Abstract
Background:
When patients do not have matched siblings or well-matched unrelated donors (MUD), hematopoietic cell transplantation (HCT) can still be successful using an HLA-mismatched unrelated donor (MMUD), in combination with post-transplant cyclophosphamide (PTCy), abatacept, or other novel approaches. This may allow clinicians to choose a suitable donor from a wide range of donor options while optimizing other donor selection characteristics, including donor age.
Objectives:
We hypothesized that allowing for 5/8 HLA match level considering high resolution matching at HLA-A, -B, -C and -DRB1, there is potential to close the donor availability gap for all patients regardless of their race/ethnicity. In this work, we estimate the likelihood of matching for all race/ethnic groups at different HLA match thresholds.
Study Design:
Our study aimed to assess the potential for identifying an available MUD or MMUD in the National Marrow Donor Program® (NMDP)/Be The Match® (BTM) donor registry for 21 detailed and 5 broad racial/ethnic groups, utilizing high-resolution HLA matching for HLA-A, B, C, and DRB1 at various levels (8/8, 7/8, 6/8, and 5/8). We used donor registry population data from the NMDP/BTM in 2020 and redistributed the donor registry data according to existing population ratios, accounting for demonstrated donor availability. Finally, we utilized a genetic model at the population level to estimate the match likelihood for detailed and broad racial/ethnic groups.
Results:
8/8 HLA match likelihoods ranging from 16–74% were obtained for various detailed race/ethnic groups with available donors ≤35 years old. When considering more mismatches within the HLA loci, registry coverage became >99% with a 5/8 HLA match level for donors of all ages or those ≤ 35 years old, with DPB1 T cell epitope permissive matching, or when searching donors outside of their race/ethnic group.
Conclusions:
Our registry models demonstrate the potential of using MMUDs at various HLA match levels to study whether this will expand access to HCT across race/ethnic groups. Furthermore, expanded donor options may erase the donor availability gap for all patients while allowing for selection of MMUDs with favorable characteristics such as younger age.
Keywords: Unrelated donors, Registry modeling, HLA match level, Match likelihoods, Donor age, DPB1 TCE match/permissive mismatch
INTRODUCTION
Allogeneic hematopoietic cell transplantation (alloHCT) can cure a variety of malignant and non-malignant disorders. The best donor option is traditionally an HLA-matched sibling donor (MSD); however, availability ranges from 13–51% based on patient age and ancestry [1, 2]. When a MSD is not available, the next best donor option is a high-resolution HLA-A, B, C and DRB1 (8/8) - matched unrelated donor (MUD) [3]. Multiple studies including the annual Center for International Blood and Marrow Transplant Research (CIBMTR) Center Specific Analysis [4, 5] supports these results. The 2022 Center Specific Analysis report collated data from >25,000 unrelated alloHCT and related alloHCT performed between 2018–2020 in the U.S. with follow up through one year after transplant. Alternative donor types can be compared and assessed using the odds ratio (OR) of overall survival (OS) at 1 year compared to matched sibling donor (MSD) as a reference with OR for MUD 0.91 (95%CI 0.82–1.02, p=0.097), haploidentical related donor (Haplo) OR 0.72 (95% CI: 0.65–0.80, p<0.001), mismatched (7/8) unrelated donor (MMUD) OR 0.73 (95% CI: 0.62–0.86, p<0.001) and umbilical cord blood (Multiple UCB≥4/6) OR 0.46 (95% CI: .35–0.59, p<0.001). This large multi-center analysis of real-world data shows a clear hierarchy for selection of donor sources that first prioritizes MSD followed by MUD and then distinct mismatched donor sources (Haplo, MMUD and umbilical cord blood) [4, 6].
Existence of a MUD varies by the ancestry of the patient due to high levels of HLA polymorphism and the composition of worldwide registries. In a prior study, Gragert et al. [1] analyzed greater than 10.5 million adult donors from Be the Match Registry (BTM) operated by the National Marrow Donor Program (NMDP) and found 16–75% likelihood of having an available 8/8 MUD with lowest among African American patients and highest in White patients. Without an available MUD, MMUD can help close the donor availability gap, increasing the likelihood of an available ≥7/8 MUD/MMUD to 66%−97%. However, the use of MMUD has historically come at a cost of greater risk of mortality, graft vs host disease (GVHD) and non-relapse mortality (NRM) [3, 7], particularly in the setting of traditional calcineurin inhibitor based GVHD prophylaxis. Novel GVHD prophylaxis strategies, such as post-transplant cyclophosphamide (PTCy) [8–10], abatacept [11] or graft engineering, [12] have shown the potential to lower the risk of alloHCT across traditional HLA barriers.
The PTCy strategy has facilitated rapid growth in the use of Haplo donors over the past decade, helping to address the donor availability gap, particularly for ethnically diverse patients [13, 14]. A recent prospective multicenter phase II trial sponsored by the NMDP and conducted by the CIBMTR (15-MMUD) demonstrated that this approach can achieve excellent results in ≤7/8 MMUD bone marrow transplants for hematological malignancies with no difference in outcomes observed between 7/8 and <7/8 MMUD in a series of 80 adult patients [9]. In addition, the 15-MMUD outcomes did not significantly differ from a contemporaneous cohort of Haplo transplants reported to the CIBMTR. The ≤7/8 MMUD PTCy approach is currently being evaluated in the context of adult alloHCT using peripheral blood stem cell (PBSC) and bone marrow (BM) grafts in the pediatric patient population through the NMDP-sponsored phase II ACCESS trial (https://clinicaltrials.gov/study/NCT04904588).
The ability to perform safe and effective alloHCT utilizing ≤7/8 MMUD has the potential to greatly widen available donor possibilities and eliminate the donor availability barrier to transplant. We sought to quantify the impact of less restrictive matching in the framework of previously established registry models. In this work, we calculated HLA match likelihoods using a population-level genetic model for all detailed and broad race/ethnic groups in the National Marrow Donor Program® (NMDP)/Be The Match® (BTM) donor registry for high-resolution HLA-A, B, C, and DRB1 8/8, 7/8, 6/8, and 5/8 match levels. We adjusted for donor availability and optimization of other characteristics for donor selection, including younger donor age (≤35 years old) and HLA-DPB1 T cell epitope (TCE) match. We hypothesized that by reducing the HLA match level requirement to a minimum of 5/8, there is potential to close the donor availability gap for all patients regardless of their race/ethnicity. Another aim of this work was to estimate within- and outside-group match likelihoods for patients with and without considering available donors of their own race/ethnic groups.
MATERIALS and METHODS
Study Population
Our modeling approach is based on sampling from HLA haplotype frequencies from 21 detailed race/ethnic groups generated from all registry donors, regardless of typing resolution through 2020 using an expectation-maximization-based algorithm [15, 16]. To address variation in typing resolution, we used these haplotype frequencies to model a registry with the same size and race/ethnic composition as the full NMDP/BTM database. The apportioning of these 23.3 million adult donors among the 21 detailed and 5 broad ethnic groups is shown in Table 1. Note that this process required re-apportioning donors that lack race/ethnic information or who self-identify as multi-racial to be re-apportioned to these 21 groups. We modeled patients’ HLA by repeatedly sampling genotypes from the same HLA haplotype frequencies (across the 4 HLA loci: HLA-A, HLA-B, HLA-C, and HLA-DRB1). This allows us to determine the match rate at high-resolution despite the fact that the registry has partial and ambiguous typing for many donors and patients.
Table 1.
