Abstract
Members of the ATP-dependent family of chromatin remodeling enzymes play key roles in the regulation of transcription, development, DNA repair and cell cycle. Each of these enzymes are multi-subunit assemblies that hydrolyze thousands of molecules of ATP in order to change nucleosome positions, disrupt DNA–histone interactions and perhaps destabilize chromatin folding. Here I review recent studies that suggest these potent machines can be ‘tamed’ by one of several mechanisms: targeting their activity to localized regions, blocking their chromatin binding activity or inhibiting their remodeling activity.
Introduction
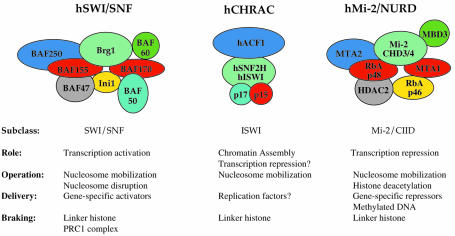
During interphase of the eukaryotic cell cycle, the bulk of DNA is assembled into highly folded, 100–400 nm nucleoprotein filaments. DNA-mediated processes can function in this environment due to the actions of highly conserved chromatin ‘remodeling’ enzymes. One class of chromatin remodeling factor comprises a family of related ATP-dependent complexes that use the energy of ATP hydrolysis to enhance the accessibility of nucleosomal DNA (Vignali et al., 2000). This family can be further subdivided into three groups based on their biochemical properties and the overall sequence similarity of their ATPase subunits: (i) the SWI–SNF group; (ii) the ISWI group; and (iii) the Mi-2/CHD group (Figure 1; Boyer et al., 2000a). Whereas many members of the ISWI-like and Mi-2-like subgroups appear dedicated to transcriptional repression pathways (Kehle et al., 1998; Deuring et al., 2000), most SWI–SNF-like enzymes play roles in the activation of transcription. In contrast to these transcriptional roles, some ISWI-based enzymes, such as ACF, may play key roles in nucleosome assembly (Ito et al., 1997), and other family members may facilitate other diverse chromatin-based processes, such as homologous recombination and DNA repair (Peterson, 1996).
Fig. 1. Three subclasses of ATP-dependent chromatin remodeling complexes. Shown here are representative members of the SWI–SNF, ISWI and Mi-2/CHD subclasses of chromatin remodeling enzymes that are found in human cells. Depicted subunit organization is for illustrative purposes only (for a review see Vignali et al., 2000).
Operating the machine
In the case of ATP-dependent remodeling enzymes, ‘chromatin remodeling’ refers to numerous in vitro ATP-dependent changes in a chromatin substrate, including disruption of histone–DNA contacts within nucleosomes, movement of histone octamers in cis and in trans, loss of negative supercoils from circular minichromosomes, and increased accessibility of nucleosomal DNA to transcription factors and restriction endonucleases (Peterson and Workman, 2000). In vivo, SWI–SNF-like enzymes can help DNA-bending proteins facilitate nucleosome sliding, as well as drive formation of Z-DNA structures (Liu et al., 2001; Lomvardas and Thanos, 2001). Recent genetic and biochemical studies have also led to the suggestion that SWI–SNF may disrupt higher-order chromatin folding (Krebs et al., 2000; Horn et al., 2002a).
How do these enzymes catalyze such diverse events? Early models for ATP-dependent remodeling focused on changes in the histone component of the nucleosome. For instance, SWI–SNF-like enzymes were proposed to use the energy of ATP hydrolysis to drive removal of one or both of the histone H2A–H2B dimers (Peterson and Tamkun, 1995). Later, Hayes and Kingston proposed an alternative model in which dimers might not be lost, but dramatically rearranged, generating a novel ‘remodeled’ nucleosome conformation (Lee et al., 1999). However, these types of models seem less likely in light of recent work showing that histone–histone cross-linking does not block or even slow the rate of remodeling (Cote et al., 1998; Boyer et al., 2000b). In contrast, there is now increasing evidence that ATP-dependent remodeling may involve (perhaps exclusively) changes in the topology of nucleosomal DNA. For instance, Owen-Hughes and colleagues demonstrated that members of all three subclasses of the ATP-dependent remodeling family use ATP hydrolysis to introduce superhelical torsion into chromatin (Havas et al., 2000). Furthermore, SWI–SNF action is blocked by the constrained topology of small, circular chromatin, suggesting that such changes in DNA topology are required for remodeling (Gavin et al., 2001). Consequently, mechanistic studies are now focused on determining how DNA topology is altered. Current models propose that ATP-dependent remodeling may involve DNA tracking activity (Havas et al., 2000), rotation of DNA along its long axis (Boyer et al., 2000b), or formation of DNA bulges or small loops (Langst and Becker, 2001; Narlikar et al., 2001).
