identified as histone H1 variants.
Fig. 1. (A) Restriction maps of pEPI-1 and pGFP-C1. Arrows indicate the positions of primers used for PCR analysis. (B) South-western analysis after nuclear fractionation using either pEPI-1 or pGFP-C1 as a probe. Only binding to fractions 2 (soluble nuclear proteins) and 5 (nuclear matrix) is shown. Lanes 1 and 2, binding of pEPI-1 to fractions 2 and 5, respectively; lanes 3 and 4, binding of pGFP-C1 to fractions 2 and 5, respectively.
As chromosomal rearrangements, which may even mask specific interactions, cannot be strictly excluded in this type of experiment, we decided to prepare the nuclear matrix fraction after crosslinking with cis-diamminedichloroplatinum II (cis-DDP), a reagent that ties matrix proteins to endogenous S/MARs with high specificity (Ferraro et al., 1996), and to study the interaction of our vector with components of the nuclear matrix. We demonstrate, by south-western analysis, that pEPI-1 has a pronounced affinity to the matrix protein hnRNP-U/SAF-A. Moreover, we show, for the first time, the binding of an S/MAR-containing vector to a matrix protein in vivo. These observations will contribute to a mechanistic explanation for the observed mitotic stability of our episomally replicating construct.
RESULTS AND DISCUSSION
In the analysis of DNA–nuclear matrix interactions, one major problem is that uncontrolled chromosomal rearrangements may occur. For example, high salt treatment may cause sliding of DNA over its attachment points or exchange of the DNA-binding proteins. Moreover, DNA-binding proteins on their own often have a low affinity and a poor recognition of DNA, and the formation of multicomponent aggregates is required for their binding (Luderus et al., 1994; Ferraro et al., 1996; Kipp et al., 2000). This also restricts the identification of S/MARs and S/MAR-binding proteins by classical in vitro binding assays. The use of a small episomally replicating vector may facilitate these analyses. Vector pEPI-1 (Figure 1A), in which an S/MAR is linked to the SV40 origin of replication, propagates episomally in the absence of the virally encoded large T-antigen and is stably maintained even without selection. Its S/MAR-free counterpart, pGFP-C1, integrates randomly into the genome (Piechaczek et al., 1999; Baiker et al., 2000; Bode et al., 2000). We have provided evidence recently that mitotic stability of pEPI-1 may be due to its association with the nuclear matrix, and have shown, by south-western analysis, that pEPI-1 specifically binds to a protein of ∼32 kDa in the nuclear matrix fraction (Baiker et al., 2000; Figure 1B). Although this protein was not recognized by commercially available antibodies against histone H1, we now performed in-depth MALDI/TOF-MS analyses, which clearly demonstrated that this protein has homology to histones H1.1 and H1.4. Histone H1 has been shown to copurify with the S/MARs and is known to preferentially bind AT-rich regions (Bode et al., 2000).
We now decided to analyze the interactions of the vector with the nuclear matrix in detail by crosslinking with cis-DDP, a reagent with an established specificity for non-histones. Using this approach, we hoped to avoid a masking of specific S/MAR factors by ubiquitous H1. The preferential action of cis-DDP for nuclear matrix–S/MAR interactions was shown previously by Ferraro et al. (1992). This specificity is provided both by the amino acid composition of this protein class and by the special structural features, which are an exclusive property of S/MAR DNA (Bubley et al., 1996). The results obtained by this particular in vivo crosslinking approach consistently confirmed that S/MARs prepared in the classical way are indeed the anchorage points in intact nuclei (Ferraro et al., 1996). Using this strategy we will (i) demonstrate that pEPI-1 copurifies with the nuclear matrix; (ii) identify the matrix protein(s) to which it binds by south-western analysis; and (iii) demonstrate that binding of the vector to a member of the hnRNP class of proteins actually occurs in vivo.
