Abstract
Many factors, including microbiome structure and activity in the drinking water distribution system (DWDS), affect the colonization potential of opportunistic pathogens. The present study aims to describe the dynamics of active bacterial communities in DWDS and identify the factors that shape the community structures and activity in the selected DWDSs. Large-volume drinking water and hot water, biofilm, and water meter deposit samples were collected from five DWDSs. Total nucleic acids were extracted, and RNA was further purified and transcribed into its cDNA from a total of 181 water and biofilm samples originating from the DWDS of two surface water supplies (disinfected with UV and chlorine), two artificially recharged groundwater supplies (non-disinfected), and a groundwater supply (disinfected with UV and chlorine). In chlorinated DWDSs, concentrations of <0.02–0.97 mg/l free chlorine were measured. Bacterial communities in the RNA and DNA fractions were analysed using Illumina MiSeq sequencing with primer pair 341F-785R targeted to the 16S rRNA gene. The sequence libraries were analysed using QIIME pipeline, Program R, and MicrobiomeAnalyst. Not all bacterial cells were active based on their 16S rRNA content, and species richness was lower in the RNA fraction (Chao1 mean value 490) than in the DNA fraction (710). Species richness was higher in the two DWDSs distributing non-disinfected artificial groundwater (Chao1 mean values of 990 and 1 000) as compared to the two disinfected DWDSs using surface water (Chao1 mean values 190 and 460) and disinfected DWDS using ground water as source water (170). The difference in community structures between non-disinfected and disinfected water was clear in the beta-diversity analysis. Distance from the waterworks also affected the beta diversity of community structures, especially in disinfected distribution systems. The two most abundant bacteria in the active part of the community (RNA) and total bacterial community (DNA) belonged to the classes Alphaproteobacteria (RNA 28 %, DNA 44 %) and Gammaproteobacteria (RNA 32 %, DNA 30 %). The third most abundant and active bacteria class was Vampirovibrionia (RNA 15 %), whereas in the total community it was Paceibacteria (DNA 11 %). Class Nitrospiria was more abundant and active in both cold and hot water in DWDS that used chloramine disinfection compared to non-chlorinated or chlorine-using DWDSs. Thirty-eight operational taxonomic units (OTU) of Legionella, 30 of Mycobacterium, and 10 of Pseudomonas were detected among the sequences. The (RT)-qPCR confirmed the presence of opportunistic pathogens in the DWDSs studied as Legionella spp. was detected in 85 % (mean value 4.5 × 104 gene copies/100 ml), Mycobacterium spp. in 95 % (mean value 8.3 × 106 gene copies/100 ml), and Pseudomonas spp. in 78 % (mean value 1.6 × 105 gene copies/100 ml) of the water and biofilm samples. Sampling point inside the system (distance from the waterworks and cold/hot system) affected the active bacterial community composition. Chloramine as a chlorination method resulted in a recognizable community composition, with high abundance of bacteria that benefit from the excess presence of nitrogen. The results presented here confirm that each DWDS is unique and that opportunistic pathogens are present even in conditions when water quality is considered excellent.
Keywords: Bacterial communities, Disinfection, Drinking water, Opportunistic pathogens, Ribosomal RNA
1. Introduction
Drinking water microbial quality may change after leaving the waterworks and before reaching the user’s tap in a drinking water distribution system (DWDS) (Potgieter et al., 2018; Hayward et al., 2022). Drinking water quality in European Union member states must fulfill microbial and chemical requirements and not contain feacal contamination indicating nor pathogenic microbes (EU, 2020). In the future, also risk assessment on the building’s water system is required in so called priority premises, such as hospitals and hotels, to assess whether the risk for the presence of Legionella has increased (EU, 2020). The selection of drinking water treatment techniques and disinfection affects the microbial content of the water going from the waterworks into the distribution system, but microbial nutrients from source water, pipe material, temperature, and the age of the water may also affect the microbiome composition and activity in water and biofilms on the inner surfaces of the pipes in many complex ways (Potgieter et al., 2018; Neu and Hammes, 2020; Leslie et al., 2021). Biofilms also give shelter to bacteria, including opportunistic pathogens, that may not be detected abundant in water when DWDS is well maintained (Waak et al., 2019; Leslie et al., 2021; Rosenqvist et al., 2023).
Disinfection residual is used to maintain good quality (WHO, 2022) and disinfection is always required when surface water is used as source water in Finland (STM, 2015). While most of the published results about bacterial communities in DWDSs concern chlorinated drinking water systems, bacterial communities in non-chlorinated DWDSs are less studied (Potgieter et al., 2018; Bruno et al., 2018; Rosenqvist et al., 2023). It is known that disinfection residual in DWDSs causes selective pressure on microbiota and decreases diversity in bacterial communities and their functional potential compared to non-disinfected water systems (Waak et al., 2019; Dai et al., 2020). Also, changes in water temperatures shifts bacterial composition (Ji et al., 2017; Dai et al., 2018). High hot water temperatures may decrease bacterial diversity compared to cold water (Zhang et al., 2021), but complex bacterial communities in full-scale hot water systems are not yet well understood.
In addition to general changes in microbial numbers and composition, disinfection may also affect the colonization of opportunistic pathogen microbes (Leslie et al., 2021; Hayward et al., 2022) by promoting the occurrence of some bacteria species more than others (Donohue et al., 2019). Several studies show that the use of chloramine may promote the occurrence of Mycobacterium species or may be less efficient against Mycobacterium spp. compared to free chlorine compounds, whereas Legionella spp. have been found to be more common in drinking water exposed to free chlorine compounds than in water disinfected using chloramine (Donohue et al., 2019; Gomez-Alvarez et al., 2012; Wang et al., 2013), although not all drinking water system studies support this finding (Buse et al., 2019; Bautista-de los Santos et al., 2016). Survival of opportunistic pathogens, such as Legionella, may be favored also by the temperature prevailing in hot water systems. Temperatures over 50 °C are needed to control growth of many bacteria, including opportunistic pathogens (Proctor et al., 2017; Dai et al., 2020; Leslie et al., 2021; Hayward et al., 2022), but this temperature is not always reached in hot water systems (Schwake et al., 2016).
The presence of viable, metabolically active bacteria belonging to genera capable of causing infection may pose a health risk to water consumers, and should be differentiated from dead cells or free DNA of pathogenic bacteria that are not able to infect humans. Recent studies have used ribosomal RNA as a target of microbial community studies to reveal the active fraction of microbes (Pitkänen et al., 2013; Inkinen et al., 2018, and 2019). An RNA-based approach gives an estimation of live (either dormant or metabolically active) members of the bacterial community, as ribosomal RNA is actively produced and regulated by living cells and it degrades more quickly than DNA after cell death (Li et al., 2017). In DWDSs, living members of bacterial communities and their activity are less studied compared to studies using DNA-based methods (Potgieter et al., 2018; Bruno et al., 2018; Waak et al., 2019; Rosenqvist et al., 2023).
The objective of this study was to characterize active and total bacterial community members and their richness and diversity in cold and hot water, and biofilms of pipes and water meters in five well-maintained full-scale DWDSs based on 16S rRNA and 16S rRNA gene analysis. We aimed to determine the effects of distance in distribution system, season of the year, and water quality parameters on the bacterial communities and their activity. Our objective was also to discover the presence and activity of opportunistic pathogens in cold and hot water and biofilms in the disinfected and non-disinfected DWDSs studied. The overarching goal was to assess the effects of raw water source, different treatment/disinfection methods, and environmental factors on microbiomes in drinking water and biofilms, providing information regarding potential future options to limit the growth potential of harmful microbes within the microbiomes of DWDSs.
