Abstract
Background:
In cystic fibrosis (CF), pathophysiologic changes in the gastrointestinal tract lead to malnutrition and altered gut microbiome. Microbiome alterations have been linked to linear growth, gut inflammation and respiratory manifestations. Elucidating these gut microbiome alterations may provide insight into future nutritional management in CF.
Methods:
Infants were followed for 12-months at four sites in the United States (US-CF) and Australia (AUS-CF). 16s rRNA gene sequencing was performed on longitudinal stool samples. Associations between microbial abundance and age, antibiotic prophylaxis, malnutrition, and breast feeding were evaluated using generalized linear mixed models. Taxonomic and predictive functional features were compared between groups.
Results:
Infants with CF (N=78) were enrolled as part of a larger study. AUS-CF infants had higher mean weight-for-age z-scores than US-CF infants (p=0.02). A subset of participants (CF N=40, non-CF disease controls N=10) provided stool samples for microbiome analysis. AUS-CF infants had lower stool alpha diversity compared to US-CF infants (p<0.001). AUS-CF infants had higher relative abundance of stool Proteobacteria compared to US-CF infants which was associated with antibiotic prophylaxis (p<0.001). Malnutrition (weight-for-age <10th percentile) was associated with depleted Lactococcus (p<0.001). Antibiotic prophylaxis (p=0.002) and malnutrition (p=0.012) were linked with predicted decreased activity of metabolic pathways responsible for short chain fatty acid processing.
Conclusions:
In infants with CF, gut microbiome composition and diversity differed between the two continents. Gut microbial diversity was not linked to growth. The relationship between malnutrition and antibiotic prophylaxis with reduced SCFA fermentation could have implications for gut health and function and warrants additional investigation.
Keywords: Infant growth, Cystic fibrosis, Gut microbiome
1. INTRODUCTION
Gut microbial colonization begins at birth, evolves over time, and is influenced by several external factors, including mode of delivery, feeding regimen, geographic location, and medication exposures (1). Normal early development of the gut microbiome is essential in the maturation of the host immune system (2). Alterations in the gut microbiome of children are recognized in diseases including malnutrition, obesity, atopy, inflammatory bowel disease, and cystic fibrosis (CF) (3). In children with CF, growth is an important marker of nutrition and has strong associations with later lung function and survival (4).
In CF, alterations in the gut environment occur due to cystic fibrosis transmembrane conductance regulator (CFTR) dysfunction leading to alterations in the gut mucus, intestinal pH, and nutrient absorption (5). These alterations create an environment that contributes to bacterial dysbiosis with downstream effects in the respiratory tract (6). Infants with CF have been shown to have a distinct pattern of gut microbiome development, dominated by Streptococcus and Veillonella, Bifidobacterium, and Bacteroides (7). In infants with CF, changes in the gut microbiome have been shown to precede Pseudomonas aeruginosa colonization in the airway and those who were breast fed had a trend towards prolonged time to first pulmonary exacerbation (7). This highlights a potential link between the gut and airway microbiome. Studies have reported increased prevalence of Proteobacteria, mostly Escherichia coli (8), with reduced Bifidobacterium and other commensals (6, 9) in the gut of infants with CF. These changes appear to be associated with increased gut inflammatory markers (10). These alterations in the gastrointestinal tract may have long-lasting effects on health; the gut microbiome in infants with CF has been linked to linear growth (11).
This study aims to elucidate these previously reported gut microbiome findings. We describe and compare the gut microbiome in a large, prospective, observational study of infants with CF born in the United States (US) and Australia (AUS). We also explore the relationships between microbiome composition, growth, and infant dietary intake. This unique study design allows for insight into potential geographic differences in the gut microbiome in infants with CF.
2. METHODS
2.1. Study population
Four sites were part of this study: Indiana University and Riley Hospital for Children (Indianapolis, USA), Washington University and St. Louis Children’s Hospital (St. Louis, USA), Royal Children’s Hospital (Melbourne, Australia) and Princess Margaret Hospital for Children (Perth, Australia). Infants with CF identified by newborn screening and diagnosed based on sweat chloride concentrations >60 mmol/L or two pathogenic CFTR variants were enrolled before 4-months of age. Results from this cohort have been previously published (12–14). The study presented here was an ancillary study with additional collection of stools samples from a sub-set of the participants to assess the gut microbiome. Non-CF disease controls were children under 24-months of age recruited at Indiana University who underwent bronchoscopy for clinical indications (Supplemental Table 1). As the non-CF control dataset was small, additional analysis was performed using a publicly available dataset of healthy infants (15) to ensure representation of a more general population. Study approval was obtained from the regulatory board at each institution, and written informed consent was obtained from parents.
Infants with CF were followed for a year and underwent infant pulmonary function testing, bronchoscopy, computed x-ray tomography of the chest (chest CT). Control infants participated in one visit. CF research visits were scheduled every one-to-three months until approximately 12 months of age (participants were allowed a window to complete all study procedures). Additional ancillary study data were obtained and included the following: growth (weight, length, and weight-for-length z-scores), feeding mode (breast milk and/or formula), time of transition to solid foods, medications, and environmental smoke exposure. Data were entered and managed using REDCap (16), hosted at the Indiana Clinical and Translational Sciences Institute.