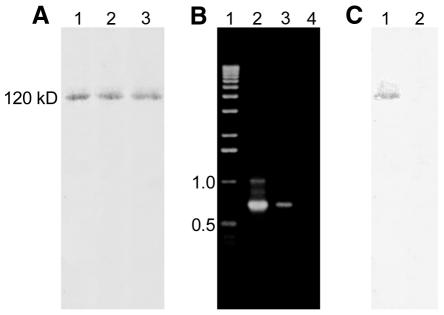
To this end, CHO cells transfected with either pEPI-1 or pGFP-C1 were crosslinked with cis-DDP and the reaction was terminated by raising the Cl– concentration in the medium. After cell lysis, the nuclear matrix DNA–protein complex was bound to hydroxyapatite (HAP) and DNA was isolated from the HAP-bound material as well as from non-bound material in the supernatant as described in the Methods (Ferraro et al., 1992). The presence of vector DNA in both fractions was traced by PCR analysis. While a PCR product was obtained using 10 ng of HAP-bound DNA derived from pEPI-1 transfected cells (Figure 2A, lane 3), the corresponding product was not obtained from pGFP-C1 transfected cells (Figure 2A, lane 4). Only by increasing the template concentration by at least 10-fold did a faint band (Figure 2A, lane 5) sometimes emerge on agarose gels. Conversely, the concentration of pEPI-1 in the supernatant (Figure 2B, lane 3) was significantly lower than that of the integrating vector pGFP-C1 (Figure 2B, lane 4), demonstrating that pEPI-1 preferentially copurifies with the nuclear matrix fraction. This result was obtained consistently regardless of whether the DNA was fragmented by restriction digestion or by sonification.
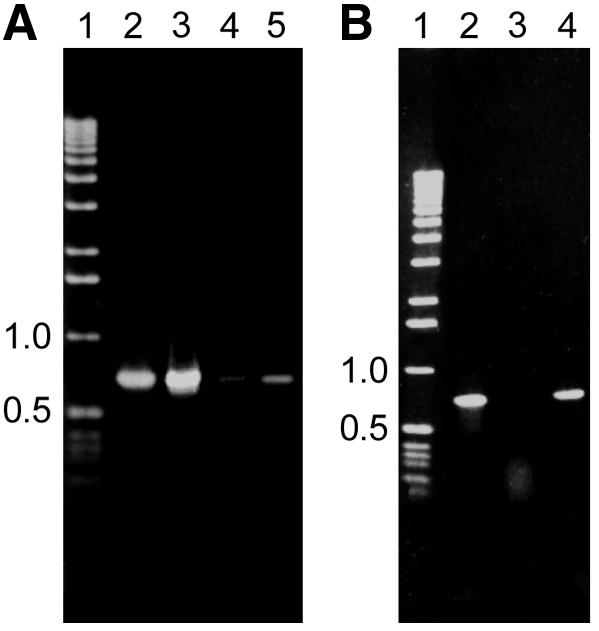
Fig. 2. PCR analysis from DNA bound to HAP or the supernatant after crosslinking of CHO cells transfected with either pEPI-1 or pGFP-C1. (A) PCR analysis of HAP-bound DNA. Lane 1, molecular weight marker; lane 2, control PCR using 1 ng of pEPI-1 vector DNA as a template; lane 3, PCR analysis from pEPI-1 transfected cells using 10 ng of DNA as a template; lane 4, PCR analysis from pGFP-C1 transfected cells using 10 ng of DNA as a template; lane 5, as lane 4 but using 100 ng of DNA as a template. (B) PCR analysis of the supernatant from pEPI-1 transfected cells using 10 ng of DNA as a template. Designations are as in (A).
Nuclear proteins either bound to HAP or occurring in the supernatant were isolated and a south-western analysis using either pEPI-1 or pGFP-C1 as a probe was performed. The banding pattern of the nuclear matrix fraction proteins was highly reproducible and was clearly distinguishable from the supernatant protein banding pattern. Although both vectors associated with the same protein components in the supernatant fraction, pEPI-1 was found to recognize a 32 kDa protein more strongly (Figure 3, lanes 1 and 3). Using MALDI/TOF-MS analysis, this protein was identified as histone H1, a histone with a well-known affinity for S/MARs.