2. Materials and methods
2.1. Water sampling
In this study, molecular methods were used to analyze the dynamics of bacterial communities of five DWDSs in Finland. The waterworks studied, described earlier by Ikonen et al. (2017) and Inkinen et al. (2019), were two artificial ground waterworks without disinfection located in the same city, two surface waterworks with UV-light disinfection combined with chlorine dioxide (ClO2) and chlorine (Cl2) or chloramine (NH2Cl) disinfection, and a ground waterworks with UV-light and chlorine (NaOCl) disinfection (systems A-E; Table 1]). Large-volume (100 liter) samples of cold drinking water after flushing of 1–2 min and hot water without flushing were collected over the four seasons of the year, twice in each season, using the dead-end ultrafiltration method (Smith and Hill, 2009). If the water contained chlorine, 1 g of sodium thiosulphate (Na2S2O3 x 5H2O) dissolved in 125 ml of sample water was added to the ultrafiltration cartridge to prevent residual disinfection after sampling. Cold water sampling points were stationed at three different distances (Table 1) from the waterworks in the same distribution line in each DWDS, except for sampling point three in DWDS E, which was on a different line in the same distribution system. Hot water samples were collected only at point two in all five systems. Building type and used pipe material in sampling points are in Supplemental Table S1.
Table 1.
Drinking water distribution systems (DWDSs) A-E and their sampling locations. Samples from cold and hot water sampling points, young pipe biofilm (Pipe) and matured water meter biofilm samples, distance from the drinking water treatment plant (DWTP), average and standard deviation of free and total chlorine concentrations and temperature. Additional 100 l cold water samples collected with water meter biofilms: N(DWDS A) = 2, N(DWDS B) = 2, and N(DWDS C) = 1. na = not applicable.
| DWDS | Sampling point |
Distance from DWTP (km) |
Sample type |
RNA samples |
DNA samples |
Free Cl (mg/l) mean ± sd |
Total Cl (mg/l) mean ± sd |
Temperature (°C) mean ± sd |
|
|---|---|---|---|---|---|---|---|---|---|
| No disinfection | A | 1 | 2 | Cold water | 8 | 8 | na | na | 12.7 ± 4.3 |
| 2 | 8 | Cold water | 8 | 8 | na | na | 10.6 ± 3.0 | ||
| 2 | 8 | Hot water | 8 | 8 | na | na | 53.8 ± 2.3 | ||
| 3 | 11 | Cold water | 8 | 8 | na | na | 9.2 ± 3.4 | ||
| na | na | Water meter | 2 | 2 | na | na | na | ||
| B | 1 | 1 | Cold water | 8 | 8 | na | na | 9.5 ± 1.8 | |
| 2 | 3 | Cold water | 8 | 8 | na | na | 10.8 ± 1.8 | ||
| 2 | 3 | Hot water | 8 | 8 | na | na | 57.5 ± 3.5 | ||
| 3 | 8 | Cold water | 8 | 8 | na | na | 8.0 ± 2.1 | ||
| na | na | Water meter | 2 | 2 | na | na | na | ||
| Chlorine + UV | C | 1 | 2 | Cold water | 7 | 7 | 0.41 ± 0.16 | 0.47 ± 0.12 | 8.9 ± 7.5 |
| 2 | 8 | Cold water | 8 | 8 | 0.38 ± 0.30 | 0.33 ± 0.09 | 11.8 ± 5.0 | ||
| 2 | 8 | Hot water | 8 | 8 | na | na | 50.6 ± 3.9 | ||
| 3 | 20 | Cold water | 7 | 7 | 0.09 ± 0.10 | 0.16 ± 0.10 | 10.1 ± 4.3 | ||
| na | na | Water meter | 2 | 1 | na | na | na | ||
| Chloramine + UV | D | 1 | 5 | Cold water | 8 | 8 | 0.06 ± 0.09 | 0.15 ± 0.06 | 10.3 ± 3.1 |
| 2 | 14 | Cold water | 8 | 8 | 0.09 ± 0.10 | 0.12 ± 0.14 | 10.5 ± 4.0 | ||
| 2 | 14 | Hot water | 8 | 8 | na | na | 54.6 ± 2.8 | ||
| 3 | 19 | Cold water | 7 | 8 | 0.07 ± 0.08 | 0.14 ± 0.13 | 8.1 ± 3.2 | ||
| na | na | Water meter | 1 | 1 | na | na | na | ||
| 2 | 14 | Pipe | 3 | 3 | na | na | na | ||
| Chlorine + UV | E | 1 | 9 | Cold water | 8 | 2 | 0.19 ± 0.04 | 0.28 ± 0.11 | 5.7 ± 1.5 |
| 2 | 26 | Cold water | 9 | 9 | 0.11 ± 0.05 | 0.12 ± 0.05 | 7.0 ± 2.1 | ||
| 2 | 26 | Hot water | 8 | 8 | na | na | 54.1 ± 4.1 | ||
| 3* | 36 | Cold water | 7 | 2 | 0.21 ± 0.08 | 0.34 ± 0.21 | 5.1 ± 1.1 | ||
| na | na | Water meter | 3 | 2 | na | na | na | ||
| 2 | 26 | Pipe | 3 | 2 | na | na | na |
Sampling point 3 is collected from another pipeline than points 1 and 2.
2.2. Biofilm sampling
Additionally, biofilm samples using pipe collectors and loose deposit with mature biofilms of water meters were collected from the DWDSs as described in Inkinen et al. (2019). Three one-year-old pipe collectors were collected from each of distribution systems D and E, for a total of six. Pipe collectors were located at point two of the water sampling points. Also, a total of ten mature biofilm/deposit samples aged 6 to 19 years old were collected from the same DWDSs as water samples but mainly from different points of the pipelines. Large-volume water samples were taken from the same sampling points as mature biofilms of water meters (Table 1).
2.3. Water quality analyses
Water quality analyses including heterotrophic plate count (HPC), total bacteria cell count determined using 4,6-diamidino-2-phenylindole staining (DAPI), and physicochemical water parameters, including measurements of free and total chlorine, pH, temperature, oxygen, and electric conductivity in the field and in the laboratory, metal concentrations, and microbial nutrient levels were determined as described earlier by Ikonen et al., 2017.
2.4. Nucleic acid extraction, sequencing, and PCR
Dead-end ultrafiltration eluates were further concentrated by filtering the samples through 0.45-μm-pore-size, 47-mm-diameter polycarbonate filters (pore size 0.4 μm, Nuclepore Polycarbonate, Whatman, Kent, UK) and freezing at −80 °C before extraction of nucleic acids as previously described (Inkinen et al., 2019). In brief, DNA and RNA were extracted using a Chemagen magnetic kit (Perkin Elmer, Waltham, MA, USA). RNA was further purified using an Ambion TURBO DNA-free DNase kit (Life Technologies, Carlsbad, CA, USA), and then the cDNA was synthesized using a random hexamer primed Superscript III system for RT-PCR (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was stored at −80 °C, while cDNA and DNA extracts were stored at −20 °C until use.
For high-throughput amplicon sequencing, the samples were barcoded and analysed using the Illumina Miseq platform (LGC Genomics GmbH, Berlin, Germany). The nucleic acids were used as templates for polymerase chain reaction amplification with the modified primer set 341F (5′-CCTACGGGNGGCWGCAG-3′) and 785R (5′-GACTACHVGGGTATCTAAKCC-3′) (Klindworth et al., 2013) (Supplemental Table S2) and targeting the V3-V4 hypervariable region of the 16S rRNA gene, together with Nextera DNA Library Kit adapters. For quality control, negative controls from sample processing steps of water and biofilm samples without template DNA and positive controls with mock microbial community were analysed. Final products were quantity and quality assessed on Qubit dsDNA (Life Technologies) and Bioanalyzer DNA (Agilent, USA) high-sensitivity assay kits, then stored at −20 °C before pooling for sequencing. Equimolar amounts of purified amplicons (100 ng) were pooled and used to construct Illumina libraries using the Ovation Rapid DR. Multiplex System 1–96 (NuGEN).