2.2. Sample collection and processing
Fecal sample collection was attempted at each study visit. Parents collected stool from a diaper, which was immediately placed in a sterile specimen cup, kept in their home freezer, and transported chilled to the next clinic visit, where it was stored at −80°C. For non-CF disease controls, fecal material was collected from the child’s diaper, placed in a sterile specimen cup and frozen at −80°C during the bronchoscopy visit.
Frozen fecal samples were shipped to Washington University for DNA extraction using QIAGEN PowerLyzer® PowerSoil kit® (catalog 12855) as per manufacturer guidelines, then sent to the University of Illinois-Chicago for 16S rRNA gene sequencing (V4 region) using the MiSeq Illumina sequencing platform with the V2–500 kit (Illumina Inc) (17).
2.3. Statistical analysis
Statistical analysis was performed using R version 4.1.0 unless specified (18). Benjamini-Hochberg was applied for multiple comparisons and post-corrected p-values <0.05 were considered significant. Participant demographics and exposures were compared using the Fisher’s exact test. Growth parameters, including weight and length, were transformed to weight, length, and weight-for-length z-scores according to the WHO Growth Charts (ages 0 to <2 years) (19). Linear mixed models (LMM) were used to determine significant differences in these metrics over time.
Sequencing data were processed through DADA2 (20) for taxonomic profiling and Picrust2 (21) for metabolic pathways prediction against the MetaCyc database (22). R package details are available in the online supplement. Alpha diversity (Shannon Index) was compared across the three groups (US-CF, AUS-CF, and non-CF disease controls) using the Kruskal-Wallis test and comparison over time (US-CF vs AUS-CF) using linear mixed modeling (LMM). To compare beta diversity, Principal Coordinates Analysis (PCoA) was performed based on the Bray-Curtis dissimilarity, followed by the permutational multivariate analysis of variance (PERMANOVA) to determine significance. For phylum analysis, relative abundance (RA) of each major phylum (defined as mean RA > 1%) was compared using the Wilcoxon test (US-CF vs non-CF, AUS-CF vs non-CF) and LMM (US-CF vs AUS-CF over time). At the genus level, RA of each major genus (defined as mean RA > 1%) was compared using the zero-inflated Wilcoxon test (US-CF vs non-CF, AUS-CF vs non-CF) and generalized linear mixed model (GLMM) (US-CF vs AUS-CF over time). Generalized linear mixed models were used to explore associations between genus abundances and predicted pathways, age, antibiotic prophylaxis, malnutrition, and any breast milk exposure in the infants with CF. Antibiotic prophylaxis was defined as daily antibiotic use over time (initiated prior to stool sample collection), and malnutrition was defined as weight-for-age <10th percentile at any time point in the study (23).
3. RESULTS
3.1. Sample Characteristics