Delivering the machine
Given their capacity to disrupt chromatin structure and to hydrolyze thousands of molecules of ATP, it should not be surprising that ATP-dependent remodeling enzymes are kept under tight rein in vivo. In the past few years, most studies have focused on the active recruitment of remodeling enzymes by DNA site-specific transcriptional activators or repressors (Peterson and Workman, 2000). For example, yeast SWI–SNF interacts with the acidic activation domains of several activators, and these contacts can target remodeling activity in vitro and in vivo. Likewise, human SWI–SNF can be targeted by a host of activators, including erythroid kruppel-like factor (EKLF), C/EBP-β, MyoD, heat shock factors and several steroid receptors. Similarly, transcriptional repressors, such as Drosophila hunchback, can recruit members of the Mi-2 subclass of remodeling enzymes. In general, the recruitment of remodeling activity by sequence-specific DNA-binding proteins seems an effective way to direct localized changes in chromatin structure.
While the direct targeting of remodeling complexes clearly plays a central role in re-programming the chromatin structure of specific loci, it may prove equally important for a cell to ‘shield’ chromosomal domains from renegade remodeling enzymes. For instance, inappropriate chromatin remodeling could cause enhanced rates of DNA recombination, disruption of chromosome condensation, or promiscuous transcriptional activation of silenced genes. One way of shielding the genome is to globally inactivate the enzyme. For example, subunits of the human SWI–SNF complex are phosphorylated during mitosis, which correlates with removal of the complex from condensing chromosomes (Sif et al., 1998). Recent studies (Shao et al., 1999; Francis et al., 2001; Horn et al., 2002b) also suggest that chromatin ‘shielding’ factors exist, which block the ATP-dependent remodeling of chromatin fibers.
Braking the machine: a global role for linker histone
Linker histones are ubiquitous components of cellular chromatin that constrain the entry/exit DNA of the nucleosome and incorporate another ∼20 bp of DNA into a particle called a chromatosome. In addition to this effect on the basic structure of the nucleosome, linker histones also stabilize the higher-order folding of nucleosomal arrays (Carruthers et al., 1998). One possibility is that the binding of linker histone to nucleosomes might inhibit the ATP-dependent changes in DNA topology that are key to remodeling activity and might serve as a means to control enzyme function. Consistent with this idea, the activity of a human SWI–SNF complex is markedly decreased on a chromatosome compared with a mononucleosome substrate (Hill and Imbalzano, 2000). To test whether linker histones might exert a general inhibitory effect on remodeling enzymes, nucleosomal arrays were reconstituted with or without linker histone, and the remodeling activities of yeast SWI–SNF, human SWI–SNF, ACF and Mi-2 complexes were determined using a quantitative restriction enzyme accessibility assay (Horn et al., 2002a). Strikingly, linker histone incorporation virtually eliminated the activity of all of these enzymes.
How do linker histones block ATP-dependent remodeling? Although linker histones constrain nucleosomal DNA, they also stabilize the higher-order folding of arrays, and thus inhibition could result from an inability of the enzyme to bind folded arrays. Indeed, at least part of the linker histone inhibition has been shown to be caused by a decreased affinity of the enzyme for the chromatin array (Horn et al., 2002b). To determine whether linker histone might also inhibit remodeling through the topological constraint of individual chromatosomes within the array, a second set of arrays was produced using trypsinized core histones. Limited trypsinization of the core histones removes the histone tails, which are essential for nucleosomal and chromatin array folding (Fletcher and Hansen, 1996; Carruthers and Hansen, 2000), but do not affect chromatosome formation (Carruthers and Hansen, 2000). Surprisingly, even on these unfolded substrates, linker histone inhibited remodeling by SWI–SNF, albeit to a lesser degree (Horn et al., 2002b). Interestingly, incorporation of linker histone onto a trypsinized array did not inhibit the binding of SWI–SNF. Based on these results and studies with individual chromatosomes (Hill and Imbalzano, 2000), linker histone appears to act at two levels to block chromatin remodeling: (i) the condensed state of linker histone-containing chromatin inhibits the binding of remodeling complexes to the fiber; and (ii) the binding of linker histone to each nucleosome is able to inhibit the catalytic function of a chromatin-bound remodeling activity. These results suggest a model in which linker histones produce a generally repressive chromatin environment that is inhibitory to ATP-dependent chromatin remodeling enzymes. Such repression could provide a means to maintain large chromosomal domains in a neutral chromatin configuration, as well as to shield chromatin that lies immediately adjacent to a domain of targeted chromatin remodeling activity (Figure 2).