Donor statistics of the detailed/broad race/ethnic groups on the Registry.
| Detailed Race/Ethnic Group | Effective Registry Size (donors of any age) | Effective Registry Size (donors of ≤35 years old) | Broad Race/Ethnic Group | Effective Registry Size (donors of any age) | Effective Registry Size (donors of ≤35 years old) |
|---|---|---|---|---|---|
| African American | 364670 (3.42) | 152179 (3.39) | African American | 433724 (4.06) | 185822 (4.14) |
| African | 31510 (0.3) | 15895 (0.35) | |||
| Caribbean Black | 33058 (0.31) | 16293 (0.36) | |||
| Black - South or Central American | 4486 (0.04) | 1455 (0.03) | |||
| South Asian Indian | 345250 (3.23) | 131675 (2.93) | Asian or Pacific Islander | 909662 (8.52) | 380760 (8.48) |
| Filipino | 87989 (0.82) | 38273 (0.85) | |||
| Hawaiian or Other Pacific Islander | 18928 (0.18) | 7486 (0.17) | |||
| Japanese | 34637 (0.32) | 7464 (0.17) | |||
| Korean | 118003 (1.11) | 44258 (0.99) | |||
| Chinese | 176462 (1.65) | 86092 (1.92) | |||
| Southeast | 53331 (0.5) | 26925 (0.6) | |||
| Asian Vietnamese | 75062 (0.7) | 38587 (0.86) | |||
| Middle Eastern or N. Coast of Africa | 440319 (4.13) | 177556 (3.95) | Caucasian | 8378155 (78.5) | 3576144 (79.61) |
| European Caucasian | 7937836 (74.37) | 3398588 (75.66) | |||
| Caribbean Hispanic | 271006 (2.54) | 122947 (2.74) | Hispanic | 879156 (8.24) | 327676 (7.29) |
| Mexican or Chicano | 214696 (2.01) | 5420 (0.12) | |||
| Hispanic - South or Central American | 393454 (3.69) | 199309 (4.44) | |||
| American Indian - South or Central Am. | 10166 (0.1) | 4573 (0.1) | Native American | 72582 (0.68) | 21803 (0.49) |
| Alaska Native or Aleut | 5135 (0.05) | 2483 (0.06) | |||
| North American Indian | 53407 (0.5) | 12399 (0.28) | |||
| Caribbean Indian | 3874 (0.04) | 2348 (0.05) |
Match Likelihood Estimation Procedure
In this study, we considered all donors on the registry at the end of 2020. To address missing and multi-race, we reapportioned donors into 21 detailed race/ethnic groups according to previous studies [1, 17]. We developed registry models for donors of all ages and donors ≤35 years old. We estimated HLA match likelihoods at 5/8–8/8 levels including a DPB1 TCE match/permissive mismatch using the population-based genetic model, 4- and 5-locus HLA haplotype frequencies, genotype pair frequencies, and DPB1 TCE group frequencies. The detail of the allele-to-TCE-group mappings and the rules for permissive mismatching of DPB1 TCE genotypes were previously described [17–19].
We also estimated match likelihoods for 5 broad race/ethnic groups by applying a simple weighting technique to the match likelihood results of 21 detailed race/ethnic groups. In the weighting scheme, at first, we calculated the proportion of each detailed race/ethnic group with respect to its broad race/ethnic group (refer to Table 1). Then to estimate the match likelihood of a broad race/ethnic group, we considered the summation of the product of match likelihood and proportioned value of all its detailed race/ethnic groups.
Finally, we computed “within group” and “outside group” match likelihoods. The “within group” HLA match likelihood reflects both the intrinsic heterogeneity of HLA and the effective registry size of that group and resembles the proportion of patients who can be served by searching for donors only within their own race/ethnic group. Since it is sometimes difficult to identify matched donors for particular patient race/ethnic groups due to high diversity or low sample size, the “outside group” match likelihood is measured to determine the proportion of patients who can find matches only outside of their race/ethnic group.
Analysis of real-world donor searches
The standard search process for a MUD is founded on a matching threshold of 6/8 alleles (at HLA-A, -B, -C and -DRB1) or more across the global list of adult donors. In 2021, the NMDP implemented an option to lower this threshold to 4/8 alleles or better to support clinical trials with this as the inclusion criteria. During the period from 2021–01-19 through 2023–01-11 the NMDP received 913 requests for searches at the lower threshold of 4/8 alleles or better. Fifteen patients were excluded that had productive searches (at least 10 donors in the US, aged 18–35 with probability of 8/8 match >=0.75). The remaining N = 898 patient searches were summarized in terms of the number of potential donors in quantiles (supplemental table S1).
RESULTS
Overall HLA Match Likelihoods
Table 1 lists the effective registry size of the 21 populations and 5 broad groups in 2020. The number in the parenthesis for a race/ethnic group indicates the percentage of donors available in that group with respect to total effective donor populations in 2020. European Caucasian and Black–South or Central American hold the largest and smallest effective registry size, respectively, in the detailed race/ethnic groups, and the same is true for the Caucasian and African American broad race/ethnic groups, respectively.
Figs. 1 and 2 list the overall match likelihoods of finding an available HLA-matched donor of any age and donors ≤35 years of age in the NMDP/BTM registry at 5/8–8/8 HLA match levels for 21 detailed, and 5 broad race/ethnic groups, respectively. The 8/8 HLA match likelihood for donors ≤35 years old ranges from 16–74% for 21 detailed race/ethnic groups whereas the range is 21–73% for 5 broad race/ethnic groups. If we consider donors of any age, the 8/8 HLA match likelihood ranges become 23–81% and 29–80% for 21 detailed and 5 broad race/ethnic groups, respectively. The ≥5/8 HLA match likelihood increases to ≥99% for all groups considering donors of all ages and ≤35 years of age.
Fig 1.
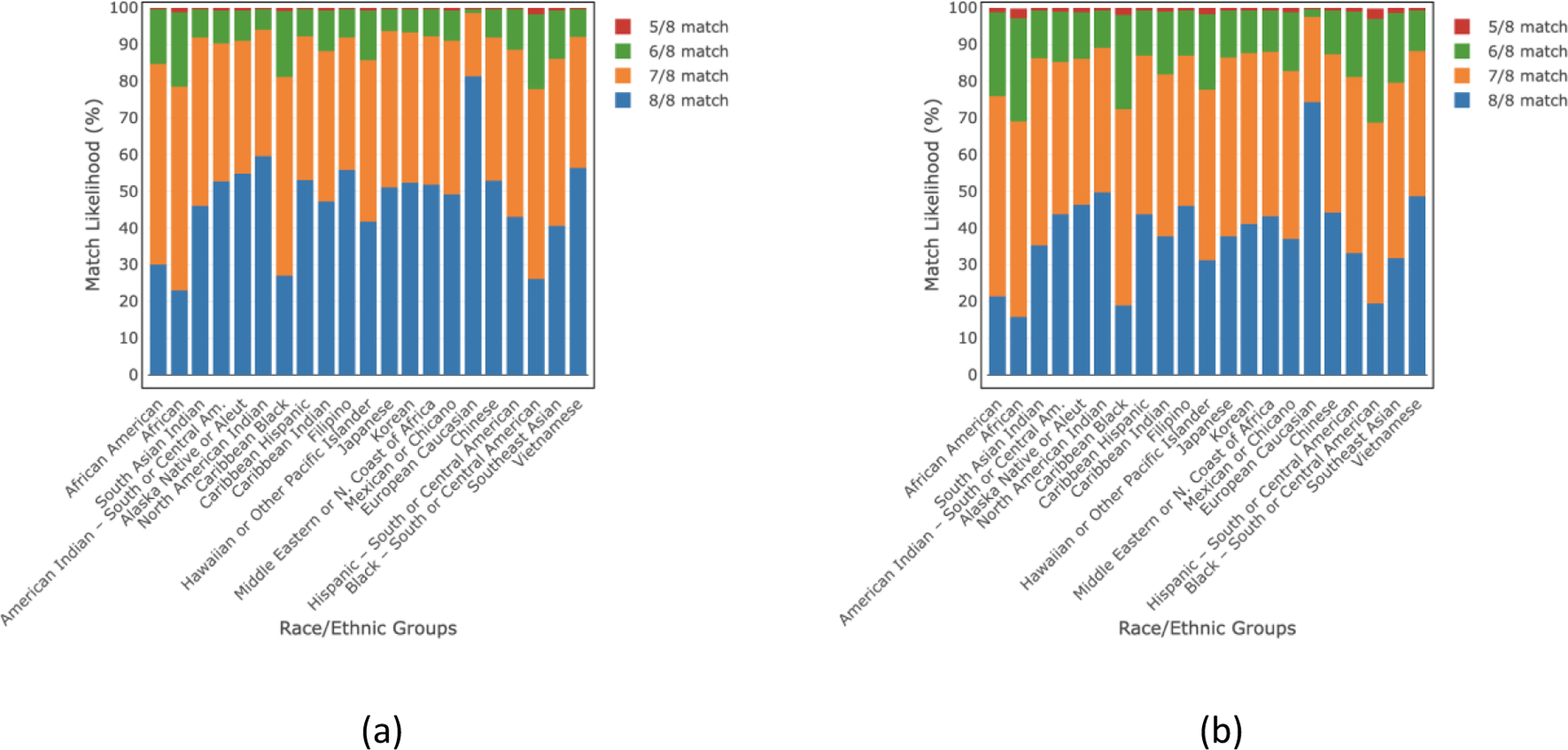
HLA match likelihoods (%) at 5/8–8/8 levels with (a) donors of all ages and (b) donors of age ≤35 years in 21 detailed race/ethnic groups.