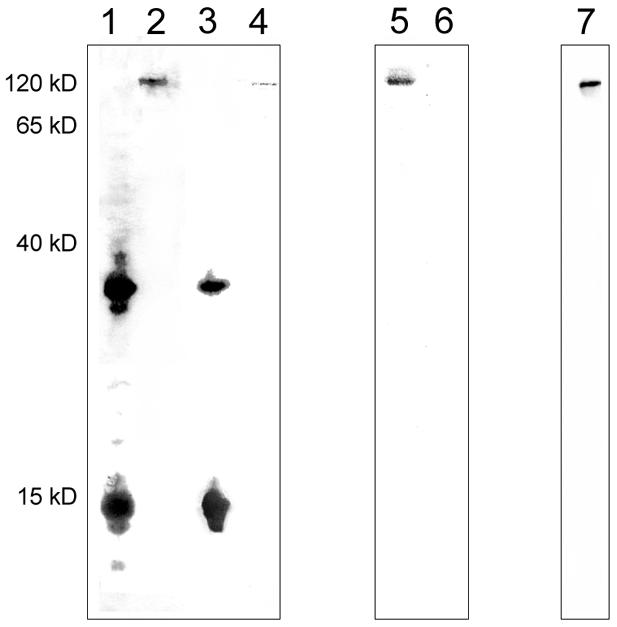
Fig. 3. South-western analysis of HAP-bound proteins and proteins of the supernatant after crosslinking with cis-DDP using either pEPI-1 or pGFP-C1 as a probe. Lane 1, pEPI-1 bound to supernatant proteins; lane 2, pEPI-1 bound to HAP-bound proteins; lane 3, pGFP-C1 bound to supernatant proteins; lane 4, pGFP-C1 bound to HAP-bound proteins. Lanes 1–4, binding was performed in the presence of a 10-fold excess of competitor DNA. Lane 5, as lane 2; lane 6, as lane 4. Lanes 5 and 6, binding was performed in the presence of a 100-fold excess of competitor DNA. Lane 7, western analysis of HAP-bound proteins using an anti-SAF-A antibody. Conditions for western and south-western analyses are described in the Methods.
Regarding the crosslinking approach, cis-DDP does not introduce crosslinks between histones and DNA. Since H1 is not part of the HAP-bound fraction obtained here, it cannot be excluded that the above H1 complexes have arisen from secondary re-association. Here, pEPI-1 binds very specifically to a protein with a molecular weight of 120 kDa in the nuclear matrix fraction (Figure 3, lanes 2 and 5). The signal intensity was independent of the amount of competitor DNA in the reaction (Figure 3, lanes 2 and 5). Only weak binding was observed when pGFP-C1 was used as a probe (Figure 3, lane 4) and was no longer detectable when the amount of unlabeled competitor DNA was raised 10-fold (Figure 3, lane 6). Interestingly, no binding of pEPI-1 to any other matrix-associated proteins was observed in these analyses. The molecular mass of the pEPI-1 associating factor and the fact that it represents a prominent nuclear matrix protein suggested that it could be the well-characterized S/MAR-binding protein SAF-A (Fackelmayer and Richter, 1994; Kipp et al., 2000). Indeed, western analysis with an anti-SAF-A antibody (Figure 3B, lane 7) clearly shows that the 120 kDa protein is homologous to SAF-A.