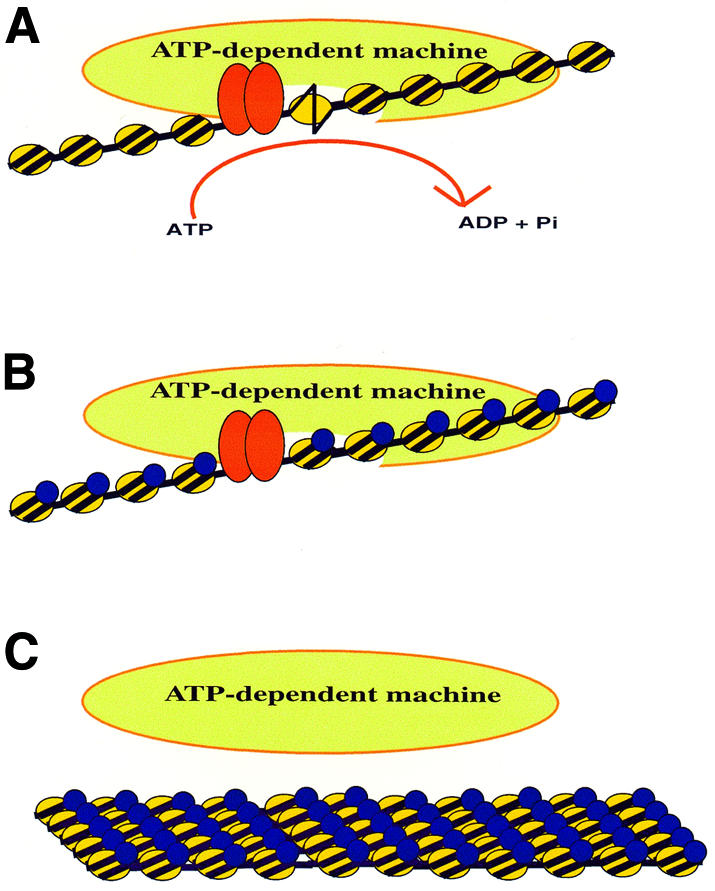
Fig. 2. Regulation of ATP-dependent chromatin remodeling. (A) Recruitment of remodeling activity by a sequence-specific DNA binding protein (red ovals). (B) Inhibition of remodeling activity by a chromatin binding protein (blue ovals) that constrains nucleosomal DNA topology. (C) Blocking the binding of a chromatin remodeling enzyme by a chromatin shielding factor (blue ovals) that stabilizes nucleosomal array oligomerization.
Braking the machine: developmental control
Some interesting parallels can be drawn between the actions of linker histone and members of the polycomb group (PcG) of proteins. PcG proteins are conserved from flies to mammals, and they are required for maintaining patterns of transcriptional repression through cell divisions (Pirrotta, 1998). In Drosophila, PcG products play essential roles during development, maintaining the transcriptional silencing of homeotic genes. In mammalian cells, products of the PcG genes are essential for control of cell proliferation. In contrast to the global localization of linker histone, PcG proteins are targeted to specific chromosomal regions by DNA sequences called polycomb response elements (PREs). How PcG proteins maintain repression of transcription is not clear, but most models propose that they exert their effects through chromatin structure. Consistent with such models, genetic studies in Drosophila have suggested that PcG proteins function by antagonizing the activity of the trithorax group of proteins, which includes subunits of the Drosophila SWI–SNF (brm) remodeling complex.