Seventy-eight infants with CF (22 US and 56 AUS) and 20 non-CF disease controls (all US) were recruited for this study. Of these, a subset provided fecal samples for microbiome analyses (17 US-CF, mean 7.88 samples; 23 AUS-CF, mean 5.26 samples and 10 non-CF disease controls, mean 1 sample). Demographic features, medication use, and dietary intake are summarized in Table 1. A notable difference is that 98% of AUS-CF infants were routinely prescribed antibiotic prophylaxis treatment (amoxicillin-clavulanate), while none of the US-CF infants received prophylaxis (p <0.001). Less than half of all the infants were exclusively breastfed, but over half were fed breast milk during the study period. All US-CF infants received infant formula, whereas only 68% of AUS-CF infants were fed formula (p <0.001).
TABLE 1:
Demographics
| Whole cohort | Sub-cohort with stool sequenced | |||||
|---|---|---|---|---|---|---|
| Non-CF Disease Control (N=20) | US-CF (N=22) | AUS-CF (N=56) | Non-CF Disease Control (N=10) | US-CF (N=17) | AUS-CF (N=23) | |
| Gender (Male) | 14 (70.0%) | 9 (40.9%) | 28 (50.0%) | 7 (70.0%) | 7 (41.2%) | 10 (43.5%) |
| Race (White) | 19 (95.0%) | 22 (100%) | 54 (96.4%) | 10 (100%) | 17 (100%) | 23 (100%) |
| Genotype | ||||||
| Homozygous Phe508del | 0 (0%) | 13 (59.1%) | 30 (53.6%) | 0 (0%) | 11 (64.7%) | 15 (65.2%) |
| Heterozygous Phe508del | 0 (0%) | 8 (36.4%) | 24 (42.9%) | 0 (0%) | 5 (29.4%) | 7 (30.4%) |
| Other | 0 (0%) | 1 (4.5%) | 2 (3.5%) | 0 (0%) | 1 (5.9%) | 1 (4.4%) |
| Anthropometrics at enrollment | ||||||
| Weight-for-age z-score | 0.11±1.01 | −1.09±1.13 | −0.79±1.30 | −0.10±1.18 | −1.33±1.15 | −0.50±0.86 |
| Length-for-age z-score | −0.47±1.17 | −0.66±1.30 | −0.70±1.72 | −0.53±1.22 | −0.81±1.42 | −0.27±1.08 |
| Weight-for-length z-score | 0.45±1.26 | −0.70±0.83 | 0.55±1.72 | 0.10±1.42 | −0.83±0.85 | −0.34±1.09 |
| Pancreatic Status | ||||||
| Pancreatic Insufficiency | - | 18 (81.8%) | 47 (83.9%) | - | 14 (82.4%) | 21 (91.3%) |
| Medication | ||||||
| Antibiotic prophylaxis | - | 0 (0%) | 55 (98.2%) | - | 0 (0%) | 22 (95.7%) |
| Pancreatic enzyme use | - | 19 (86.4%) | 48 (85.9%) | - | 15 (88.2%) | 21 (91.3%) |
| H2 blocker/Proton pump inhibitor | - | 14 (66.7%) | 22 (39.2%) | - | 10 (58.8%) | 11 (47.8%) |
| Diet | ||||||
| Breast milk exclusive | - | 10 (45.5%) | 21 (37.5%) | - | 8 (47.1%) | 9 (39.1%) |
| Ever had breast milk | - | 12 (54.5%) | 35 (63.5%) | - | 9 (52.9%) | 14 (60.9%) |
| Ever had infant formula | - | 22 (100%) | 43 (67.8%) | - | 17 (100%) | 20 (87.0%) |
| Site (n) | ||||||
| Indianapolis | 20 (100%) | 15 (68.2%) | 0 (0%) | 10 (100%) | 13 (76.5%) | 0 (0%) |
| St. Louis | 0 (0%) | 7 (31.8%) | 0 (0%) | 0 (0%) | 4 (23.5%) | 0 (0%) |
| Melbourne | 0 (0%) | 0 (0%) | 33 (58.9%) | 0 (0%) | 0 (0%) | 16 (69.6%) |
| Perth | 0 (0%) | 0 (0%) | 23 (41.1%) | 0 (0%) | 0 (0%) | 7 (30.4%) |
N (%) or mean +/−
Growth parameters for the whole cohort (n=78) of US-CF infants (n=22) compared to AUS-CF infants (n=56) are shown in Figure 1A. AUS-CF infants had a significantly greater increase in weight-for-age z-score over time (p=0.02). The two groups were not significantly different in length-for-age and weight-for-length z-scores over time. In the sub-set of infants who provided stool samples (US-CF n=17, AUS-CF n=23), there was a significant difference in all growth parameters between the two groups (Figure 1B).