Fig 2.
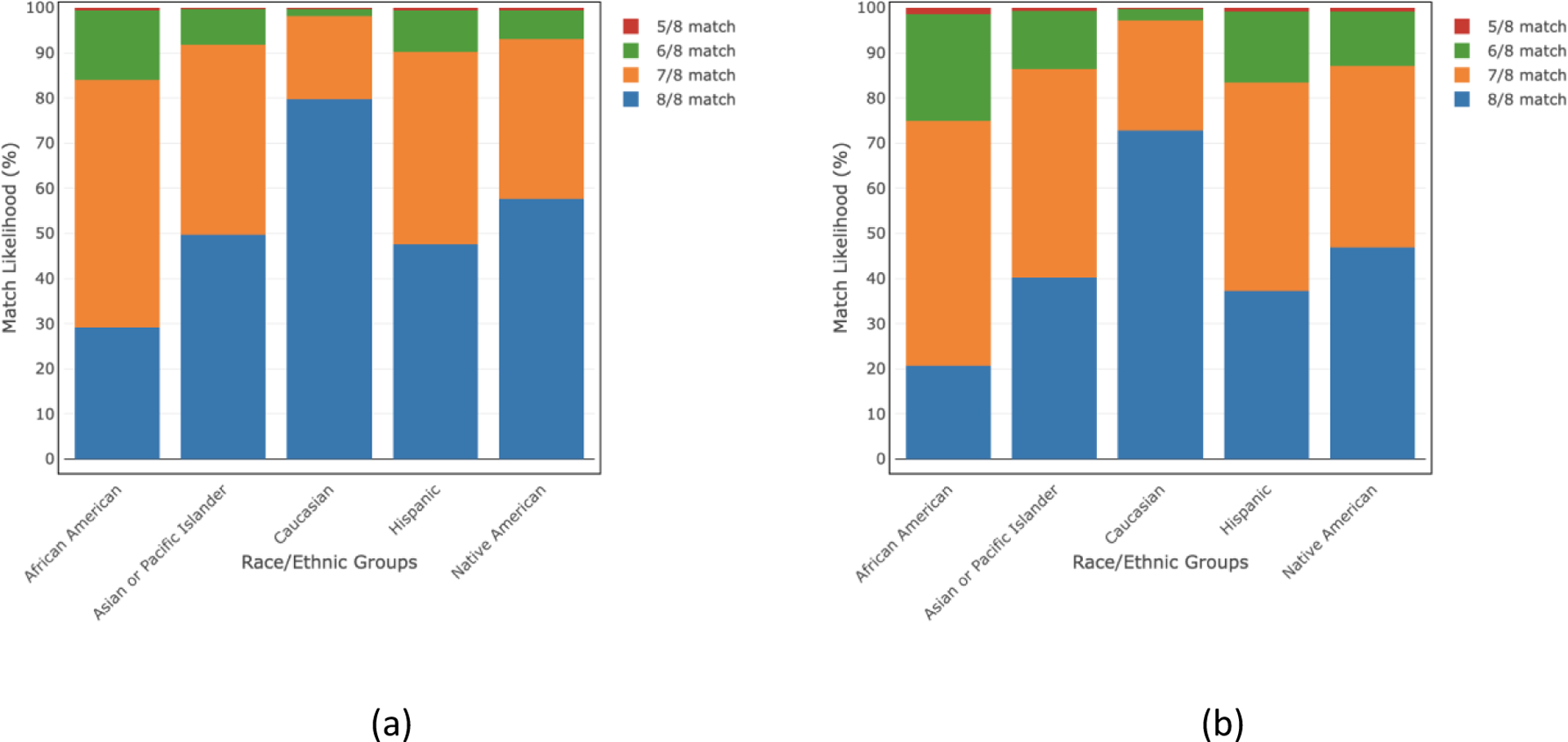
HLA match likelihoods (%) at 5/8–8/8 levels with donors of all ages and donors of age ≤35 years in 5 broad race/ethnic groups.
We also estimated the match likelihood for donors of any age and donors ≤35 years of age for all detailed and broad race/ethnic groups while considering DPB1 TCE match/permissive mismatches (Figs. 3 and 4, respectively). For DPB1 TCE match/permissive mismatches, the 8/8 match likelihood for donors of age ≤35 ranges from 13–68% for the detailed race/ethnic groups and 18–66% for the broad race/ethnic groups. These ranges become 19–76% and 25–74% for all detailed and broad race/ethnic groups, respectively when we consider donors of any age scenario. The ≥5/8 HLA match likelihood is ≥99% for all groups considering donors ≤35 years of age and all ages. All match likelihoods for both donors of all ages and donors ≤35 years old are also listed in the supplemental tables S2–S5.
Fig 3.