Recently, Kingston and colleagues have purified a PcG-containing complex, PRC1, from Drosophila embryos (Shao et al., 1999). PRC1 is enormous (2–6 MDa) and contains a host of polypeptide subunits, including four known PcG proteins (Pc, Psc, Ph and dRING1), the DNA-binding protein Zeste and several TBP-associated factors (Saurin et al., 2001). PRC1 does not appear to have catalytic activities (e.g. ATPase activity), although several subunits of PRC1 bind DNA (Francis et al., 2001). Strikingly, pre-incubation of PRC1 with a nucleosomal array eliminates the remodeling activity of human SWI–SNF (Shao et al., 1999). Inhibition was not observed when SWI–SNF bound the array first, nor did PRC1 inhibit SWI–SNF when the two complexes were pre-incubated in solution prior to addition to the array. These results suggest that PcG products, once recruited to a locus by a PRE, might maintain patterns of transcriptional repression by shielding chromatin domains from remodeling enzymes.
Do linker histones and PRC1 use a similar mechanism to block chromatin remodeling? Linker histones block the binding of a SWI–SNF-like enzyme to chromatin, as well as the activity of a chromatin-bound enzyme. It is not yet clear whether PRC1 can block the catalytic activity of SWI–SNF, although several subunits of PRC1 can interact with DNA, and could conceivably constrain nucleosomal DNA topology enough to have such an effect. In contrast, PRC1 prevention of chromatin binding by SWI–SNF has been clearly demonstrated by chromatin immunoprecipitation assays (Francis et al., 2001). In the case of linker histones, inhibition of SWI–SNF binding correlates best with the ability of linker histones to drive the oligomerization of nucleosomal arrays (Horn et al., 2002b). This type of chromatin higher-order structure is believed to mimic fiber–fiber interactions that are important for formation of chromonema fibers in vivo (Fletcher and Hansen, 1996; Carruthers et al., 1998). It is unknown whether PcG proteins also create folded structures like oligomerized arrays, although PRC1 does inhibit SWI–SNF at substoichiometric levels with respect to nucleosomes (Shao et al., 1999), consistent with a condensation mechanism. Furthermore, PRC1-containing chromatin is similar to oligomerized chromatin in that it can exclude SWI–SNF, but not restriction enzymes (Carruthers et al., 1998; Francis et al., 2001). These observations raise the interesting possibility that stabilization of fiber–fiber interactions may be a common feature of chromatin shielding proteins (Figure 2C).
Perspectives
Over the past few years, attention has begun to shift towards studies that center on the regulation of ATP-dependent remodeling machines. The targeting of remodeling enzymes by gene-specific transcriptional repressors or activators is now a well-documented method for controlling the local concentration of ATP-dependent remodeling complexes. In contrast, there are only two examples of what I have coined here as ‘chromatin shielding’ activities that appear to protect chromatin domains from remodeling. One interesting candidate for a new member of this group is yeast Sir3p, which plays a key role in establishing heterochromatic structures at yeast telomeres and at the silent mating type loci (Gasser and Cockell, 2001). Recent in vitro studies indicate that Sir3p can drive formation of chromatin structures that share several hallmarks of oligomerized nucleosomal arrays (Georgel et al., 2001). Perhaps one function of the heterochromatic state of Sir-containing chromatin is to shield domains from the potent activity of ATP-dependent remodeling machines. Clearly, as we learn more about how ATP-dependent remodeling enzymes recognize their chromatin substrate and alter DNA–histone contacts, we are likely to uncover a multitude of ways in which these activities can be controlled.

Craig L. Peterson
Acknowledgments
Acknowledgements
I thank Peter Horn and Tony Imbalzano for critical comments on the manuscript.