Figure 1. Growth measures between US-CF and AUS-CF infants across time.
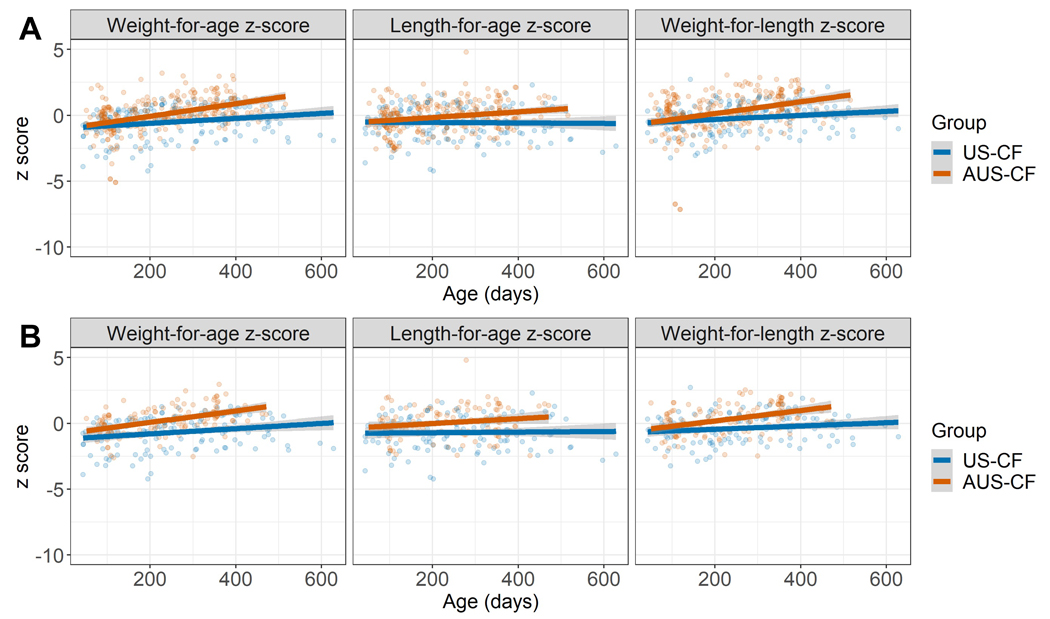
Scatter plots with fitted linear regression lines of US-CF and AUS-CF infants. (A) Whole cohort. For weight-for-age z-score, AUS-CF infants (n=56) had more weight gain than US-CF infants (n=22) over time, p=0.024. There was not a significant difference in length-for-age z-score, p=0.069 or weight-for-length z-score, p=0.107. (B) Sub-cohort with stool sequenced. Weight-for-age z-score (waz), length-for-age z-score (laz) and weight-for-length z-score (wlz) differs between US-CF infants (n=17) and AUS-CF infants (n=23). (waz p=0.00382; laz p=0.027087; wlz p=0.0167)
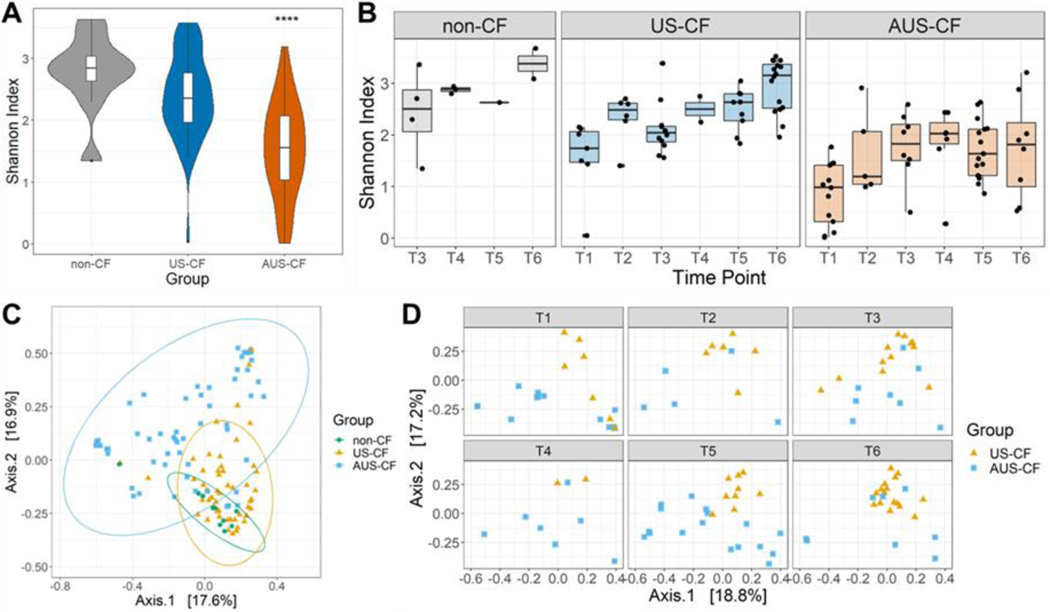
3.2. Gut Microbial Diversity
We evaluated the overall gut microbiome diversity and changes over time in infants with CF. Alpha diversity was represented using Shannon diversity. Compared to non-CF disease controls, US-CF infants had similar Shannon diversity to controls, whereas AUS-CF infants had lower Shannon diversity (p<0.001) (Figure 2A), a difference that persisted over time (Figure 2B). There were marked differences in beta diversity (p=0.0001) between all three groups (Figure 2C), which again continued over time in the US-CF group as compared to the AUS-CF infants (p=0.008) (Figure 2D). Data from the publicly available healthy controls is displayed in Figure S1. Fifty infants with samples as a newborn, at 4 months and 12 months of life were analyzed. These infants had increasing Shannon diversity over the first year of life (Figure S1A), with similar relative abundance of the major phylum and genus as the non-CF controls (Figure S1B–C). In our cohort of CF infants, Shannon diversity did not correlate with measures of growth including weight-for-age z-score, length-for-age z-score or weight-for-length z-score (Figure S2)
Figure 2. Microbiome Diversity.
(A) Overall comparison of Shannon diversity among non-CF, US-CF and AUS-CF (comparison across the three groups, p < 3.0e-9; non-CF vs US-CF: p = 0.439; non-CF vs AUS-CF: p = 2.66e-05). (B) Boxplot for Shannon diversity of non-CF, US-CF, and AUS-CF at different age time points (T1: 0–6 months; T2: 6–9 months; T3: 9–12 months; T4: 12–15 months; T5: 15–18 months; T6: 18–24 months). US-CF vs AUS-CF, p = 1.60e-5. (C) PCoA plot of beta diversity with overall comparison across the three groups, p = 1e-04. (D) Cross-sectional beta diversity, US-CF vs AUS-CF: p = 0.008
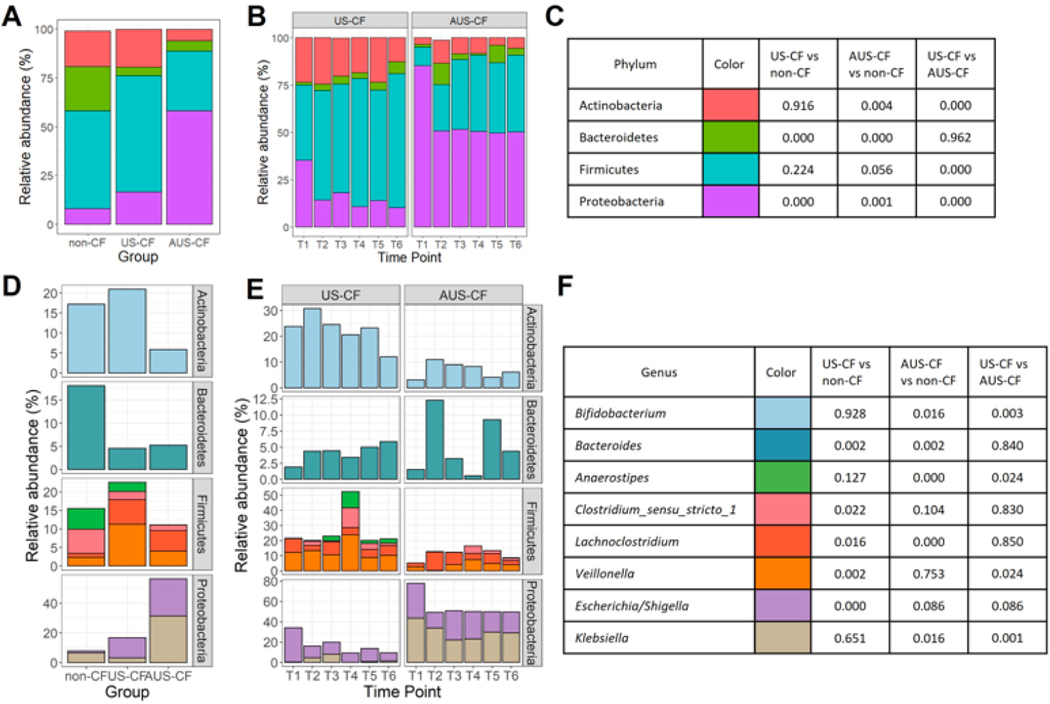
3.3. Gut Microbiome at the Phylum and Genus Level
Four major phyla were found in the microbiome: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria in each of the infant groups. Compared to non-CF disease controls, both US-CF and AUS-CF had a higher proportion of Proteobacteria and lower Bacteroidetes (Figure 3A). Proteobacteria abundance was different between the two CF infant groups, with AUS-CF infants having higher proportions of Proteobacteria than US-CF infants (Figure 3B, 3C). Also, AUS-CF had lower proportions of Actinobacteria and Firmicutes than US-CF infants, differences that persisted over time (Figure 3B, 3C).