The occurrence and abundance of Mycobacterium spp., Legionella spp., Pseudomonas spp., and occurrence of species Mycobacterium intracellulare, Mycobacterium avium, Legionella pneumophila, Legionella pneumophila serogroup 1in samples were measured with RNA and DNA extracts (including extraction and filtration blanks). The TaqMan RT-qPCR assays were performed as previously described (Pitkänen et al., 2013). The qPCR reactions were performed using a QuantStudio 6 real-time PCR system (Applied Biosystems) on 20 μl volume using TaqMan Environmental PCR Master Mix (Life Technologies) with primers and probes at final concentrations of 0.2 μM (IDT Technologies, Inc.; for primer and probe sequences, see Supplemental Table S2). The cycling conditions included 95 °C for 10 min enzyme activation and pre-denaturation followed by 40 cycles of 95 °C for 15 s denaturation and 60 °C for 60 s annealing. Standard curves were generated using artificial gene fragments (gBlocks, IDT Technologies, Inc.) containing the sequences for each of the targeted genes.
In qPCR, undiluted and 10-diluted cDNA and DNA samples were used as qPCR templates to detect PCR polymerase inhibition. For samples in which PCR inhibition was detected, qPCR data was generated using the results from diluted samples. Background signals, if detected in filtration blanks, were subtracted from all the results to generate final qPCR data per assay. In most cases the limit of detection (LOD) was set at 3 copies per reaction. The copy number per 100 ml of water was calculated for those samples with values above the limit of quantification (LOQ) (i.e., as determined by the lowest value within the quantification range). The final RT-qPCR and qPCR, equivalent LOD (eLOD) and equivalent LOQ (eLOQ) values were calculated after taking into account the volume of sample filtered, factors associated with the various processing steps of the RNA and DNA manipulations, and the dilutions used for each sample analysed (Supplemental Table S3).
2.5. Next generation sequencing data preprocessing and analysis
Raw reads were used after primer removal as input in Cutadapt software (Martin, 2011) to remove the Illumina TruSeq adapters.Reads without adapters were further quality trimmed with Trimmomatic software (Bolger et al., 2014). Then the forward and reverse reads were merged using Flash2 software (Magoč and Salzberg, 2011). The merged reads were quality processed using split_libraries_fastq.py script on the QIIME platform and demultiplexed (Caporaso et al., 2010). The reads were checked for chimeras (artificial sequences due to PCR error) using vsearch algorithm and chimeric sequences were filtered out. After chimera removal, the reads were clustered at 97 % using uclust_ref and an operational taxonomic unit (OTU) picking step was performed with an open reference OTU picking approach. Further downstream processing was performed by removal of chloroplast and mitochondrial OTUs (Inkinen et al., 2019). Sequence processing of the samples included negative (N = 41) and positive (N = 9) controls. Negative controls of sample processing were used to define a minimum read count for a sample to be included in the analysis and the count was 918 sequences. Samples with less than that were excluded. Taxonomy of sequences was obtained using database GTDB R207 (released in April 2022). The datasets generated during the study including bacteria sequences are available in the Short Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject PRJNA509718.
2.6. Phylogenetic analysis of opportunistic pathogens
Phylogenetic trees of Legionella, Mycobacterium, and Pseudomonas were constructed using reference strain sequences from a database, sequence libraries from the DWDSs studied, and an outgroup (Coxiella burnetii for Legionella, Nocardia asteroides for Mycobacterium, and Acinetobacter calcoaceticus for Pseudomonas). The 16S rRNA gene sequences of clinically relevant Legionella, Mycobacterium, and Pseudomonas species were collected from the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide) for reference data. The corresponding sequences with the same OTU IDs were picked from the sequence libraries studied based on the Greengenes database using the closed reference approach. The picked sequences were aligned to the 16S rRNA gene region by mothur using align.seqs command with gg_13_8_99.align and were trimmed for equal length. All the sequences were aligned using ClustalW and used for the phylogenetic tree construction by Neighbour Joining algorithm using 1000 bootstrap values in MEGA5.2.
2.7. Statistical analyses
Statistical calculations for alpha diversities were performed using Chao1, Shannon, and Simpson indices. The alpha diversities (microbial community differences within samples) were calculated using alpha_rarefaction.py script with QIIME. Beta diversities (microbial community differences between the samples) and the summary of taxonomy in samples were calculated using MicrobiomeAnalyst. Before statistical analysis RNA and DNA sequence libraries were filtered in MicrobiomeAnalyst using default settings to remove low count sequences and normalized using default settings with total sum scaling. Beta diversities were visualized using Bray-Curtis dissimilarity index. Pairwise comparison between sample groups from different DWDSs and sampling points were calculated using permutational multivariate analysis of variance (PERMANOVA). Microsoft Excel was additionally used for preparing figures. Further statistical comparison of diversity and abundance of active members and opportunistic pathogens in different sample groups were calculated using IBM SPSS Statistics. Canonical correspondence analysis (CCA) was used to show relations between bacterial communities in DWDSs A-E and physico-chemical parameters, i.e., temperature, pH, turbidity, absorbance 254 nm and 420 nm, electric conductivity (EC), aluminum (Al), copper (Cu), iron (Fe), manganese (Mn) concentration, chlorine, and microbially available nutrients, i.e., acetate carbon, assimilable organic carbon (AOC), and microbially available phosphorus (MAP); as well as between community composition and microbiological water quality parameters, i.e., total bacteria cell count (DAPI) and heterotrophic plate count (HPC).
3. Results and discussion
3.1. The structural differences in the bacterial communities
A total of 4 255 115 sequencing reads and 5 836 OTUs were identified as bacteria after removing samples with sequence counts of less than 918 (Supplemental Table S4). More than two sequence reads were recorded for 5 183 identified OTUs. Average and maximum read count per sample were 12 441 and 58 206 sequences, respectively.
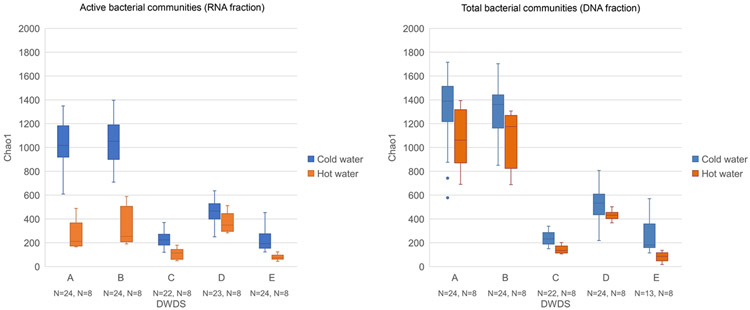
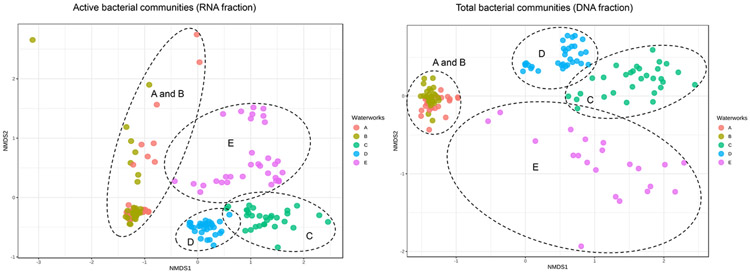
Species richness of active bacterial community (RNA fraction) was significantly higher in the cold water samples from the two non-disinfected DWDSs using artificially recharged groundwater (Chao1 diversity index mean values of 1 029; N = 24 and 1 042; N = 24 at DWDS A and B, respectively) as compared to the samples from the three disinfected DWDSs (Chao1 mean values 232; N = 22 at DWDS C, 471; N = 23 at D, and 219; N = 24 at E) (Kruskal-Wallis pairwise comparison, P < 0.05) (Fig. 1), similar to the findings of earlier genome-based studies (Bautista-de los Santos et al., 2016; Dai et al., 2020). Additionally, species richness was significantly higher in chloraminated DWDS D, compared to the two DWDSs (C and E) that used free chlorine compounds for chlorination (Kruskal-Wallis pairwise comparison, P < 0.05). In total bacteria communities (DNA fraction), differences of bacterial diversity between non-disinfected and disinfected DWDSs were similar but diversity in non-disinfected DWDSs was even higher compared to disinfected (Fig. 1). Source water and other treatment methods varied between the DWDSs and they may also have contributed to the differences. The difference in active bacterial community structures (RNA samples) between non-disinfected artificial ground waterworks and disinfected groundwater and surface water was clear in the beta-diversity analyses (Fig. 2). The DWDS bacteria from non-chlorinated artificial ground waterworks (A and B) were closer to each other compared to DWDS bacteria in chlorinated groundwater (E) or in surface waterworks (C and D). Similar clustering was also observed based on DNA samples. Furthermore, early studies of these DWDSs found a higher diversity of eukaryotic and archaea communities in non-disinfected DWDSs (A-B) compared to disinfected systems (C-E) (Inkinen et al., 2019 and 2021). In contrast, the abundance of functional genes based on metagenomic analysis was higher in disinfected DWDSs samples than in non-disinfected samples (Gomez-Alvarez et al., 2023).