So far, proteins specifically binding to S/MARs have been characterized mostly by their affinity to defined sequences in vitro. The episomally replicating S/MAR-containing vector pEPI-1 provides a convenient possibility to identify proteins binding to S/MARs in vivo. Therefore, we performed an immunoprecipitation of the HAP-bound DNA–protein complex with an anti-SAF-A antibody. Following crosslinking of cells, transfected with either pEPI-1 or pGFP-C1, DNA–protein complexes were bound to HAP. This binding was reversed by incubation in 0.4 M sodium phosphate pH 7.2, the resulting supernatant was subjected to immunoprecipitation with SAF-A antibodies, and DNA and proteins were isolated from the precipitate. Western analysis demonstrated that SAF-A was immunoprecipitated from cells transfected with pEPI-1 and pGFP-C1 (Figure 4A, lanes 2 and 3). In contrast, when DNA was isolated from the immunoprecipitate and a PCR reaction performed with vector-specific primers, a PCR product was consistently obtained from pEPI-1 transfected cells (Figure 4B, lane 3) but never from pGFP-C1 transfected cells (Figure 4B, lane 4). No pEPI-1 specific PCR fragment could be amplified from the supernatant. As a control, we performed an immunoprecipitation with a non-specific antibody (anti-Oxytricha βTP, anti-rabbit IgG). In this case neither SAF-A (Figure 4C, lane 2) nor vector DNA was detectable in the precipitate. When DNA from pEP-1 transfected cells was digested with restriction enzymes, releasing the S/MAR sequence prior to HAP binding, a vector-specific PCR product could no longer be amplified from the immunoprecipitate. These results clearly demonstrate that pEPI-1 binds to SAF-A in vivo and suggest strongly that this binding is due to the S/MAR sequence present in this construct.
Fig. 4. (A) Western analysis using an anti-SAF-A antibody from proteins isolated from immunoprecipitates with an anti-SAF-A antibody of HAP-bound DNA–protein complexes after cis-DDP crosslinking of cells transfected with either pEPI-1 or pGFP-C1. Lane 1, total nuclear proteins from untransfected CHO cells; lane 2, proteins isolated from pEPI-1 transfected cells; lane 3, proteins isolated from pGFP-C1 transfected cells. (B) PCR analysis after immunoprecipitation with an anti-SAF-A antibody of HAP-bound DNA–protein complexes after cis-DDP crosslinking of cells transfected with either pEPI-1 or pGFP-C1. Lane 1, molecular weight marker; lane 2, control PCR using 1 ng of pEPI-1 vector DNA as template; lane 3, PCR analysis of immunoprecipitate from 5 × 107 pEPI-1 transfected cells; lane 4, PCR analysis of immunoprecipitate from 5 × 107 pGFP-C1 transfected cells. (C) Western analysis with an anti-SAF-A antibody of proteins after immunoprecipitation with non-specific antibodies (anti-Oxytricha βTP). Lane 1, proteins from the supernatant after immunoprecipitation; lane 2, proteins from immunoprecipitate. A similar result was obtained when immunoprecipitation was performed with an anti-rabbit IgG.
SAF-A (Romig et al., 1992) is a prominent protein of the nuclear matrix that specifically binds to S/MARs ranging from yeast to human through the highly conserved ‘SAF-box’. This box recognizes S/MARs through a multitude of weak interactions that collectively result in high-specificity binding (Kipp et al., 2000). Besides its apparent function in the organization of eukaryotic chromatin, SAF-A has been found to be identical to hnRNP-U (Fackelmayer and Richter, 1994), a constituent of hnRNP particles involved in the packaging and processing of RNA (Dreyfuss et al., 1993).
In a south-western analysis, we demonstrated that pEPI-1 specifically binds SAF-A. Applying advanced crosslinking and immunoprecipitation strategies, we could demonstrate not only that this vector is indeed bound to SAF-A in vivo but also that only this and no other hnRNP protein can be traced at this level of sensitivity. Such a conclusion was not possible by using standard nuclear fractionation techniques without crosslinking, as in this case the results were dominated by the ubiquitous, but possibly non-specific, association with histone H1. As shown in Figure 3, we can observe preferential binding of H1 to the S/MAR-containing vector (Figure 3, compare lanes 1 and 3) but not in the HAP-bound fraction that contains the crosslinked S/MAR–protein complexes.
The mitotic stability of pEPI-1 was ascribed previously to its association with the nuclear matrix (Baiker et al., 2000). We now provide direct experimental evidence for this hypothesis, which is considered a first important step towards a mechanistic explanation for the stable propagation of this vector. Of the two prominent S/MAR-associating components, lamin B1 and hnRNP-U/SAF-A (Ferraro et al., 1996), only the latter component could be detected, arguing for the specificity of this approach.