Figure 3.
Stacked bar plot of mean relative abundance of major phylum by (A) groups and (B) time points (T1: 0–6 months; T2: 6–9 months; T3: 9–12 months; T4: 12–15 months; T5: 15–18 months; T6: 18–24 months). (C) Color legend and p-values for (A) and (B). (D) Stacked bar plot of mean relative abundance of significantly changed major genus across groups and (E) over time. (F) Color legends and p-values for (D) and (E).
The major genera across the three groups were compared (Figure 3D, 3E, 3F) with each genus represented by a color and significant values are displayed for between group comparisons (Figure 3F). For Proteobacteria, the most abundant genera that accounted for differences between the three groups were Escherichia/Shigella (lower abundance in non-CF disease controls vs US-CF infants) and Klebsiella (higher abundance in AUS-CF infants vs US-CF and non-CF disease control infants). For the phylum Bacteroidetes, it was Bacteroides that had the highest abundance in non-CF disease controls vs both US-CF and AUS-CF infants, while differences in Actinobacteria was primarily driven by Bifidobacterium (lower abundance in AUS-CF infants vs US-CF or non-CF disease control infants). For Firmicutes, the main genera were Lachnoclostridium (higher abundance in both groups of CF infants vs non-CF disease controls) and Veillonella (highest abundance in US-CF infants).
Using Picrust2, the functional nature of the bacteria present in the gut of the infants was assessed, and predicted MetaCyc pathways were analyzed for beta diversity (Figure S3). There were marked differences (p=0.0001) of microbial community functional elements when comparing non-CF disease controls, AUS-CF, and US-CF infants when analyzed using the PCoA plot (Figure S3A). Between the CF cohorts, US-CF infants differed from AUS-CF infants across time points (p<0.001) with axis-1 accounting for 60.5% of the variation present (Figure S3B). The top 20 functional pathways that contributed to the axis-1 variation for AUS-CF and US-CF infants are shown in Figure S3C. The two most prevalent in AUS-CF infants were pathways related to fatty acid elongation and oxidation (FAO-PWY and FASYN-ELONG-PWY).
3.4. Association of Gut Microbiome with Clinical Factors
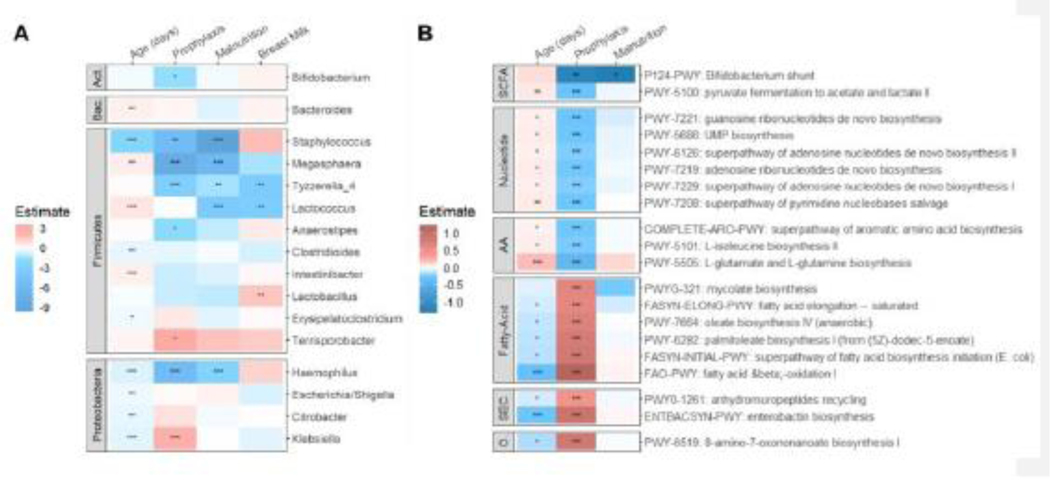
We examined the associations between clinical factors and environmental exposures with gut microbial composition, specifically evaluating the link between microbial feature abundance (genus and predicted metabolic pathways) with age, antibiotic prophylaxis, malnutrition, and breast milk exposure in the infants with CF (Figure 4). As CF clinical care strongly focuses on growth and attempts to limit malnutrition, we used malnutrition as the clinical variable for growth in our analyses (defined as weight-for-age <10th %tile). Breast milk exposure was defined as the infant ever having breast milk. Six US-CF and two AUS-CF infants fulfilled the criteria for malnutrition. Figure 4A shows the estimated coefficient of the association between genus abundance and each of these clinical factors. Age in days was associated with enrichment of Firmicutes and depletion in Proteobacteria. Klebsiella was abundant in infants treated with antibiotic prophylaxis, whereas other genera were depleted, including Staphylococcus, Megasphaera, Tyzerella and Haemophilus. Megasphaera, Tyzerella, Haemophilus, Staphylococcus, and Lactococcus were also reduced in infants with CF with malnutrition, while Lactobacillus was enriched in infants who had ever had breast milk.