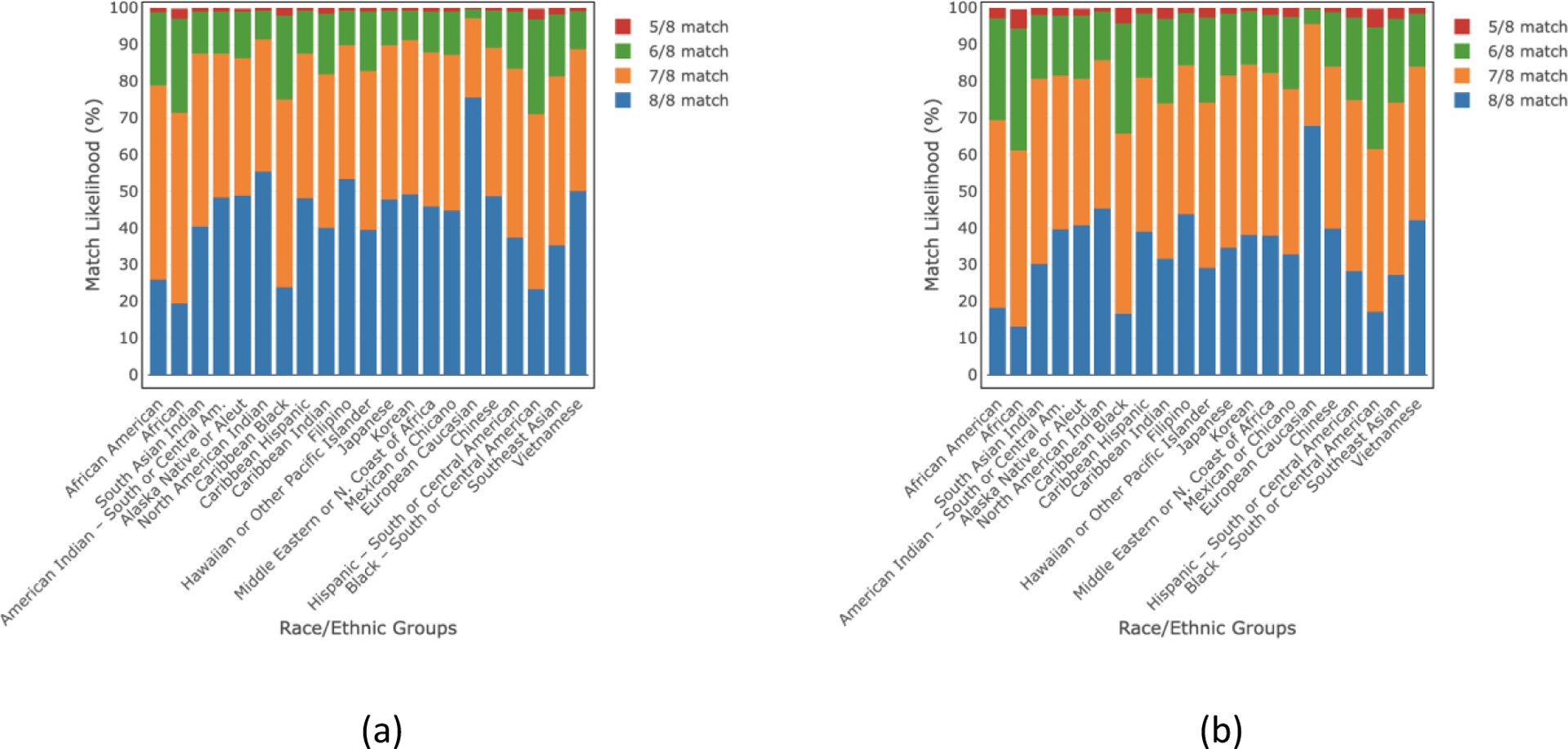
HLA match likelihoods (%) at 5/8–8/8 levels including a DPB1 TCE match/permissive mismatch for (a) donors of all ages and (b) donors of age≤35 years in 21 race/ethnic groups.
Fig 4.
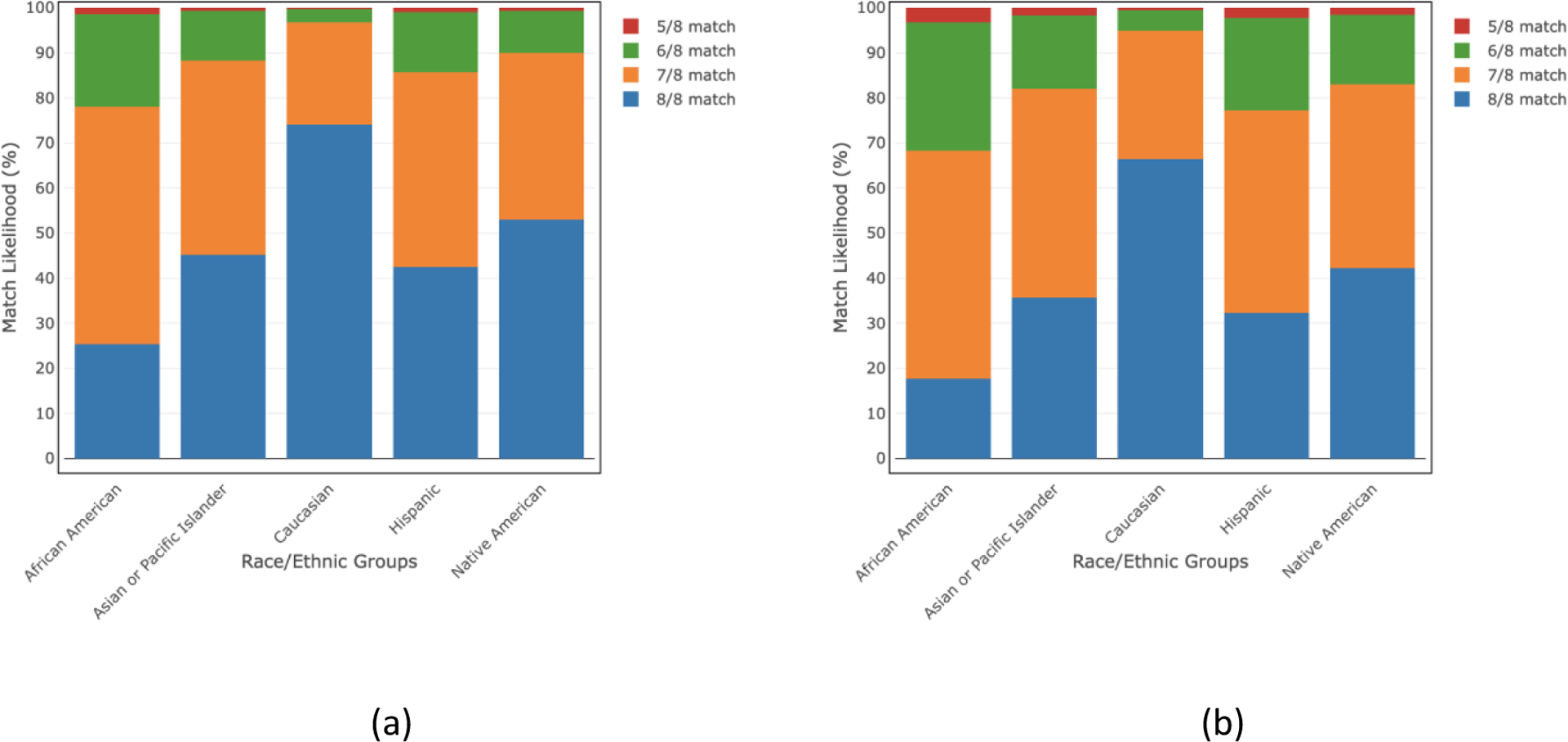
HLA match likelihoods (%) at 5/8–8/8 levels including a DPB1 TCE match/permissive mismatch for donors of all ages and donors of age≤35 in 5 broad race/ethnic groups.
Within and outside group HLA Match Likelihoods
Figs. 5 and 6 demonstrate the estimated HLA match likelihoods for detailed race/ethnic groups, indicating the likelihood of finding a suitable donor within their own group and the potential for exploring donor options outside their respective race/ethnic group. For within-group match likelihoods, at the ≥5/8 HLA match level, we found these to be >91% and >76% for donors of any age and donors of age ≤35 years, respectively. Considering ≥5/8 HLA match level, the outside group match likelihoods became >99% both for donors of any age and donors of age ≤35 years. Within and outside group HLA match likelihoods are also provided in tabular format in the supplemental tables S6–S7.
Fig 5.