Fig. 1.
Alpha diversity of active (RNA fraction) and total (DNA fraction) bacterial communities by Chao1 index in cold and hot water in DWDSs A-E. Circles are outlier results.
Fig. 2.
Non-metric multidimensional scaling (NMDS) plots of beta diversity of bacterial community compositions of RNA and DNA water samples using Bray-Curtis dissimilarity index in DWDSs A-E.
Significantly higher bacterial community richness was noted in cold water samples (Chao1 mean 696; N = 224) than in samples from hot water systems (Chao1 mean 394; N = 80) (Mann-Whitney U test, P < 0.05). The higher alpha diversity in cold water compared to hot water samples was seen more strongly in non-disinfected DWDSs than in disinfected (Fig. 1). When comparing samples only from sampling point two, where hot water samples were collected, a significant difference in alpha diversity (Kruskal-Wallis pairwise comparison, P < 0.05) were noted in DWDSs A (non-disinfected artificial groundwater) between cold (Chao1 mean 1 091; N = 8) and hot water (264; N = 8) as well as in DWDS B between cold (Chao1 mean 1 112; N = 8) and hot water (322; N = 8) in active RNA fraction. In DWDS E (disinfected groundwater) in sampling point 2 difference was noted in both RNA fraction between cold (Chao1 mean 312; N = 9) and hot water (82; N = 8)) and in DNA fraction between cold (Chao1 mean 303; N = 9) and hot water (79; N = 8) (Kruskal-Wallis pairwise comparison, P < 0.05). The active part (RNA fraction) of the bacterial community was less diverse compared to the total DNA community. For comparison, most of the published research of bacterial communities have used DNA-based methods (Potgieter et al., 2018; Bruno et al., 2018; Waak et al., 2019). In cold water, significantly lower species richness was noted when bacterial communities were analysed according to RNA fraction (Chao1 mean 606; N = 117) (Mann-Whitney U test, P < 0.05) compared to DNA fraction (Chao1 mean 794; N = 107). Also, in hot water systems, species richness was significantly lower in RNA fraction (Chao1 mean 229; N = 40) than in DNA fraction (Chao1 mean 559; N = 40) (Mann-Whitney U test, P < 0.05). Similar differences between the sample groups were also seen when alpha diversity was measured as Shannon and Simpson indexes (Supplemental Figure S1). However, in DWDS A, cold water sampling point 3 showed a less diverse bacterial community than in hot water system (measured as Chao1 index). Furthermore, in DWDS C, the alpha diversity of the bacterial community in cold water at point 3, measured as Shannon and Simpson indexes, was less diverse than in hot water. Water temperature (cold vs. hot water system) shaped the bacterial community composition in each DWDSs. This was also seen in beta-diversity analysis (Fig. 3). However, in DWDSs B and D the water temperature was a less dominant factor as compared to the difference between groups of active bacteria (RNA) and total bacteria (DNA). Total bacterial communities in drinking water distribution systems based on DNA methods have been more often described in publications (Potgieter et al., 2018; Bruno et al., 2018; Waak et al., 2019; Rosenqvist et al., 2023) compared to hot water systems and active RNA-based communities. Earlier studies show that raising the temperature in hot water systems shifts bacterial composition (Ji et al., 2017; Dai et al., 2018) and bacterial communities are less diverse in hot tap water than in cold tap water (Zhang et al., 2021).
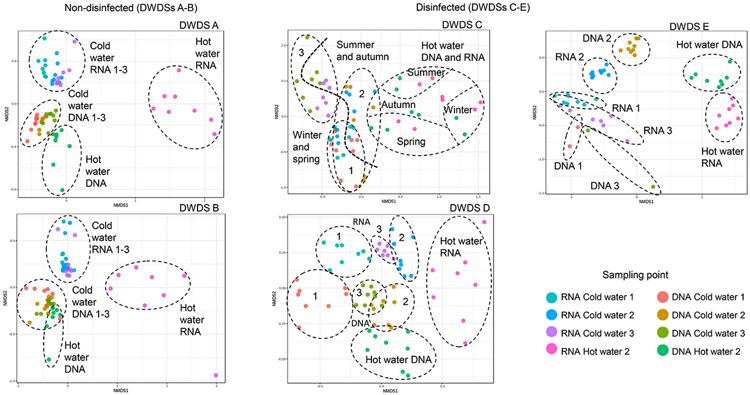
Fig. 3.
Non-metric multidimensional scaling (NMDS) plots of beta diversity of bacterial community compositions measured using Bray-Curtis dissimilarity index in each five studied drinking water distribution systems A-E. Samples from three sampling points (1–3) in cold water distribution systems and hot water samples from sampling point 2 are indicated by different colors.
Changes in the bacterial community structure between the sampling points 1–3 inside each distribution system were also noted in beta-diversity analysis (Fig. 3). The biggest difference was between the first and the third sampling point in disinfected DWDS C (Bray-Curtis Index, DNA: R2=0.60, RNA: R2=0.48) and DWDS D (DNA: R2=0.56, RNA: R2=0.43) (P < 0.005, PERMANOVA). In DWDS E, points 1 and 3 clustered closely to each other, but sampling point 2 differed from sampling point 1 (RNA: R2=0.50, P = 0.001, PERMANOVA). In non-disinfected DWDSs A and B not as clear separation of sampling points was seen in NMDS figure (Fig. 3), but the sample group from DWDS A cold water sampling point 3 was separated from point 1 by Bray-Curtis dissimilarity index (DNA: R2=0.49, RNA: R2=0.53, P = 0.001, PERMANOVA). Chlorine concentrations may explain separation in DWDS E as they were higher at sampling points 1 and 3 as compared to sampling point 2 (Table 1). Aging of water, i.e. the distance from the water treatment plant has been noted to change the bacterial community structure especially in chloraminated DWDS by Potgieter et al. (2018), and to change the eukaryotic and archaea communities in the DWDSs studied here (Inkinen et al., 2019 and 2021). The effect of season on structure of bacterial communities was limited. In DWDS C it had some effect on beta diversity of community structure (Bray-Curtis Index, RNA: R2=0.31–0.38, DNA: R2=0.15–0.26, P < 0.005, PERMANOVA) and alpha diversity as it was higher in spring (Chao1 mean 277; N = 16) than in summer (185; N = 16) and autumn (172, N = 12) (Kruskal-Wallis, P < 0.05) but not in winter (Chao1 mean 252; N = 16). Also, in DWDS B season affected only a little on beta diversity (RNA: R2=0.22–0.31, DNA: R2=0.25–0.29, P < 0.005, PERMANOVA).
Biofilms from water meters (Chao1 mean 383; N = 18, Supplemental Table S5.) and pipe collectors (Chao1 mean 172; N = 11) show significantly less diversity (Kruskal-Wallis test, P < 0.05) than water samples (Kruskal-Wallis test, P < 0.05). The exception was DWDS E where the Chao1 indexes of the two water meters were higher (Chao1 mean 408; N = 5) than those of the water samples (cold water: Chao1 mean 233; N = 37, and hot water: Chao1 mean 80; N = 16). Less diverse communities had earlier been observed in biofilms compared to water samples, and more clearly in chloraminated water than in water without disinfectant residues (Waak et al., 2019). Bacterial communities of biofilm samples of DWDSs A-E clustered separately in beta-diversity analysis (Supplemental Figure S2). Additionally, young biofilms collected from pipe collectors clustered separately from mature biofilms of water meters (Supplemental Figure S2). A similar clustering of biofilm samples separate from water samples was seen in eukaryotic and archaea communities in the same DWDSs in earlier studies (Inkinen et al., 2019 and 2021).