References
- Boyer L.A., Logie, C., Bonte, E., Becker, P.B., Wade, P.A., Wolffe, A.P., Wu, C., Imbalzano, A.N. and Peterson, C.L. (2000a) Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem., 275, 18864–18870. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Shao, X., Ebright, R.H. and Peterson, C.L. (2000b) Roles of the histone H2A–H2B dimers and the (H3/H4)2 tetramer in nucleosome remodeling by the SWI–SNF complex. J. Biol. Chem., 275, 11545–11552. [DOI] [PubMed] [Google Scholar]
- Carruthers L.M. and Hansen, J.C. (2000) The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem., 275, 37285–37290. [DOI] [PubMed] [Google Scholar]
- Carruthers L.M., Bednar, J., Woodcock, C.L. and Hansen, J.C. (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry, 37, 14776–14787. [DOI] [PubMed] [Google Scholar]
- Cote J., Peterson, C.L. and Workman, J.L. (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl Acad. Sci. USA, 95, 4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Fletcher T.M. and Hansen, J.C. (1996) The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr., 6, 149–188. [DOI] [PubMed] [Google Scholar]
- Francis N.J., Saurin, A.J., Shao, Z. and Kingston, R.E. (2001) Reconstitution of a functional core polycomb repressive complex. Mol. Cell, 8, 545–556. [DOI] [PubMed] [Google Scholar]
- Gasser S.M. and Cockell, M.M. (2001) The molecular biology of the SIR proteins. Gene, 279, 1–16. [DOI] [PubMed] [Google Scholar]
- Gavin I., Horn, P.J. and Peterson, C.L. (2001) SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell, 7, 97–104. [DOI] [PubMed] [Google Scholar]
- Georgel P.T., Palacios DeBeer, M.A., Pietz, G., Fox, C.A. and Hansen, J.C. (2001) Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl Acad. Sci. USA, 98, 8584–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havas K., Flaus, A., Phelan, M., Kingston, R., Wade, P.A., Lilley, D.M.J. and Owen-Hughes, T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133. [DOI] [PubMed] [Google Scholar]
- Hill D.A. and Imbalzano, A.N. (2000) Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry, 39, 11649–11656. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Crowley, K., Carruthers, L.M., Hansen, J.C. and Peterson, C.L. (2002a) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nature Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Carruthers, L.M., Logie, C., Hill, D.A., Solomon, M.J., Wade, P.A., Imbalzano, A.N., Hansen, J.C. and Peterson, C.L. (2002b) Regulation of ATP-dependent chromatin remodeling by linker histone phosphorylation. Nature Struct. Biol., submitted. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger, M., Pazin, M.J., Kobayashi, R. and Kadonaga, J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle, D., Treuheit, S., Christen, B., Kennison, J.A., Bienz, M. and Muller, J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Fry, C.J., Samuels, M. and Peterson, C.L. (2000) Global role for chromatin remodeling enzymes in mitotic gene expression. Cell, 102, 587–598. [DOI] [PubMed] [Google Scholar]
- Langst G. and Becker, P.J. (2001) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell, 8, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Lee K.M., Sif, S., Kingston, R.E. and Hayes, J.J. (1999) hSWI/SNF disrupts interactions between the H2A N-terminal tail and nucleosomal DNA. Biochemistry, 38, 8423–8429. [DOI] [PubMed] [Google Scholar]
- Liu R., Liu, H., Chen, X., Kirby, M., Brown, P.O. and Zhao, K. (2001) Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell, 106, 309–318. [DOI] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos, D. (2001) Nucleosome sliding via TBP DNA binding in vivo. Cell, 106, 685–696. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J., Phelan, M.L. and Kingston, R.E. (2001) Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell, 8, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. (1996) Multiple SWItches to turn on chromatin? Curr. Opin. Genet. Dev., 6, 171–175. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Tamkun, J.W. (1995) The SWI–SNF complex: a chromatin remodeling machine? Trends Biochem. Sci., 20, 143–146. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman, J.L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1998) Polycombing the genome: PcG, trxG, and chromatin silencing. Cell, 93, 333–336. [DOI] [PubMed] [Google Scholar]
- Saurin A.J., Shao, Z., Erdjument-Bromage, H., Tempst, P. and Kingston, R.E. (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature, 412, 655–660. [DOI] [PubMed] [Google Scholar]
- Shao Z., Raible, F., Mollaaghababa, R., Guyon, J.R., Wu, C.T., Bender, W. and Kingston, R.E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell, 98, 37–46. [DOI] [PubMed] [Google Scholar]
- Sif S., Stukenberg, P.T., Kirschner, M.W. and Kingston, R.E. (1998) Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev., 12, 2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Hassan, A.H., Neely, K.E. and Workman, J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]