Figure 4.
GLMM analysis of microbial feature abundance (A-genus, B-pathway) with variables age, antibiotic prophylaxis, malnutrition, and breast milk exposure. The estimated coefficients with significance are plotted as a heatmap. (Act: Actinobacteria; Bac: Bacteroidetes; SCFA: short chain fatty acid; AA: amino acid; SEC: secondary metabolites; O: other biosynthesis. Significance levels: ****: p <0.0001; ***: p <0.001, **: p < 0.01; *: p < 0.05).
In Figure 4B, the top 20 predicted metabolic pathways that differed between US-CF and AUS-CF infants are displayed, which are clearly separated by antibiotic prophylaxis and age. In reciprocal fashion, pathways that were depleted with antibiotic prophylaxis were enriched with age and vice versa. Malnutrition was associated with depleted Bifidobacterium shunt, a pathway of fermentation to short-chain fatty acids acetate and lactate. Antibiotic prophylaxis was associated with reduced biosynthesis of nucleosides and nucleotides, as well as certain amino acids, but these pathways were more enriched with increased age. Fatty acid biosynthesis pathways were predicted to be increased in infants receiving antibiotic prophylaxis.
4. DISCUSSION
This prospective observational study compared the composition of the gut microbiome in infants with CF from two countries longitudinally and examined its relationship to growth, malnutrition, diet, and antibiotic prophylaxis. Australian and US infants with CF were similar in height and weight at enrollment, but the former cohort gained more weight over time. The majority (98%) of Australian infants were treated with daily antibiotic prophylaxis, which had marked effects on the gut microbiome. Australian infants had less diverse gut microbiome at baseline and over time when compared to US infants. The gut microbiome was dominated by Proteobacteria in Australian infants with CF, with decreased concentrations of Actinobacteria and Firmicutes compared with US infants. Escherichia, Shigella, and Klebsiella accounted for the majority of the Proteobacteria in these infants. Though no significant correlation was seen between Shannon diversity and growth in the infants with CF, in those with malnutrition, the gut microbiome was predicted to be depleted of similar bacteria to those on antibiotic prophylaxis.
Differences in the gut microbiome between infants with CF living in Australia versus the US may have been related in part to geographic differences (24). US infants with CF had Shannon diversity that was like their non-CF disease control counterparts, which were all from the US. In comparison to age, antibiotic prophylaxis, and malnutrition, the type of diet seemed to have less of an influence on species abundance. There was a predominance of Proteobacteria in Australian infants with a concomitant decrease in Bifidobacterium. Escherichia, Shigella, and Klebsiella accounted for most of the Proteobacteria in these Australian infants. Compared to non-CF disease controls, both CF infant groups had higher abundance of Proteobacteria. The core bacteria in our cohort of infants with CF was similar to prior studies that reported an abundance of Bacteriodes, Veillonella, and Bifidobacterium (25). Though prior studies showed increasing Escherichia in infants with CF over time, this genus was relatively stable in our cohort (25). Fecal dysbiosis with decreased abundance of Bacteroidetes and increased abundance of Proteobacteria has been linked to poor linear growth in infants with CF (11). In our cohort, AUS-CF infants, despite higher proportions of Proteobacteria, had better weight gain over time than their US-CF counterparts. Also, we did not see a link between Shannon diversity and linear growth (or weight gain) in our cohort. However, the effects of gut dysbiosis are likely not just important locally in the gut, but potentially have more far-reaching effects on other organ systems including the respiratory tract (6, 7). With a greater understanding of these relationships and the directionality, the gut microbiome could be a target for future therapeutic interventions.
Nutrition in early life has long been a focus in CF clinical care as it has significant links to future lung function (4). Australian infants had better growth over time than US infants, consistent with past studies that used CF patient registry data to compare children in the US and Australia (26). In that study, they noted that these differences remained even after controlling for age, gender, genotype and diagnosis after newborn screen (26). In our cohort, we found growth differences started early in life as weight-for-age measures diverged in the first six months and over time; the mechanism for this difference is not known. One notable difference in treatment was the use of prophylactic antibiotics in Australia, which were started early and may account for some of this divergence. Systemic antibiotics have been shown to promote growth. A meta-analysis examining the effects of antibiotics in non-CF children reported positive effects on growth in children in low- and middle-income countries (27), thought to be related to treatment of subclinical infection or modulation of the microbiome (27). In the current study, feeding regimens did not overtly differ between the two continents, i.e., breast milk vs infant formula, though we were not able to account for the mother’s diet while breastfeeding, which could introduce additional variability.