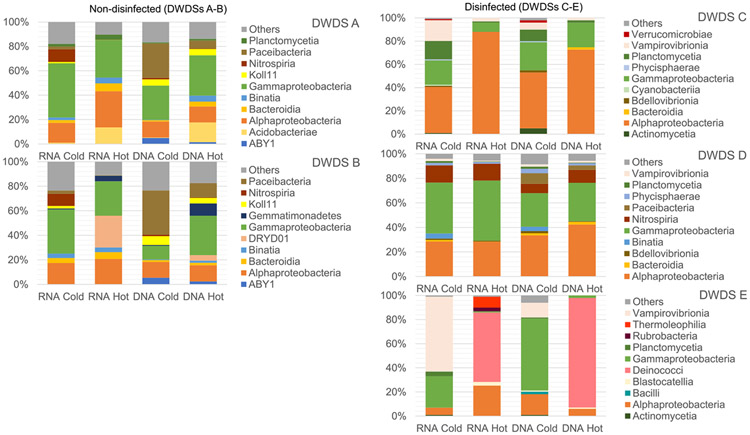
The two most abundant bacteria in the active part of the community (RNA) and the total bacterial community (DNA) in water samples belonged to classes Alphaproteobacteria (RNA 28 %, DNA 44 %) and Gammaprotebacteria (RNA 32 %, DNA 30 %). The third most abundant and active bacteria class was Vampirovibrionia (formerly Melainabacteria) (15 %), whereas in total community (DNA) the third most abundant class was Paceibacteria (11 %). The Alpha- and Gammaproteobacteria, and Vampirovibrionia (formerly Melainabacteria) had earlier been found in drinking water environments using DNA-based methods (Bruno et al., 2018; Zhang et al., 2021), and they were active in these DWDSs based on RNA fraction.
Non-disinfected groundwater DWDSs A and B had seven classes in common in the top ten most abundant bacterial classes in water samples (Fig. 4). Abundance of unassigned bacteria (group koll 11) and minor groups other than the top ten most abundant classes were higher in the waters in non-disinfected DWDSs A and B. Vampirovibrionia was seen in all disinfected DWDSs C, D, E, but was not in the top ten in non-disinfected DWDSs A and B. The relative abundance of Vampirovibronia was highest in cold water in DWDS E for both RNA and DNA fractions. Vampirovibrionia was also an active member (RNA fraction) in biofilm samples collected from water meters from DWDS E (Supplemental Figure S3). Class Nitrospiria was more abundant and active in cold and hot water in DWDS D with chloramine disinfection, compared to non-chlorinated DWDSs A and B that had Nitrospiria only in cold water and in greater abundance in active RNA fraction than in DNA fraction (Fig. 4). In the biofilm samples, Nitrospiria was the most abundant in DWDS D but was also active in the other DWDSs, except DWDS C (Supplemental Figure S3). Nitrospiria plays a role in nitrification in the bacterial ecosystem as it oxidizes nitrite into nitrate. The nitrification process may decrease water quality, cause corrosion, decrease disinfectant residue, and increase growth of bacteria (Hossain et al., 2022). Bdellovibrionia was active in cold water in chlorinated DWDS C and in cold and hot water in chloraminated water in DWDS D. Some species of Bdellovibrio (such as B. bacteriovorus and B. exovorus) belonging to Bdellovibrionia have been found to be natural predators of Gram-negative human pathogens and have also been found in chloraminated drinking water and in disinfection residue-free DWDS (Bautista-de los Santos et al., 2016; Atterbury and Tyson, 2021; Rosenqvist et al., 2023). In hot water systems, Acidobacteriae were present and active in DWDS A and Deinococci in DWDS E. Acidobacteriae have been found in hot water system studies (Ji et al., 2017). Deinococci class has been found to increase in temperatures as high as 51 °C in water systems (Dai et al., 2018). Also, Blastocatellia, Rubrobacteria, and Thermoleophilia were abundant active members of the bacterial community in the hot water in DWDS E. These bacteria have been observed in cold and hot tap water based on DNA (Zhang et al., 2021). Other bacteria detected in this study – Cyanobacteria, Nitrospiria, Paceibacteria, and Planctomycetia – have also been found in drinking water (Bruno et al., 2018; Dai et al., 2020; Rosenqvist et al., 2023). However, Paceibacteria, belonging to phylum Patescibacteria, were not as abundant active members of DWDSs A, B, and D, being found more in the DNA fraction.
Fig. 4.
The ten most abundant bacterial classes in the studied drinking water distribution systems A-E in RNA and DNA fractions in cold water and hot water samples. A group called Others consist of the minor bacterial classes other than the top ten.
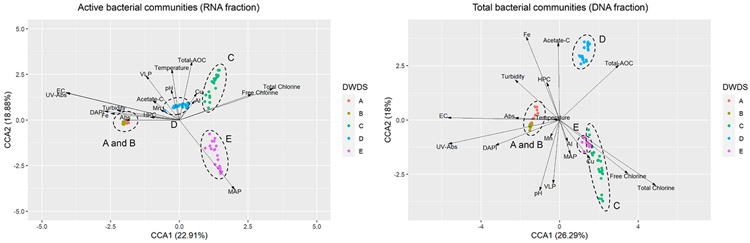
Environmental factors of cold water correlate with the bacterial communities of DWDSs differently, as seen in canonical correspondence analysis (Fig. 5). Iron (Fe) correlated more with non-disinfected artificial groundwater DWDSs A-B compared to disinfected DWDSs C-E. Copper (Cu) and aluminum (Al) correlated with DWDSs C-D using disinfected water originating from surface waterworks based on CCA. A previous study found a higher abundance of metal resistance genes in disinfected than in non-disinfected systems (Tiwari et al., 2022). Microbially assimilable phosphorus (MAP) correlated with DWDS E (disinfected groundwater supply). However, in DNA fraction these chemical parameters do not correlate as strongly with bacterial community results (Fig. 5).
Fig. 5.
Canonical correspondence analysis (CCA) showing relationships between active RNA fraction and total DNA fraction of bacterial communities (colored dots) and physico-chemical and microbiological parameters of cold water samples in DWDSs A-E. Arrows are for temperature, pH, turbidity, absorbance 254 nm (UV-Abs) and 420 nm (Abs), electric conductivity (EC), aluminum (Al), copper (Cu), iron (Fe), manganese (Mn), free and total chlorine, assimilable organic carbon (AOC), acetate carbon, microbially available phosphorus (MAP), total bacteria cell count (DAPI), heterotrophic plate count (HPC), and virus-like particle (VLP).
3.2. Activity of opportunistic pathogens in cold and hot water systems
Of the opportunistic pathogens, 22 OTUs of Legionella, 16 of Mycobacterium and eight of Pseudomonas were detected in total from all five DWDSs using closed reference OTU picking comprising 0.9 % of all the bacteria from water and biofilm samples total in RNA and DNA fractions. Additionally, 16 OTUs of Legionella, 14 of Mycobacterium, and two of Pseudomonas were detected using de novo picking. All OTUs of opportunistic pathogens that were detected in biofilm samples were present in water samples as well, but not all bacteria present in water samples were detected in biofilm samples. The (RT)-qPCR confirmed the presence of opportunistic pathogens in the five DWDSs studied as Legionella spp. was detected in 85 %, Mycobacterium spp. in 95 %, and Pseudomonas aeruginosa in 78 % of the water and biofilm samples.