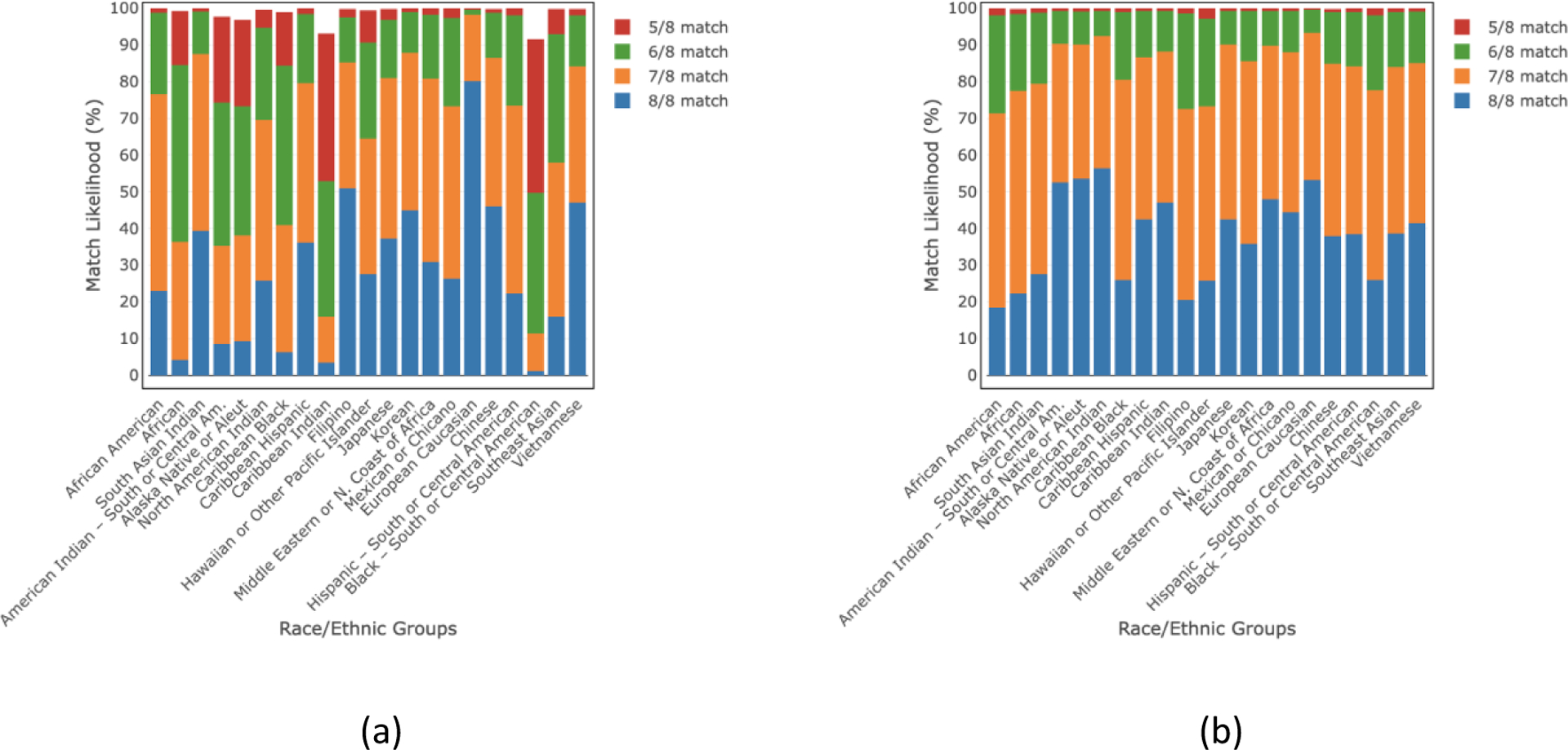
(a) “Within group” and (b) “outside group” HLA match likelihoods (%) at 5/8–8/8 levels for donors of all ages in 21 race/ethnic groups.
Fig 6.
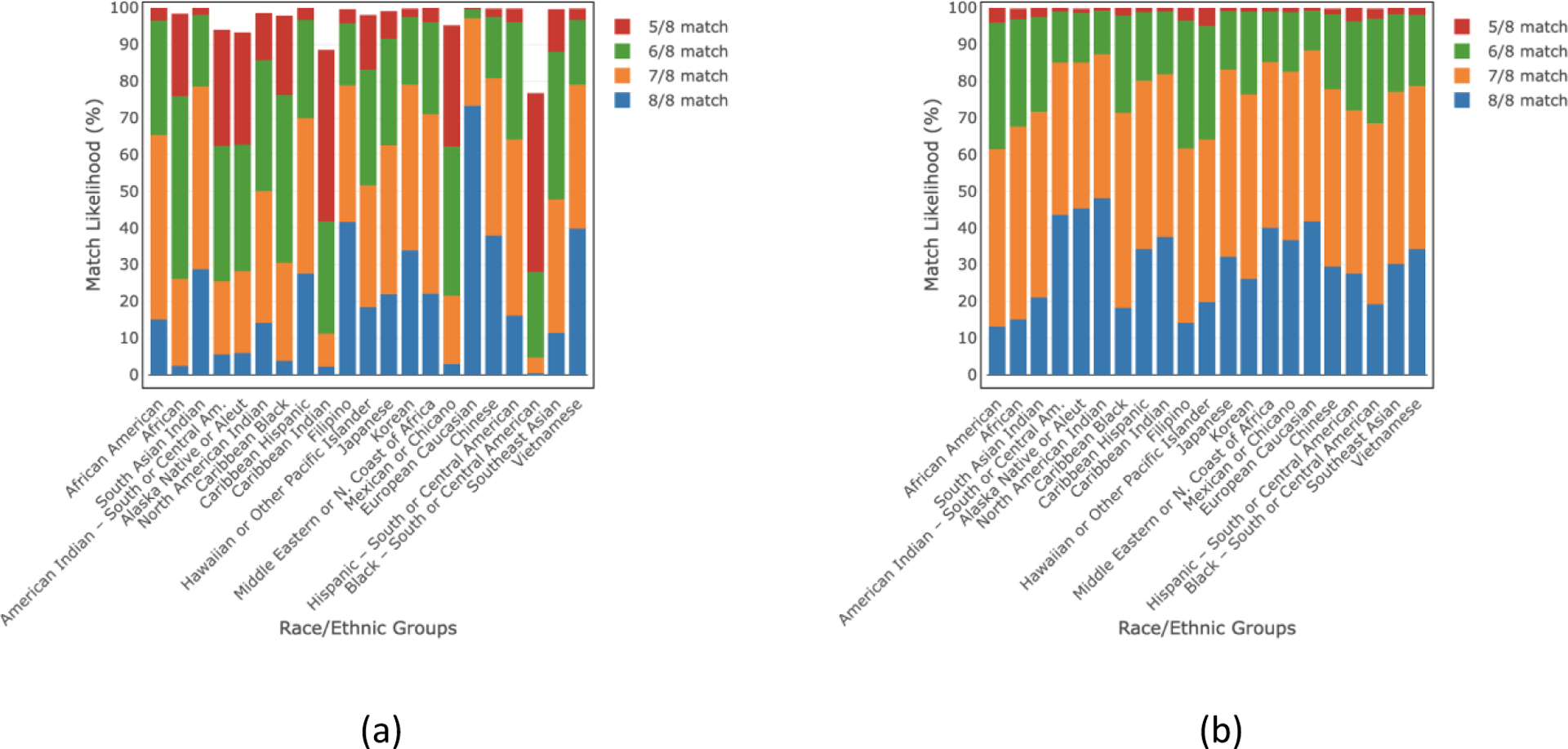
(a) “Within group” and (b) “outside group” HLA match likelihoods (%) at 5/8–8/8 levels for donors of age ≤35 years in 21 race/ethnic groups.
Real world 5 of 8 searches
The race/ethnic distribution for this patient included large fractions of minorities (27% Black/African American, 17% Hispanic/Latino, 9% Asian/Pacific Islander) (supplemental table S1). Analysis of real-world 5/8 searches for N = 898 patients searching from Jan 2021 - Jan 2023 shows that all patients have many donors to choose from. Figure 7 shows the distribution in the number of matched donors from 7/8 alleles to 5/8 alleles restricting to donors aged <=35 with <=0.75 probability of match. The median number of donors is 9, 330 and 6654 for 7/8, 6/8 and 5/8, respectively and we found that all but one patient had a match at 5/8. As the match category ranges from 7/8 to 4/8, the donor options list grows by a factor of roughly 20x per additional HLA allele mismatch (supplemental Fig. S1, supplemental table S1). Even when restricting to US donors and younger donors (aged 18–35) the distributions are such that, when extending to 4/8, all patients there have hundreds to thousands of donors to choose from (supplemental Fig. S2).
Fig 7.