A large variety of hnRNP proteins exist that, in addition to their role in transcript packaging, function in a variety of other pathways that influence gene expression and, possibly, replication. A prominent additional role that has been ascribed to hnRNP/SAF-A is the nuclear retention of associated molecules (Krecic and Swanson, 1999), whereby it may well be in the position to provide the maintenance function required for stable episomal replication. In this context, it should be noted that S/MAR functions are associated with all eukaryotic circular episomes and that S/MARs have been discussed previously as maintenance elements (Bode et al., 2001). Our study sheds light on the mechanism by which this can occur: pEPI-1 is specifically associated, via SAF-A, with the chromosome scaffold in vivo, enabling a co-segregation with the chromosomes during mitotic division. In addition to providing a binding site for SAF-A, the S/MAR in this vector will likely recruit nuclear components that substitute for some other functions of the large T-antigen, most probably to helix destabilization that leads to the formation of an ‘open complex’ at the origin of replication necessary for the assembly of the replicating machinery.
METHODS
Cells and vectors. CHO cells were grown, transfected and selected as described earlier. The vectors used were pEPI-1 (Piechaczek et al., 1999) and pGFP-C1 (Clontech; Figure 1A). While pEPI-1 replicates episomally in the absence of selection pressure in CHO cells, its S/MAR-free counterpart pGFP-C1 consistently integrates into the genome in a random fashion (Piechaczek et al., 1999).
Nuclear fractionation. Nuclear fractionation followed the procedure described previously (Fey et al., 1986). In this procedure, nuclear proteins are fractionated according to their resistance to salt treatment, resulting in a matrix preparation in the last fraction (fraction 5, Figure 1B, lanes 2 and 4).
Crosslinking with cis-DDP. cis-DDP is a reagent that links matrix proteins to S/MARs with high specificity in low [Cl–] media (Ferraro et al., 1996), thus preventing chromosomal rearrangements during matrix preparation. Cells (108) were collected by centrifugation and the cis-DDP crosslinking was performed as described previously (Ferraro et al., 1992). Crosslinking is immediately stopped by access of [Cl–] during the steps performed below.
Isolation of proteins after crosslinking (Ferraro et al., 1992). Following the crosslinking step, the cells were collected and resuspended in 80 ml of lysis buffer (5 M urea, 2 M guanidine hydrochloride, 2 M NaCl, 1 mM PMSF). The lysate was added to 1.6 g of HAP (Bio-Rad), pre-equilibrated with lysis buffer and incubated on orbitron for 1 h at 4°C. The pellet was washed three times with ice-cold lysis buffer. The crosslink between protein and HAP-bound DNA was reversed by the addition of 80 ml ice-cold reverse lysis buffer (1 M thiourea, 2 M guanidine hydrochloride, 2 M NaCl, 1 mM PMSF) and incubated on orbitron for 2 h at 4°C. After centrifugation for 30 min at 6200 g, DNA-bound proteins could be recovered from the supernatant. This supernatant was dialyzed against distilled water containing 1 mM PMSF for 24 h at 4°C and concentrated by centrifugation through Vivaspin 20 (Sartorius, 10 000 MWCO).