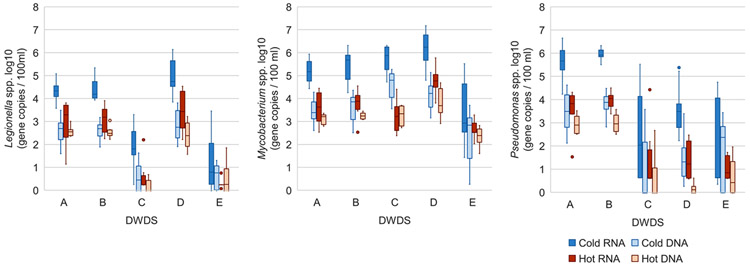
In cold water samples, Legionella spp. was significantly more abundant in RNA fraction (mean 201 gene copies / 100 ml, N = 117) compared to DNA fraction (mean 127 gene copies / 100 ml, N = 107) (Kruskal-Wallis test, P < 0.001) measured by RT-qPCR and qPCR methods, respectively. In RNA fraction, the abundance of Legionella was significantly higher in cold water than in hot water (mean RNA 131 gene copies / 100 ml, N = 40) (Kruskal-Wallis test, P < 0.001), indicating that the hot water temperature was efficient in controlling the activity of Legionella at the sampling locations studied. This difference was seen in all DWDSs A-E but the difference was not so clear at DWDS E (disinfected groundwater) (Fig. 6). The mean temperature of the hot water samples was above 50 °C, which may explain the lower abundance of opportunistic pathogens, including Legionella, which decreases at such high temperatures (Dai et al., 2018). To control the growth of Legionella in hot water, the temperature should be kept above 50 °C (preferably 55 °C) (ESGLI, 2017).
Fig. 6.
Legionella, Mycobacterium, and Pseudomonas genera in cold and hot water samples in RNA and DNA fractions in each DWDS A-E analysed using (RT-)qPCR method (gene copies / 100 ml) at logarithmic scale. Circles are outliers.
Like Legionella, Mycobacterium spp. was significantly more abundant in RNA fraction in cold water (mean 219 gene copies/100 ml, N = 117) compared to hot water samples (mean 110 gene copies/100 ml, N = 40) (Kruskal-Wallis test, P < 0.001). In DNA fraction, a difference between cold water (mean 124 gene copies/100 ml, N = 107) and hot water (mean 75 gene copies/100 ml, N = 40) was also detected (Kruskal-Wallis test, P = 0.005). A difference in abundance of Mycobacterium between cold and hot water was observed in all DWDSs A-E (Fig. 6). High hot water temperatures here may cause the lower abundance compared to cold water, as described with Legionella.
In cold water samples, Pseudomonas spp. was significantly more abundant in RNA fraction (mean 201 gene copies/100 ml, N = 117) compared to DNA fraction (mean 134 gene copies/100 ml, N = 107) (Kruskal-Wallis test, P < 0.001). Also, Pseudomonas abundance was significantly higher in cold water than in hot water (mean RNA 129 gene copies/100 ml, N = 40) but only in RNA fraction (Kruskal-Wallis test, P < 0.001), indicating that Pseudomonas was more active in cold water compared to hot water: as with Legionella, this was probably caused by high water temperatures. This was seen in all DWDSs A-E but not as clearly in disinfected systems C-E (Fig. 6). There were small but not significant differences in Pseudomonas spp. numbers between hot water DNA (mean 83 gene copies/100 ml, N = 40) and RNA with higher RNA numbers, as well as between cold and hot water in DNA samples with higher numbers in cold water.
The detection frequency of opportunistic pathogens was higher in RNA fraction than in DNA fraction in most cases, when (RT)-qPCR method targeted at the opportunistic pathogen species was used (Fig. 6). No such difference in detection frequency was seen when an amplicon sequencing method targeted at the 16S RNA gene was used (Supplemental Figure S6). This might be because amplicon sequencing is not a quantitative method but produces the relative abundance of members in the bacterial community (Bautista-de los Santos et al., 2016). Opportunistic pathogens detected with the RNA method more likely describe the living cells, since RNA is synthesized by active cells and RNA degrades faster than DNA does (Pitkänen et al., 2013; Li et al., 2017). Living cells may possibly start growing under favorable conditions, causing health risks for consumers.
3.3. Abundance of opportunistic pathogens changed between DWDSs
In RNA fraction in cold water, Pseudomonas spp. were most abundant in non-disinfected DWDSs A (mean 7.8 × 105 gene copies/100 ml, N = 24) and B (mean 1.1 × 106 gene copies/100 ml, N = 24) based on the (RT)-qPCR method (Fig. 6). However, waterworks with disinfection did not necessarily have lower numbers of opportunistic pathogens than waterworks without disinfection. DWDS D, a chloramine using surface water supply, had the highest gene copy numbers of Legionella spp. (mean 2.4 × 105 gene copies/ml, N = 23) and Mycobacterium spp. (mean 3.3 × 106 gene copies/100 ml, N = 23).
Although numbers of Legionella were highest in chloraminated DWDS D they were lowest in DWDSs C (mean 326 gene copies/100 ml, N = 22) and E (mean 168 gene copies/100 ml, N = 23), with free chlorine disinfection. In contrast, earlier studies have found chloramine to be more effective against Legionella than chlorine disinfection (Donohue et al., 2019; Gomez-Alvarez et al., 2012). Additionally, Legionella pneumophila was found in two samples from location 2 in non-disinfected DWDS A in cold water and from location 3 in cold water in non-disinfected DWDS B, as well as in the water sample collected from the water meter sampling location in DWDS B (Table 2). Legionella pneumophila serogroup 1 was found only in DWDS A from location 2 in four cold water samples and in one hot water sample.
Table 2.
Number of samples (N) where opportunistic pathogen species from genera Legionella and Mycobacterium were detected in DWDSs A-E by quantitative PCR method. ND=Not detected, WM=water meter. For (RT)-qPCR assay details, see Supplemental Tables S2 and S3.
|
Legionella pneumophila (studied from the DNA fraction) |
Mycobacterium species (studied by 16S rRNA and 16S rRNA gene assays) |
|||||
|---|---|---|---|---|---|---|
| DWDS | N (total) | L. pneumophila detected | L. pneumophila serogroup 1 detected | N (total) | M. avium detected | M. intracellulare detected |
| A | 38 | 2 (cold water) | 4 (cold water) 1 (hot water) | 75 | 6 (RNA, cold water) | ND |
| B | 38 | 2 (cold water) | ND | 76 | 4 (RNA, cold water) | ND |
| C | 33 | ND | ND | 67 | ND | ND |
| D | 36 | ND | ND | 70 | ND | ND |
| E | 25 | ND | ND | 63 | ND | 1 (DNA, biofilm WM) |
Ten of the detected Legionella OTUs were determined to belong to genus Legionella by QIIME software (circles in phylogenetic tree, Supplemental Figure S5) and 12 of the OTUs were identified by QIIME only at family level, belonging to family Legionellaceae and confirmed as Legionella using NCBI BLAST (triangles in the phylogenetic tree). The most abundant OTU in water samples (OTU ID 3479378) had more RNA reads (5 196) than DNA reads (1 032). The second most abundant Legionella (OTU ID 311942) had 669 RNA and 710 DNA reads. Both of these OTUs were close to many Legionella sequences in NCBI database but did not match any Legionella species with 100 % similarity. Other Legionella OTUs had read counts of less than one thousand. Legionella pneumophila is the most well-known pathogenic Legionella species, but other Legionella species may cause infections as well (Chauhan and Shames, 2021).
Mycobacteria were most abundant at DWDSs C and D, which distribute disinfected water from surface water works. Mycobacteria are known to be more resistant to many disinfection chemicals, including chlorine, than other bacteria (Bautista-de los Santos et al., 2016; Gomez-Alvarez et al., 2012). Similarly, Kotlarz et al. (2019) observed higher numbers of mycobacteria in drinking water that used surface water. DWDSs C and D had nutrient AOC corresponding with bacterial communities in canonical correspondence analysis (CCA, Fig. 5). Mycobacteria prevalence is found to correlate strongly with the concentration of assimilable organic carbon in the water leaving the waterworks (Torvinen et al., 2004). Opportunistic pathogens may, however, grow and cause a health risk even in water with low nutrient contents (Vital et al., 2012; Wang et al., 2013), and they were also found in this study in low nutrient ground water and artificial ground water. Mycobacterium avium was found in cold water samples in RNA fraction in DWDSs A and B. M. avium was found at locations 1 and 2 in DWDS A, and at locations 2 and 3 in DWDS B (Table 2). M. avium was also found in cold water samples gathered from the same location as water meter samples. Mycobacterium intracellulare was found in only one DNA water meter biofilm sample showing that opportunistic pathogen species may be present in biofilms even though not detected in water.