Using Picrust2, we examined predicted associations between the gut microbiome composition and malnutrition in these infants with CF. Malnutrition was associated with depleted Bifidobacterium shunt, a pathway of fermentation to short-chain fatty acids (acetate and lactate). Short-chain fatty acids are produced in the colon by bacteria fermentation of non-digestible foods like fiber and resistant starches (28) and stimulate T-cell differentiation (29), specifically regulatory T-cells. The depletion of this pathway in our malnourished infants with CF is consistent with previous studies, where acetate levels were found to be lower in malnourished children (30) and correlated to BMI z-score (31). In our study, fatty acid pathways were predicted to be enriched in those that received antibiotic prophylaxis. Several of the fatty acid pathways, PWY-6282, FASYN-INITIAL-PWY, FASYN-ELONG-PWY, and FAO-PWY, are known to be used by Escherichia, a genus that accounted for a large portion of the increased Proteobacteria seen in Australian infants with CF. Also, antibiotic prophylaxis was predicted to be associated with reduced biosynthesis of nucleoside and nucleotides, as well as certain amino acids. These findings may represent slower microbiome growth (slower DNA replication and protein synthesis) and thus may explain the decreased diversity seen in Australian infants initially and over the course of the study. These pathways depleted with antibiotic prophylaxis became less depleted over time. This may reflect lower adherence to the antibiotic regimen over time or represent an ability to restore the microbiome despite outside influences. It is difficult to determine if this was simply delayed maturation of the gut microbiome due to antibiotic prophylaxis (AUS-CF infants showing slower increase in diversity over time) or unrelated.
This study has a few limitations. The sample size was low with relatively few infants participating and a larger number of those being from Australia. Whether Proteobacteria enrichment was due to antibiotic prophylaxis or continental differences was difficult to determine, since all but one of the infants in the Australian group received antibiotic prophylaxis during the study. However, historic samples from the Royal Children’s Hospital, collected before initiation of antibiotic prophylaxis, had markedly greater diversity, suggesting antibiotic prophylaxis, rather than geography or local diet, explains differences between AUS-CF and US-CF infants (32). During the time of this trial, some of the Australian infants were participating in another blinded clinical trial examining the use of enteral Azithromycin, which would have further contributed to their antibiotic exposure. Fecal sample collection was added after the main study began enrollment, and not all infants had fecal samples collected at every visit. Sample collection occurred every one-to-three months and was coordinated with clinic visits, but the first sample was obtained after the initiation of antibiotic prophylaxis in Australian infants. These factors introduce potential variability into the analyses. Malnutrition was seen in only a small number of infants. Non-CF disease control infants were only recruited from one US site and were a convenience sample; these infants were undergoing bronchoscopy for clinical indications. The non-CF disease control infants were not age matched and had only one sample collected. However, the non-CF infants were grouped into appropriate age time periods for microbiome comparisons. Furthermore, since these were infants undergoing bronchoscopy for clinical indications, they may not represent the general non-CF infant population. To account for this limitation, additional analysis using a publicly available dataset of healthy infants was analyzed with results similar to the non-CF controls. Lastly, our metabolic analysis was based on Picrust2 prediction. These are associations and causality cannot be determined. In the future, more direct studies of bacterial metabolism may be helpful, like metabolomics or measuring production of substrates like short chain fatty acids.
In conclusion, in infants with CF, gut microbiome composition and diversity were affected by age, antibiotic prophylaxis and differed between the two continents. Gut microbial diversity was not linked to growth. However, functional examination of metabolic pathways identified associations between the gut microbiome and age, antibiotic use, as well as malnutrition in infants with CF. Clinical practices appear to contribute significantly to the differences noted, and therefore the infant gut microbiome should be an important consideration for future management of CF.
Supplementary Material
Highlights.
AUS-CF infants had higher weight for age z-scores than US-CF infants over time
AUS-CF infants had more Proteobacteria, related to continent or antibiotic prophylaxis
Malnutrition and daily antibiotics were linked to changes in the microbiome with predicted functional changes
Acknowledgments
We would like to acknowledge the participants and families that dedicated their time and efforts to this research along with the research coordinators who made the study possible.
Financial support:
The authors were supported by the National Institutes of Health (NIH) grant HL116211 and National Health and Medical Research Council award, NHMRC Ranganathan 1043768, CFF DESCHA15D0, DESCHA16D0, DESCHA18K0. The views expressed do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention by trade names, commercial practices, or organizations imply endorsement by the US government.
Footnotes
Credit authorship contribution statement
There are no previous publications or submissions with any overlapping information presented in this report. Moreover, this work is not and will not be submitted to any other journal while under consideration. Neither author has an actual or perceived conflict of interest concerning the information presented in the paper. Ashley Deschamp and Yang Chen composed the first draft, and did not receive an honorarium or grant to write the manuscript. All authors listed on the manuscript have reviewed and approved the content of the submission, and take full responsibility for the information provided.
Deschamp AR: Conceptualization, Methodology, Investigation, Data curation, Writing-original draft, Visualization, Writing-review and editing. Chen Y: Formal analysis, Visualization, Writing-original draft, Writing-review and editing. Wang WF: Investigation, Writing-review and editing. Rasic M: Investigation, Writing-review and editing. Hatch J: Data curation, Writing-review and editing. Sanders DB: Investigation, Resources, Writing-review and editing. Ranganathan SC: Conceptualization, Methodology, Investigation, Resources, Funding Acquisition, Writing-review and editing. Ferkol T: Conceptualization, Methodology, Investigation, Resources, Funding Acquisition, Writing-review and editing. Perkins D: Resources, Investigation, Writing-review and editing. Finn P: Resources, Investigation, Writing-review and editing. Davis SD: Conceptualization, Methodology, Investigation, Resources, Funding Acquisition, Writing-review and editing.