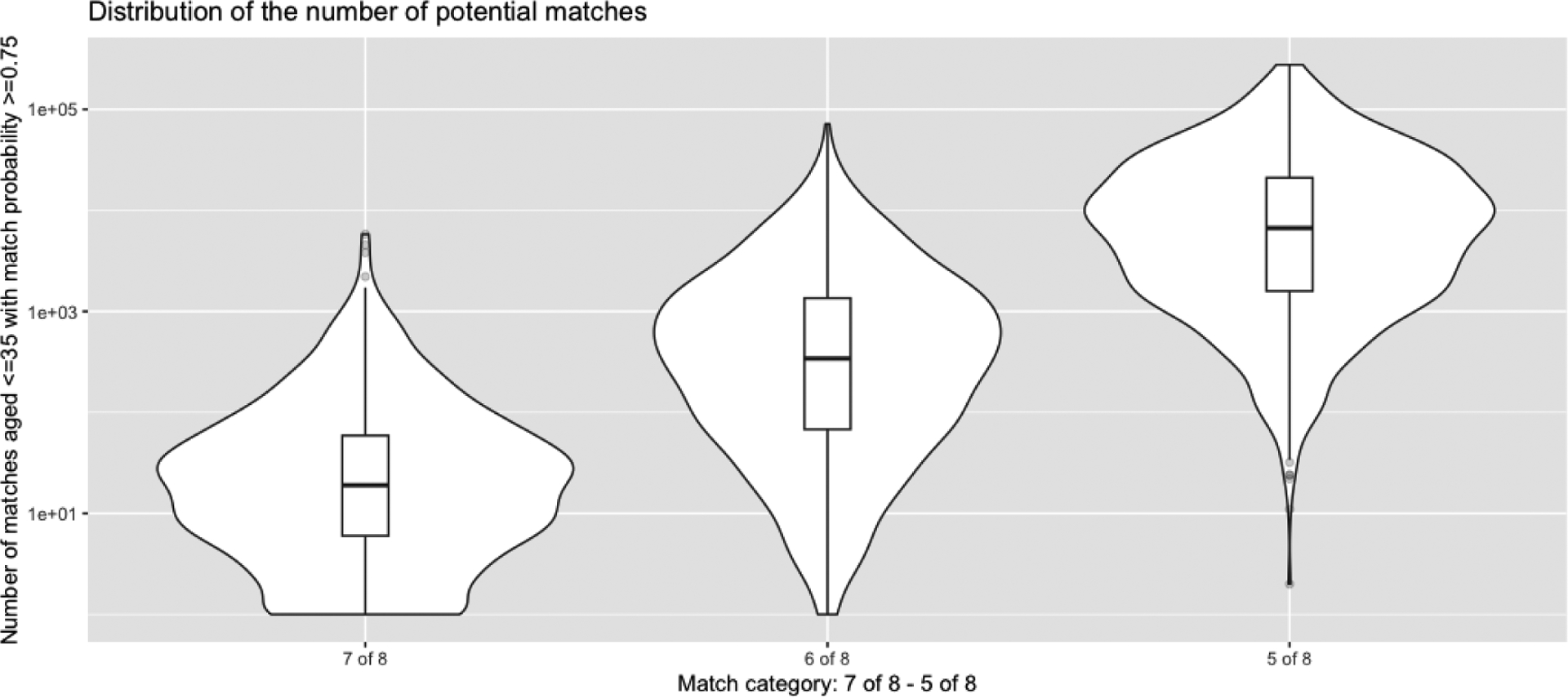
Distribution of the number of 5 of 8 – 7 of 8 or better matches from N = 898 searches from Jan 2021 to Jan 2023 restricting to donors aged <=35 with a match probability of at least 0.75.
DISCUSSION
Here we estimated the likelihood of identifying a suitable unrelated donor (URD) available in the NMDP/BTM donor registry by considering HLA-A, B, C, and DRB1 high resolution matching at 8/8, 7/8, 6/8, and 5/8 levels. We found that considering the ≥5/8 HLA match level, almost all (>99%) patients will be able to find a donor regardless of their ancestry, with each additional mismatch growing the pool of donors by roughly 20x. Even when the analysis was confined to donors aged ≤35 years, with DPB1 TCE match/permissive mismatches, and an outside race/ethnic group, donor availability remained >99% for virtually all patients in need when matching at the ≥5/8 HLA match level. The findings strongly suggest the ability to completely close the donor availability gap for all patients when they receive innovative transplant approaches that permit HLA-mismatching, such as PTCy or ex vivo T-cell depletion [20, 21].
In a seminal observational study published in 2007, the NMDP analyzed the impact of utilizing HLA mismatched transplant sources on overall survival, GVHD, and other clinical outcomes in a study involving 3,857 recipients and URDs who had high resolution HLA typing [3]. Mismatching at both the HLA antigen and allele level resulted in approximately 9–10% absolute decrement in overall survival for every level of HLA-mismatch below 8/8. This study impacted clinical practice and led many transplant centers to develop more stringent criteria for HLA matching recipients with URDs, resulting in further skewing of MUD recipient demographics toward Caucasians due to the relationship between ancestry and likelihood of identifying HLA matched URDs. In recent studies, more than 85% of the recipients of MUD HCT are white with less than 5% black or African American patients receiving URD HCT when donor matching at the 8/8 level is required [22, 23]. Many studies followed demonstrating the positive impact of matching at other HLA genes such as HLA-DP, DQ, and other low expression loci, further widening the gap in access to a potentially lifesaving transplant based on race and ethnicity [7, 24]. Notably, these data were generated predominantly in patients that receive calcineurin inhibitor based GVHD prophylaxis. The advent of PTCy as a GVHD prophylaxis regimen has enabled the use of HLA-mismatched donors, predominantly haplo related donors, as it prevents the most severe forms of acute and chronic GVHD while not increasing the risk of relapse [8]. The growth of haploidentical related HCT and to some degree the continued usage of unrelated donor umbilical cord blood grafts has partially filled the gap in access to HCT for racially/ethnically diverse patients [25]. Recently, the NMDP and other groups have prospectively studied PTCy in the MMUD setting and have generated encouraging phase II data suggesting results similar to those observed following haploidentical related donor transplants [9]. The 15-MMUD study (https://clinicaltrials.gov/study/NCT02793544) sponsored by NMDP used MMUD mismatched bone marrow grafts matched at the 4–7/8 level (39% 4–6/8) and demonstrated one-year overall survival above 70% following both reduced intensity and myeloablative conditioning regardless of match level. The ongoing NMDP ACCESS study (https://clinicaltrials.gov/study/NCT04904588) is evaluating the use of MMUD mobilized peripheral blood grafts based on encouraging data generated at the City of Hope [10]. To date, accrual to this study has been very brisk, suggesting it is addressing an unmet need in the field. Donor eligibility on ACCESS includes matching at the 4–7/8 level, like the 15-MMUD study. MMUD HCT had in general had been in decline in the US through 2018, but recent CIBMTR data, along with unpublished NMDP operations data, demonstrate a 15–20% annual growth in the use of MMUD HCT from 2020 through 2022 [13]. A secondary outcomes analysis of the BMT CTN 1702 study also recently demonstrated a shift in preference at US transplant centers from 2019 to 2022 toward greater use of MMUDs for adult recipients, driven mainly by the use of PTCy [26]. While the majority of the growth in MMUD has involved donors mismatched at the 7/8 level, NMDP operational data demonstrate more than 40% growth in the use of donors matched at 5–6/8 alleles (S Devine, unpublished observation). These recent shifts in US transplant practice prompted us to perform this analysis, given the potential for MMUD availability to close the gap in access to HCT. To date, we have not observed worse overall or GVHD free, relapse free survival (GRFS) between 7/8 and <7/8 MMUD when using PTCy based GVHD prophylaxis, similar to the haploidentical related setting, and with abatacept, no differences between 7/8 and 8/8 URD HCT outcomes [9, 11]. However, overall patient numbers are very low and definitive evidence of similarity awaits the generation of larger numbers of MMUD transplants.