One of the Mycobacterium OTUs (OTU ID 1062748) detected was close to M. fortuitum and another one (OTU ID 2651333) was close (87 %) to M. haemophilum (Supplemental Figure S6). The three most abundant OTUs in the water samples were OTU 543570 with 1 598 RNA and 5 170 DNA reads, OTU 1062748 (close to M. fortuitum) with 3 371 RNA and 2 194 DNA reads, and OTU 902334 with 1 065 RNA and 123 DNA reads. Other closed-reference picked Mycobacterium OTUs had less than one thousand read counts. Additionally, there were two de novo picked Mycobacterium OTUs with read counts of over one thousand.
Pseudomonas species were most abundant in non-disinfected DWDSs A and B, which distribute artificial groundwater. The most abundant OTU belonging to genus Pseudomonas in the water samples was OTU 288207, with a total of 3 691 reads, which was not close to any known opportunistic pathogen in phylogenetic analysis (Supplemental Figure S7). Legionella spp., Mycobacterium spp., and Pseudomonas spp. were also present in RNA and DNA fractions in biofilm or loose deposit samples (Supplemental Table S6) in five DWDSs A-E.
This study shows that there are opportunistic pathogens in non-disinfected and disinfected drinking water and biofilms in active (RNA) fraction and total (DNA) fraction, and their abundance varies between DWDSs with different disinfection strategies showing that opportunistic pathogens are also present in water distribution systems of good quality. Potential health risks exist if the water quality changes so that it provides an opportunity for pathogen bacteria to multiply, i.e. if the water temperature rises in cold water or falls in hot water.
4. Conclusions
Not all members of the communities in cold and hot water systems were active as active RNA fraction was less diverse than DNA fraction of total bacterial community.
Active communities in the studied good quality drinking waters were more diverse in non-disinfected water distribution systems compared to chloraminated systems. The least diverse communities were in systems where free chlorine was used as a disinfectant.
The structure of active bacterial communities varied between cold and hot water systems. Nitrospiria was active in non-disinfected system only in cold water but in chloraminated systems both in cold and hot water. Deinococci were active in hot water in chlorinated groundwater systems and in non-disinfected hot water systems.
Beta diversity of bacterial communities changed between the sampling points in distribution systems, representing the age of the water within each DWDS, and more in disinfected systems with disinfectant residue than in non-disinfected systems.
Opportunistic pathogens were detected in cold and hot water systems with qPCR method. Legionella and Mycobacterium genera were most abundant in chloraminated and Pseudomonas in non-disinfected systems, but their abundance using amplicon sequencing was <1 % of all bacteria.
Supplementary Material
Acknowledgement
The authors thank all personnel of the waterworks that participated in the sampling campaign and the personnel at the laboratory, namely Ms. Tarja Rahkonen, Ms. Tiina Heiskanen, and Ms. Marjo Tiittanen from the Water Microbiology Laboratory, Finnish Institute for Health and Welfare, Kuopio, Finland, who performed the analysis and nucleic acid extraction of the samples, as well as Jatta Heikkinen for help in the sampling. In Memoriam of Dr. Jaana Kusnetsov: the authors are thankful for her leading contribution on Legionella study during project period. Kenneth Quek is acknowledged for grammatical advice on the article. This document has been reviewed in accordance with U.S. Environmental Protection Agency (EPA) policy and approved for publication. The research presented was not performed or funded by the EPA and was not subject to the EPA’s quality system requirements. Any opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the EPA; therefore, any mention of trade names, manufacturers or products does not imply an endorsement by the United States Government or the EPA. EPA and its employees do not endorse any commercial products, services, or enterprises.
Funding
This work was supported by the Academy of Finland (DWDSOME, project number 275549). This work was also supported by Maa- ja vesitekniikan tuki ry (grant numbers 4342 and 4171), the Erkki Paasikivi Foundation (grant number 20220604), and the Betty Väänänen Fund from the Kuopio Naturalists’ Society (KLYY), awarded to SS.
Footnotes
CRediT authorship contribution statement
Sallamaari Siponen: Formal analysis, Investigation, Data curation, Writing – original draft, Visualization, Writing – review & editing. Balamuralikrishna Jayaprakash: Methodology, Software, Formal analysis, Visualization, Writing – review & editing. Anna-Maria Hokajärvi: Methodology, Investigation, Data curation, Writing – review & editing. Vicente Gomez-Alvarez: Formal analysis, Investigation, Writing – review & editing. Jenni Inkinen: Investigation, Data curation, Writing – review & editing. Ivan Ryzhikov: Software, Visualization, Writing – review & editing. Pia Räsänen: Methodology, Formal analysis, Investigation, Writing – review & editing. Jenni Ikonen: Investigation, Writing – review & editing. Anna Pursiainen: Investigation, Writing – review & editing. Ari Kauppinen: Investigation, Writing – review & editing. Mikko Kolehmainen: Conceptualization, Investigation, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing. Jussi Paananen: Conceptualization, Investigation, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing. Eila Torvinen: Conceptualization, Investigation, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing. Ilkka T. Miettinen: Conceptualization, Investigation, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing. Tarja Pitkänen: Conceptualization, Investigation, Resources, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2023.120858.
Data availability
The bacteria sequence data generated in this study is available in the Short Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject PRJNA509718.
References
- Atterbury RJ, Tyson J, 2021. Predatory bacteria as living antibiotics - where are we now? Microbiology 167 (1). 10.1099/mic.0.001025. Epub 2021 Jan 19. [DOI] [PubMed] [Google Scholar]
- Bautista-de los Santos QM, Schroeder JL, Sevillano-Rivera MC, Sungthong R, Ijaz UZ, Sloan WT, Pinto AJ, 2016. Emerging investigators series: microbial communities in full-scale drinking water distribution systems – a meta-analysis. Environ. Sci.: Water Res. Technol 2, 631–644. 10.1039/C6EW00030D. [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno A, Sandionigi A, Bernasconi M, Panio A, Labra M, Casiraghi M, 2018. Changes in the drinking water microbiome: effects of water treatments along the flow of two drinking water treatment plants in a urbanized Area, Milan (Italy). Front. Microbiol 10.3389/fmicb.2018.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buse HY, J Morris B, Struewing IT, Szabo JG, 2019. Chlorine and monochloramine disinfection of legionella pneumophila colonizing copper and polyvinyl chloride drinking water biofilms. Appl. Environ. Microbiol 85 (7) 10.1128/AEM.02956-18 e02956–18Erratum in: Applied Environmental Microbiology 2022 Apr 12;88(7):e0022322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Shames SR, 2012. Pathogenicity and virulence of Legionella: intracellular replication and host response. Virulence 12 (1), 1122–1144. 10.1080/21505594.2021.1903199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Rhoads WJ, Edwards MA, Pruden A, 2018. Shotgun metagenomics reveals taxonomic and functional shifts in hot water microbiome due to temperature setting and stagnation. Front. Microbiol 10.3389/fmicb.2018.02695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Sevillano-Rivera MC, Calus ST, Bautista-de Los Santos QM, Eren AM, van der Wielen PWJJ, Ijaz UZ, Pinto AJ, 2020. Disinfection exhibits systematic impacts on the drinking water microbiome. Microbiome 8, 42. 10.1186/s40168-020-00813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue MJ, Vesper S, Mistry J, Donohue JM, 2019. Impact of chlorine and chloramine on the detection and quantification of Legionella pneumophila and Mycobacterium species. Appl. Environ. Microbiol 85 (24) 10.1128/AEM.01942-19, 2019 Nov 27e01942–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EU, 2020. Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the quality of water intended for human consumption, pp. 1–62. OJ L 435 of 23 December 2020. [Google Scholar]