Declaration of Competing Interest
S Davis served on the Vertex advisory board
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Min YW, Rhee PL. The Role of Microbiota on the Gut Immunology. Clin Ther. 2015;37(5):968–75. [DOI] [PubMed] [Google Scholar]
- 3.Albenberg L, Kelsen J. Advances in Gut Microbiome Research and Relevance to Pediatric Diseases. J Pediatr. 2016;178:16–23. [DOI] [PubMed] [Google Scholar]
- 4.Yen EH, Quinton H, Borowitz D. Better nutritional status in early childhood is associated with improved clinical outcomes and survival in patients with cystic fibrosis. J Pediatr. 2013;162(3):530–5.e1. [DOI] [PubMed] [Google Scholar]
- 5.De Lisle RC, Borowitz D. The cystic fibrosis intestine. Cold Spring Harb Perspect Med. 2013;3(9):a009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers GB, Narkewicz MR, Hoffman LR. The CF gastrointestinal microbiome: Structure and clinical impact. Pediatr Pulmonol. 2016;51(S44):S35–s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoen AG, Li J, Moulton LA, O’Toole GA, Housman ML, Koestler DC, et al. Associations between Gut Microbial Colonization in Early Life and Respiratory Outcomes in Cystic Fibrosis. J Pediatr. 2015;167(1):138–47.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manor O, Levy R, Pope CE, Hayden HS, Brittnacher MJ, Carr R, et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep. 2016;6:22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duytschaever G, Huys G, Bekaert M, Boulanger L, De Boeck K, Vandamme P. Dysbiosis of bifidobacteria and Clostridium cluster XIVa in the cystic fibrosis fecal microbiota. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2013;12(3):206–15. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman LR, Pope CE, Hayden HS, Heltshe S, Levy R, McNamara S, et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis. 2014;58(3):396–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden HS, Eng A, Pope CE, Brittnacher MJ, Vo AT, Weiss EJ, et al. Fecal dysbiosis in infants with cystic fibrosis is associated with early linear growth failure. Nat Med. 2020;26(2):215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittman JE, Wylie KM, Akers K, Storch GA, Hatch J, Quante J, et al. Association of Antibiotics, Airway Microbiome, and Inflammation in Infants with Cystic Fibrosis. Annals of the American Thoracic Society. 2017;14(10):1548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders DB, Deschamp AR, Hatch JE, Slaven JE, Gebregziabher N, Corput MK, et al. Association between early respiratory viral infections and structural lung disease in infants with cystic fibrosis. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deschamp AR, Hatch JE, Slaven JE, Gebregziabher N, Storch G, Hall GL, et al. Early respiratory viral infections in infants with cystic fibrosis. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roswall J, Olsson LM, Kovatcheva-Datchary P, Nilsson S, Tremaroli V, Simon MC, et al. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell host & microbe. 2021;29(5):765–76.e3. [DOI] [PubMed] [Google Scholar]
- 16.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics. 2009;42(2):377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing. . R Foundation for Statistical Computing, Vienna, Austria. 2021. [Google Scholar]
- 19.WHO. Child growth Standards, Anthro version 3.2.2 2011. [Available from: http://www.who.int/childgrowth/software/en/. [Google Scholar]
- 20.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42(Database issue):D459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konstan MW, Butler SM, Wohl ME, Stoddard M, Matousek R, Wagener JS, et al. Growth and nutritional indexes in early life predict pulmonary function in cystic fibrosis. J Pediatr. 2003;142(6):624–30. [DOI] [PubMed] [Google Scholar]
- 24.Mobeen F, Sharma V, Tulika P. Enterotype Variations of the Healthy Human Gut Microbiome in Different Geographical Regions. Bioinformation. 2018;14(9):560–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio. 2012;3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin B, Schechter MS, Jaffe A, Cooper P, Bell SC, Ranganathan S. Comparison of the US and Australian cystic fibrosis registries: the impact of newborn screening. Pediatrics. 2012;129(2):e348–55. [DOI] [PubMed] [Google Scholar]
- 27.Gough EK, Prendergast AJ, Mutasa KE, Stoltzfus RJ, Manges AR. Assessing the Intestinal Microbiota in the SHINE Trial. Clin Infect Dis. 2015;61 Suppl 7:S738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Annison G, Illman RJ, Topping DL. Acetylated, propionylated or butyrylated starches raise large bowel short-chain fatty acids preferentially when fed to rats. J Nutr. 2003;133(11):3523–8. [DOI] [PubMed] [Google Scholar]
- 29.Furusawa Y, Obata Y, Hase K. Commensal microbiota regulates T cell fate decision in the gut. Semin Immunopathol. 2015;37(1):17–25. [DOI] [PubMed] [Google Scholar]
- 30.Monira S, Hoq MM, Chowdhury AK, Suau A, Magne F, Endtz HP, et al. Short-chain fatty acids and commensal microbiota in the faeces of severely malnourished children with cholera rehydrated with three different carbohydrates. Eur J Clin Nutr. 2010;64(10):1116–24. [DOI] [PubMed] [Google Scholar]
- 31.Riva A, Borgo F, Lassandro C, Verduci E, Morace G, Borghi E, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol. 2017;19(1):95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.