Isolation of DNA after crosslinking (Ferraro et al., 1992). Following crosslinking, the DNA was fragmented either by sonification as described earlier or by restriction digestion. In the latter case, crosslinked cells were collected by centrifugation, resuspended in 1 ml of TE and digested for 2 h at 37°C with five restriction enzymes not cutting the vectors (200 U each, EcoRV, PvuI, XhoI, NotI and ClaI) and one enzyme (EcoRI, 200 U) linearizing them. In some cases, the S/MAR sequence was deleted from pEPI-1 by digestion with EcoRI and BglII. The cells were then collected by centrifugation, resuspended in 80 ml of lysis buffer and bound to HAP as described above. Dissociation of DNA and DNA–protein complexes from HAP was achieved by incubation in 80 ml of 0.4 M sodium phosphate buffer pH 7.2 on orbitron for 2 h at 4°C. After centrifugation for 30 min at 6200 g and 4°C, the supernatant was dialyzed against TE for 20 h at 4°C. To precipitate protein-containing complexes, SDS was added to a final concentration of 1%, incubated for 10 min at 37°C, then KCl was added to a final concentration of 0.1 M, incubated for 10 min on ice and centrifuged for 10 min at 3000 g and 4°C. The pellet was resuspended in 20 ml of TE containing 0.1 M KCl and incubated for 10 min at 65°C followed by 10 min incubation on ice and centrifugation for 10 min at 3000 g. This step was repeated twice, the precipitate was resuspended in 5 ml of 10 mM Tris pH 8.0, 10 mM EDTA, 100 mM NaCl, 0.4 mg proteinase K and incubated for 24 h at 37°C. DNA was extracted with phenol–chloroform and precipitated with ethanol.
PCR analysis. The presence of vector DNA in the various fractions and after immunoprecipitation was detected by PCR analysis. PCR conditions and primers used were as described previously (Baiker et al., 2000).
Immunoprecipitation. Immunoprecipitation of the HAP-bound DNA–protein complex with anti-SAF-A antibody (Fackelmayer and Richter, 1994) was conducted as follows. Following crosslinking, lysates from ∼108 cells were bound to HAP as described above. Binding was reversed by incubation in 0.4 M sodium phosphate buffer pH 7.2, HAP was removed by centrifugation and the supernatant dialyzed against TE and concentrated in Vivaspin 20 to a final volume of ∼200 µl. PBS (10×) to a final concentration of 1× PBS was added. Anti-SAF-A antibody (12 µl/ml) was added and incubated on ice overnight. The resulting precipitate was collected by centrifugation for 2 h at 13 000 g and 4°C and washed twice with PBS. The precipitate was resuspended in 40 µl of TE and the DNA was isolated as described above. Aliquots of the DNA were used to amplify the vector DNA by PCR. As a control, immunoprecipitation was performed with antibodies of different specificities (anti-Oxytricha βTP, anti-rabbit IgG).
Gel electrophoresis, south-western and western analysis. DNA was separated on 1% agarose gels in TAE. Proteins were fractionated on 12.5% SDS–polyacrylamide gels (Laemmli, 1970) and transferred to nylon membranes by electroblotting. Western analysis was performed as described previously (Towbin et al., 1979) using the anti-SAF-A antibody (dilution 1:500) and the anti-rabbit IgG (Dako, Hamburg, Germany; dilution 1:1000). South-western analysis followed the protocol of Miskimins et al. (1985) with 0.2 µg of DIG-labeled (Kessler et al., 1990) pEPI-1 or pGFP-C1. Binding with pEPI-1 was performed in the presence of a 10 resp. 100-fold excess of unlabeled pGFP-C1 and binding of pGFP-C1 in the presence of a 10 resp. 100-fold excess of unlabeled pEPI-1.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Carlo Turano (Roma) for valuable advice and Jan Postberg (Witten) for his help with the illustrations. This work was supported by the Deutsche Forschungsgemeinschaft and the Alfried Krupp von Bohlen and Halbach foundation. B.H.C.J. is a recipient of a scholarship from the Studienstiftung des Deutschen Volkes.