- ESGLI, 2017. European technical guidelines for the prevention, control and investigation, of infections caused by Legionella species. European Centre for Disease Prevention and Control. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Revetta RP, Santo Domingo JW, 2012. Metagenomic analyses of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol 78 (17), 6095–6102. 10.1128/AEM.01018-12, 2012 SepEpub 2012 Jun 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Siponen S, Kauppinen A, Hokajärvi AM, Tiwari A, Sarekoski A, Miettinen IT, Torvinen E, Pitkänen T, 2023. A comparative analysis employing a gene- and genome-centric metagenomic approach reveals changes in composition, function, and activity in waterworks with different treatment processes and source water in Finland. Water Res. 229, 119495 10.1016/j.watres.2022.119495, 2023ISSN 0043-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward C, Ross KE, Brown MH, Bentham R, Whiley H, 2022. The presence of opportunistic premise plumbing pathogens in residential buildings: a literature review. Water 14 (7), 1129. 10.3390/w14071129. [DOI] [Google Scholar]
- Hossain S, Chow CWK, Cook D, Sawade E, Hewa GA, 2022. Review of nitrification monitoring and control strategies in drinking water system. Int. J. Environ. Res. Public Health 19 (7), 4003. 10.3390/ijerph19074003, 2022 Mar 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen JM, Hokajärvi AM, Heikkinen J, Pitkänen T, Ciszek R, Kolehmainen M, 2017. Drinking water quality in distribution systems of surface and ground waterworks in Finland. J. Water Security 3, 1–10. 10.15544/jws.2017.004. [DOI] [Google Scholar]
- Inkinen J, Ahonen M, Mäkinen R, Pursiainen A, Pitkänen T, Jayaprakash B, Jorge W, Santo Domingo JW, Salonen H, Elk M, Keinänen-Toivola MM, 2018. Bacterial community changes in copper and PEX drinking water pipeline biofilms under extra disinfection and magnetic water treatment. J. Appl. Microbiol 124, 611–624. 10.1111/jam.13662, 2018. [DOI] [PubMed] [Google Scholar]
- Inkinen J, Jayaprakash B, Siponen S, Hokajärvi A, Pursiainen A, Ikonen J, 2019. Active eukaryotes in drinking water distribution systems of ground and surface waterworks. Microbiome 7, 1–17. 10.1186/s40168-019-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkinen J, Siponen S, Jayaprakash B, Tiwari A, Hokajärvi AM, Pursiainen A, Ikonen J, Kauppinen A, Miettinen IT, Paananen J, Torvinen E, Kolehmainen M, Pitkänen T, 2021. Diverse and active archaea communities occur in non-disinfected drinking water systems-Less activity revealed in disinfected and hot water systems. Water Res. X 12, 100101. 10.1016/j.wroa.2021.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Rhoads WJ, Edwards MA, Pruden A, 2017. Impact of water heater temperature setting and water use frequency on the building plumbing microbiome. ISME J. 11 (6), 1318–1330. 10.1038/ismej.2017.14, 2017 JunEpub 2017 Mar 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO, 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids. Res 41 (1), e1. 10.1093/nar/gks808, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz N, Raskin L, Zimbric M, Errickson J, LiPuma JJ, Caverly LJ, 2019. Retrospective analysis of nontuberculous mycobacterial infection and monochloramine disinfection of municipal drinking water in Michigan. mSphere 4 (4). 10.1128/mSphere.00160-19, 2019 Jul 3e00160–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie E, Hinds J, Hai FI, 2021. Causes, factors, and control measures of opportunistic premise plumbing pathogens—a critical review. Appl. Sci 11 (10), 4474. 10.3390/app11104474. [DOI] [Google Scholar]
- Li R, Tun HM, Jahan M, Zhang Z, Kumar A, Dilantha Fernando WG, Farenhorst A, Khafipour E, 2017. Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep 7, 5752. 10.1038/s41598-017-02516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, Salzberg SL, 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. 10.1093/bioinformatics/btr507, 2011 Nov 1Epub 2011 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10–12, 2011. [Google Scholar]
- Neu L, Hammes F, 2020. Feeding the building plumbing microbiome: the importance of synthetic polymeric materials for biofilm formation and management. Water 12 (6), 1774. 10.3390/w12061774. [DOI] [Google Scholar]
- Pitkänen T, Ryu H, Elk M, Hokajärvi AM, Siponen S, Vepsäläinen A, Räsänen P, Santo Domingo JW, 2013. Detection of fecal bacteria and source tracking identifiers in environmental waters using rRNA-based RT-qPCR and rDNA-based qPCR assays. Environ. Sci. Technol 47 (23), 13611–13620. 10.1021/es403489b. [DOI] [PubMed] [Google Scholar]
- Potgieter S, Pinto A, Sigudu M, du Preez H, Ncube E, Venter S, 2018. Long-term spatial and temporal microbial community dynamics in a large-scale drinking water distribution system with multiple disinfectant regimes. Water Res. 139, 406–419. 10.1016/j.watres.2018.03.077, 2018 Aug 1Epub 2018 Mar 30. [DOI] [PubMed] [Google Scholar]
- Proctor CR, Dai D, Edwards MA, Pruden A, 2017. Interactive effects of temperature, organic carbon, and pipe material on microbiota composition and Legionella pneumophila in hot water plumbing systems. Microbiome. 5 (1), 130. 10.1186/s40168-017-0348-5, 2017 Oct 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenqvist T, Danielsson M, Schleich C, et al. , 2023. Succession of bacterial biofilm communities following removal of chloramine from a full-scale drinking water distribution system. Npj Clean Water 6, 41. 10.1038/s41545-023-00253-x, 2023. [DOI] [Google Scholar]
- Smith C, Hill VR, 2009. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl. Environ. Microbiol 75 (16), 5284–5289. 10.1128/AEM.00456-09, 2009 Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STM, 2015. Decree 1352/2015. Decree of the Ministry of Social Affairs and Health Relating to the Quality and Monitoring of Water Intended For Human Consumption. Ministry of Social Affairs and Health, Finland. http://www.finlex.fi/fi/laki/alkup/2015/20151352. [Google Scholar]
- Schwake DO, Garner E, Strom OR, Pruden A, Edwards MA, 2016. Environ. Sci. Technol. Lett 3 (9), 311–315. 10.1021/acs.estlett.6b00192, 2016. [DOI] [Google Scholar]
- Tiwari A, Gomez-Alvarez V, Siponen S, Sarekoski A, Hokajärvi AM, Kauppinen A, Torvinen E, Miettinen IT, Pitkänen T, 2022. Bacterial genes encoding resistance against antibiotics and metals in well-maintained drinking water distribution systems in Finland. Front. Microbiol 12, 803094 10.3389/fmicb.2021.803094, 2022 Feb 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torvinen E, Suomalainen S, Lehtola MJ, Miettinen IT, Zacheus O, Paulin L, Katila M-L, Martikainen PJ, 2004. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl. Environ. Microbiol 70 (4), 1973–1981. 10.1128/AEM.70.4.1973-1981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Hammes F, Egli T, 2012. Competition of Escherichia coli O157 with a drinking water bacterial community at low nutrient concentrations. Water Res. 46, 6279–6290. [DOI] [PubMed] [Google Scholar]
- Waak MB, Hozalski RM, Hallé C, LaPara TM, 2019. Comparison of the microbiomes of two drinking water distribution systems—with and without residual chloramine disinfection. Microbiome 7, 87, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pryor MA, Edwards MA, Falkinham JO III, Pruden A, 2013. Effect of GAC pre-treatment and disinfectant on microbial community structure and opportunistic pathogen occurrence. Water Res. 47, 5760–5772. [DOI] [PubMed] [Google Scholar]
- WHO, 2022. Guidelines for drinking-water quality: Fourth edition incorporating the first and second addenda [Internet]. World Health Organization, Geneva. [PubMed] [Google Scholar]
- Zhang C, Qin K, Struewing I, Buse H, Santo Domingo J, Lytle D, Lu J, 2021. The bacterial community diversity of bathroom hot tap water was significantly lower than that of cold tap and shower water. Front. Microbiol 12, 625324 10.3389/fmicb.2021.625324, 2021 Apr 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacteria sequence data generated in this study is available in the Short Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject PRJNA509718.