A well-matched donor source, ideally an HLA-matched sibling, remains the current standard as best donor choice for most blood cancer patients, and when such donors are not available, HCT with well-matched URD stands as the preferred alternative at most US transplant centers [4]. An optimal i.e., 8/8 matched donor is not readily available for all patients that need a URD HCT. When the odds of identifying an 8/8 URD are poor, the alternative donor choices include haploidentical related, MMUD, and unrelated UCB. However, a recent study revealed inferior patient outcomes (i.e., lower rates of overall survival compared to haploidentical transplantation) with cord blood usage [27]. There are currently insufficient data to inform selection of a haploidentical related versus a MMUD. Since haploidentical transplantation considers stem cells derived from half-matched donors, the choice of suitable donors is limited by family size. Many patients have developed donor specific HLA-antibodies (DSA) due to prior pregnancy or frequent blood product transfusions [28]. DSA are a concern in any mismatched transplant setting but theoretically MMUD provides more choice to avoid DSA due the higher number of donors available. Younger donors are preferred due to associations with better outcomes [25, 29–31]. The biological mechanism remains unknown. A younger donor is now becoming the preferred choice in haploidentical HCT if feasible, but again MMUD availability allows for a greater number of young donors, and these numerically expanded donor options may allow selecting MMUDs beyond just age including favorable attributes such as ABO match, CMV status, and CCR5-delta32 genotype [32–34].
There are some limitations to this work. In the total number of available donors, around 7.5 million donors came from 5 broad race/ethnic groups and 11.3 million donors were from 4 multi-race, unknown, others, and declined groups. We distributed these donors to the 21 detailed race/ethnic group using a reapportioned scheme discussed earlier. A large proportion of donors from these 9 race/ethnic groups were added to the 21 detailed race/ethnic groups that may significantly augment unavoidable bias to the HLA match likelihood estimations in the population-based genetic model. The HLA match likelihood results for donors age ≤35 were obtained from the population-based genetic model using the same HLA frequency data as for donors of any age. Therefore, it assumes the same genetic distribution of donors in these different age groups. However, we know that in recent years, NMDP and other registry recruiting efforts have been focused on younger and more diverse donors, and in the younger generations, there are more people of mixed ancestry [35]. This could lead to a difference in the genetic distribution of younger donors than in the full donor set as a whole and as a result, the estimated HLA match likelihoods for donors of ≤35 years old may not reflect the actual match likelihoods of these younger donors. Also, there is a possibility that the Hardy-Weinberg equilibrium [36, 37] assumption of the population-based genetic modeling approach does not hold true in all U.S. populations at the broad level and most at the detailed level.
The analyses of our study included 23.3 million donors from an HLA-imputed donor database though currently, around 41 million pre-imputed donors are possible to obtain based on NMDP/BTM HapLogic match predictions [38]. In the future, we plan to refresh all HLA match likelihood and validation results by considering up-to-date and most suitable donor databases. In addition, we will perform all these analyses while considering other donor selection attributes such as CMV status, ABO, CCR5-delta32 genotype, and selective mismatching at HLA-A, B, C, and DRB1. Also, we will generate HLA frequencies built only on younger donors to find more precise results from the population-based genetic model for those younger donors.
These data have several potential uses. They demonstrate that if innovations in GVHD prophylaxis permit MMUD HCT down to the 5/8 level as in the haploidentical related setting, a suitable donor and graft source can be found for all patients regardless of racial or ethnic background, the presence of DSA, or the lack of suitable family donors. These analyses can also assist in estimating an approximate number of URDs that need to be recruited for all race/ethnic groups to meet the demand of patients. Though we evaluated the HLA match likelihoods using donor statistics in 2020, our study can help project an increase in the probability of identifying URDs in the mismatched setting with an anticipated number of donors for all race/ethnic groups, or even with the current registry. Finally, the data encourage continued pursuit of clinical research to improve transplant outcomes for patients under all types of HLA-mismatched HCT, related and unrelated, so that we can continue making progress toward achieving the global goal of health equity for any patient in need of HCT.
Supplementary Material
Highlights:
Prior unrelated donor registry modeling demonstrated a gap in availability of HLA-A, B, C and DRB1 or 8/8 matched donors for all patients in need.
The study assessed unrelated donor likelihoods for different race/ethnic groups in the United States, considering high-resolution HLA matching.
When considering high resolution HLA matches down to the 5/8 level, registry coverage exceeds 99%, expanding HCT access across all race/ethnic groups.
ACKNOWLEDGMENTS
Financial disclosure:
The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); 75R60222C00011 from the Health Resources and Services Administration (HRSA); N00014-20-1-2705, N00014-21-1-2954 and N00014-23-1-2057; Support is also provided by Be the Match Foundation, the Medical College of Wisconsin, the National Marrow Donor Program, Gateway for Cancer Research, Pediatric Transplantation and Cellular Therapy Consortium. Opinions, findings, conclusions, or recommendations expressed in this article are those of the authors and are not necessarily endorsed by the Office of Naval Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no competing interests.
REFERENCES
- 1.Gragert L, Eapen M, Williams E, Freeman J, Spellman S, Baitty R, Hartzman R, Rizzo JD, Horowitz M, Confer D: HLA match likelihoods for hematopoietic stem-cell grafts in the US registry. New England Journal of Medicine 2014, 371(4):339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besse K, Maiers M, Confer D, Albrecht M: On modeling human leukocyte antigen–identical sibling match probability for allogeneic hematopoietic cell transplantation: estimating the need for an unrelated donor source. Biology of Blood and Marrow Transplantation 2016, 22(3):410–417. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, Fernandez-Vina M, Flomenberg N, Horowitz M, Hurley CK: High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood, The Journal of the American Society of Hematology 2007, 110(13):4576–4583. [DOI] [PubMed] [Google Scholar]
- 4.Spellman SR: Hematology 2022—what is complete HLA match in 2022? Hematology 2022, 2022(1):83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blood CfI, Research MT: Methodology employed for annual report on hematopoietic cell transplant center-specific survival rates.