REFERENCES
- Adachi Y., Kas, E. and Laemmli, U.K. (1989) Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J., 8, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiker A., Maercker, C., Piechaczek, C., Schmidt, S.B., Bode, J., Benham, C. and Lipps, H.J. (2000) Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nature Cell Biol., 2, 182–184. [DOI] [PubMed] [Google Scholar]
- Bode J., Benham, C., Knopp, A. and Mielke, C. (2000) Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr., 10, 73–90. [PubMed] [Google Scholar]
- Bode J., Fetzer, C.P., Nehlsen, K., Scinteie, M., Hinrichsen, B.H., Baiker, A., Piechazcek, C., Benham, C. and Lipps, H.J. (2001) The Hitchhiking Principle: optimizing episomal vectors for use in gene therapy and biotechnology. Int. J. Gene Ther. Mol. Biol., 6, 33–46. [Google Scholar]
- Bubley G.J., Xu, J., Kupiec, N., Sanders, D., Foss, F., O’Brien, M., Emi, Y., Teicher, B.A. and Patiemo, S.R. (1996) Effect of DNA conformation on cisplatin adduct formation. Biochem. Pharmocol., 51, 717–721. [DOI] [PubMed] [Google Scholar]
- Cook G.A., Wilkinson, D.A., Crossno, J.T.,Jr, Raghow, R. and Jennings, L.K. (1999) The tetraspanin CD9 influences the adhesion, spreading, and pericellular fibronectin matrix assembly of Chinese hamster ovary cells on human plasma fibronectin. Exp. Cell Res., 251, 356–371. [DOI] [PubMed] [Google Scholar]
- Dickinson L.A. and Kohwi-Shigematsu, T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol., 15, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis, M.J., Pinol-Roma, S. and Burd, C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Fackelmayer F.O. and Richter, A. (1994) Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry, 33, 10416–10422. [DOI] [PubMed] [Google Scholar]
- Ferraro A., Grandi, P., Eufemi, M., Altieri, F. and Turano, C. (1992) Crosslinking of nuclear proteins to DNA by cis-diamminedichloroplatinum in intact cells. Involvement of nuclear matrix proteins. FEBS Lett., 307, 383–385. [DOI] [PubMed] [Google Scholar]
- Ferraro A., Cervoni, L., Eufemi, M., Altieri, F. and Turano, C. (1996) Comparison of DNA–protein interactions in intact nuclei from avian liver and erythrocytes: a cross-linking study. J. Cell. Biochem., 62, 495–505. [DOI] [PubMed] [Google Scholar]
- Fey E.G., Krochmalnic, G. and Penman, S. (1986) The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol., 102, 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Kas, E. and Laemmli, U.K. (1989) Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J. Mol. Biol., 210, 573–585. [DOI] [PubMed] [Google Scholar]
- Kessler C., Holtke, H.J., Seibl, R., Burg, J. and Muhlegger, K. (1990) Non-radioactive labeling and detection of nucleic acids. I. A novel DNA labeling and detection system based on digoxigenin: anti-digoxigenin ELISA principle (digoxigenin system). Biol. Chem. Hoppe Seyler, 371, 917–927. [DOI] [PubMed] [Google Scholar]
- Kipp M., Göhring, F., Ostendorp, T., van Drunen, C.M., van Driel, R., Przybylski, M. and Fackelmayer, F.O. (2000) SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol., 20, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson, M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Luderus M.E., de Graaf, A., Mattia, E., den Blaauwen, J.L., Grande, M.A., de Jong, L. and van Driel, R. (1992) Binding of matrix attachment regions to lamin B1. Cell, 70, 949–959. [DOI] [PubMed] [Google Scholar]
- Luderus M.E., den Blaauwen, J.L., de Smit, O.J., Compton, D.A. and van Driel, R. (1994) Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol., 14, 6297–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins W.K., Roberts, M.P., McClelland, A. and Ruddle, F.H. (1985) Use of a protein-blotting procedure and a specific DNA probe to identify nuclear proteins that recognize the promoter region of the transferrin receptor gene. Proc. Natl Acad. Sci. USA, 82, 6741–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczek C., Fetzer, C., Baiker, A., Bode, J. and Lipps, H.J. (1999) A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res., 27, 426–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz A. and Fackelmayer, F.O. (1996) Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res., 24, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romig H., Fackelmayer, F.O., Renz, A., Ramsperger, U. and Richter, A. (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J., 11, 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin, T. and Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Kas, E., Gonzalez, E. and Laemmli, U.K. (1993) SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J., 12, 3237–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]