- 6.Ciurea SO: Clinical Transplant-Related Long-Term Outcomes of Alternative Donor Allogeneic Transplantation (CTRL-ALT-D). In: 2019 TCT| Transplantation & Cellular Therapy Meetings of ASBMT and CIBMTR: 2019. Tandem Meetings. [Google Scholar]
- 7.Pidala J, Lee SJ, Ahn KW, Spellman S, Wang H-L, Aljurf M, Askar M, Dehn J, Fernandez Viña M, Gratwohl A: Nonpermissive HLA-DPB1 mismatch increases mortality after myeloablative unrelated allogeneic hematopoietic cell transplantation. Blood, The Journal of the American Society of Hematology 2014, 124(16):2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luznik L, O’Donnell PV, Symons HJ, Chen AR, Leffell MS, Zahurak M, Gooley TA, Piantadosi S, Kaup M, Ambinder RF: HLA-haploidentical bone marrow transplantation for hematologic malignancies using nonmyeloablative conditioning and high-dose, posttransplantation cyclophosphamide. Biology of Blood and Marrow Transplantation 2008, 14(6):641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw BE, Jimenez-Jimenez AM, Burns LJ, Logan BR, Khimani F, Shaffer BC, Shah NN, Mussetter A, Tang X-Y, McCarty JM: National Marrow Donor Program–sponsored multicenter, phase II trial of HLA-mismatched unrelated donor bone marrow transplantation using post-transplant cyclophosphamide. Journal of clinical oncology 2021, 39(18):1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al Malki MM, Tsai N-C, Palmer J, Mokhtari S, Tsai W, Cao T, Ali H, Salhotra A, Arslan S, Aldoss I: Posttransplant cyclophosphamide as GVHD prophylaxis for peripheral blood stem cell HLA-mismatched unrelated donor transplant. Blood Advances 2021, 5(12):2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins B, Qayed M, McCracken C, Bratrude B, Betz K, Suessmuth Y, Yu A, Sinclair S, Furlan S, Bosinger S: Phase II trial of costimulation blockade with abatacept for prevention of acute GVHD. Journal of clinical oncology 2021, 39(17):1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bleakley M, Heimfeld S, Loeb KR, Jones LA, Chaney C, Seropian S, Gooley TA, Sommermeyer F, Riddell SR, Shlomchik WD: Outcomes of acute leukemia patients transplanted with naive T cell–depleted stem cell grafts. The Journal of clinical investigation 2015, 125(7):2677–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolon YT, Atshan R, Allbee-Johnson M, Estrada-Merly N, Lee SJ: Current use and outcome of hematopoietic stem cell transplantation: CIBMTR US summary slides. Center for International Blood and Marrow Transplant Research 2022. [Google Scholar]
- 14.Auletta J, Kou J, Chen M, Shaw B: Current use and outcome of hematopoietic stem cell transplantation: CIBMTR US summary slides. Center for International Blood and Marrow Transplant Research 2021. [Google Scholar]
- 15.Kollman C, Maiers M, Gragert L, Müller C, Setterholm M, Oudshoorn M, Hurley CK: Estimation of HLA-A,-B,-DRB1 haplotype frequencies using mixed resolution data from a National Registry with selective retyping of volunteers. Human immunology 2007, 68(12):950–958. [DOI] [PubMed] [Google Scholar]
- 16.Gragert L, Madbouly A, Freeman J, Maiers M: Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Human immunology 2013, 74(10):1313–1320. [DOI] [PubMed] [Google Scholar]
- 17.Gragert L, Spellman SR, Shaw BE, Maiers M: Unrelated Stem Cell Donor HLA Match Likelihood in the US Registry Incorporating HLA-DPB1 Permissive Mismatching. Transplantation and Cellular Therapy 2023, 29(4):244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crivello P, Zito L, Sizzano F, Zino E, Maiers M, Mulder A, Toffalori C, Naldini L, Ciceri F, Vago L: The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biology of Blood and Marrow Transplantation 2015, 21(2):233–241. [DOI] [PubMed] [Google Scholar]
- 19.Zino E, Frumento G, Marktel S, Sormani MP, Ficara F, Terlizzi SD, Parodi AM, Sergeant R, Martinetti M, Bontadini A: A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood 2004, 103(4):1417–1424. [DOI] [PubMed] [Google Scholar]
- 20.McCurdy SR, Luznik L: How we perform haploidentical stem cell transplantation with posttransplant cyclophosphamide. Hematology 2014, the American Society of Hematology Education Program Book 2019, 2019(1):513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy AB, Li Y, Talano J-A, Elgarten CW, Seif AE, Wang Y, Johnson B, Monos DS, Kadauke S, Olson TS: Unrelated donor α/β T cell–and B cell–depleted HSCT for the treatment of pediatric acute leukemia. Blood Advances 2022, 6(4):1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luznik L, Pasquini MC, Logan B, Soiffer RJ, Wu J, Devine SM, Geller N, Giralt S, Heslop HE, Horowitz MM: Randomized phase III BMT CTN trial of calcineurin inhibitor–free chronic graft-versus-host disease interventions in myeloablative hematopoietic cell transplantation for hematologic malignancies. Journal of Clinical Oncology 2022, 40(4):356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolaños-Meade J, Hamadani M, Wu J, Al Malki MM, Martens MJ, Runaas L, Elmariah H, Rezvani AR, Gooptu M, Larkin KT: Post-Transplantation Cyclophosphamide-Based Graft-versus-Host Disease Prophylaxis. New England Journal of Medicine 2023, 388(25):2338–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Viña MA, Klein JP, Haagenson M, Spellman SR, Anasetti C, Noreen H, Baxter-Lowe LA, Cano P, Flomenberg N, Confer DL: Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood, The Journal of the American Society of Hematology 2013, 121(22):4603–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fingrut WB, Gyurkocza B, Davis E, Flynn J, Chinapen S, Naputo KA, Quach S, Cho C, Giralt SA, Jakubowski AA: Racial disparities in access to alternative donor allografts persist in the era of “donors for all”. Blood Advances 2022, 6(20):5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehn J: A novel donor search and selection algorithm facilitates a comparable incidence of transplant for patients regardless of baseline search prognosis: Report from BMTCTN 1702 trial. 49th annual meeting of the EBMT. The Lancet Haematology 2023, 10(6):e400–e401. [Google Scholar]
- 27.Fuchs EJ, O’Donnell PV, Eapen M, Logan B, Antin JH, Dawson P, Devine S, Horowitz MM, Horwitz ME, Karanes C: Double unrelated umbilical cord blood vs HLA-haploidentical bone marrow transplantation: the BMT CTN 1101 trial. Blood 2021, 137(3):420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciurea SO, Cao K, Fernandez-Vina M, Kongtim P, Malki MA, Fuchs E, Luznik L, Huang X-J, Ciceri F, Locatelli F: The European Society for Blood and Marrow Transplantation (EBMT) consensus guidelines for the detection and treatment of donor-specific anti-HLA antibodies (DSA) in haploidentical hematopoietic cell transplantation. Bone marrow transplantation 2018, 53(5):521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw BE, Logan BR, Spellman SR, Marsh SG, Robinson J, Pidala J, Hurley C, Barker J, Maiers M, Dehn J: Development of an unrelated donor selection score predictive of survival after HCT: donor age matters most. Biology of Blood and Marrow Transplantation 2018, 24(5):1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariotti J, Raiola AM, Evangelista A, Carella AM, Martino M, Patriarca F, Risitano A, Bramanti S, Busca A, Giaccone L: Impact of donor age and kinship on clinical outcomes after T-cell–replete haploidentical transplantation with PT-Cy. Blood advances 2020, 4(16):3900–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeZern AE, Franklin C, Tsai H-L, Imus PH, Cooke KR, Varadhan R, Jones RJ: Relationship of donor age and relationship to outcomes of haploidentical transplantation with posttransplant cyclophosphamide. Blood advances 2021, 5(5):1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt AH, Sauter J, Baier DM, Daiss J, Keller A, Klussmeier A, Mengling T, Rall G, Riethmüller T, Schöfl G: Immunogenetics in stem cell donor registry work: The DKMS example (Part 2). International journal of immunogenetics 2020, 47(2):139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDermott DH, Conway SE, Wang T, Ricklefs SM, Agovi MA, Porcella SF, Tran HTB, Milford E, Spellman S, Abdi R: Donor and recipient chemokine receptor CCR5 genotype is associated with survival after bone marrow transplantation. Blood, The Journal of the American Society of Hematology 2010, 115(11):2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little AM, Akbarzad-Yousefi A, Anand A, Diaz Burlinson N, Dunn PP, Evseeva I, Latham K, Poulton K, Railton D, Vivers S: BSHI guideline: HLA matching and donor selection for haematopoietic progenitor cell transplantation. International Journal of Immunogenetics 2021, 48(2):75–109. [DOI] [PubMed] [Google Scholar]
- 35.Kollman C, Spellman SR, Zhang M-J, Hassebroek A, Anasetti C, Antin JH, Champlin RE, Confer DL, DiPersio JF, Fernandez-Viña M: The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood, The Journal of the American Society of Hematology 2016, 127(2):260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beatty PG, Boucher KM, Mori M, Milford EL: Probability of finding HLA-mismatched related or unrelated marrow or cord blood donors. Human immunology 2000, 61(8):834–840. [DOI] [PubMed] [Google Scholar]
- 37.Mori M, Graves M, Milford EL, Beatty PG: Computer program to predict likelihood of finding and HLA-matched donor: methodology, validation, and application. Biology of Blood and Marrow Transplantation: Journal of the American Society for Blood and Marrow Transplantation 1996, 2(3):134–144. [PubMed] [Google Scholar]
- 38.Davis E, Devlin S, Cooper C, Nhaissi M, Paulson J, Wells D, Scaradavou A, Giralt S, Papadopoulos E, Kernan NA: Validation of an algorithm to predict the likelihood of an 8/8 HLA-matched unrelated donor at search initiation. Biology of Blood and Marrow Transplantation 2018, 24(5):1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


