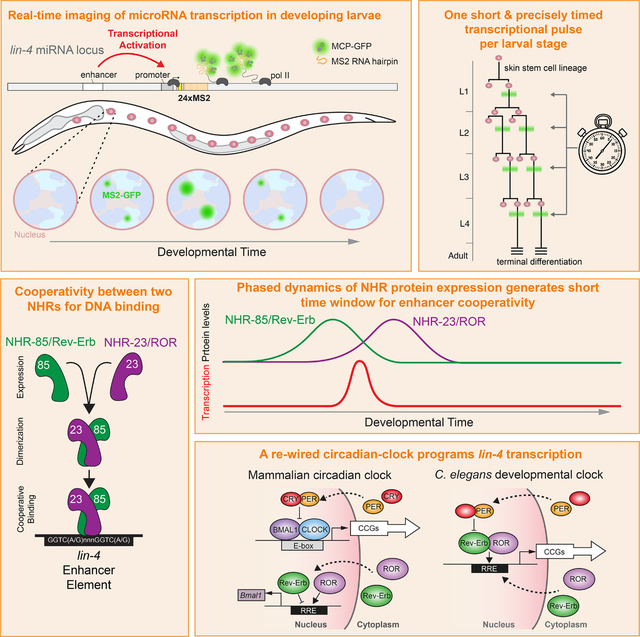
SUMMARY
Development relies on the exquisite control of both the timing and levels of gene expression to achieve robust developmental transitions. How cis- and trans-acting factors control both aspects simultaneously is unclear. We show that transcriptional pulses of the temporal patterning microRNA lin-4 are generated by two nuclear hormone receptors in C. elegans, NHR-85 and NHR-23, whose mammalian orthologs, Rev-Erb and ROR, function in the circadian clock. While Rev-Erb and ROR antagonize each other to control once-daily transcription in mammals, NHR-85/NHR-23 heterodimers bind cooperatively to lin-4 regulatory elements to induce a single pulse of expression during each larval stage. Each pulse’s timing, amplitude, and duration are dictated by the phased expression of these NHRs and the C. elegans Period ortholog, LIN-42, that binds to and represses NHR-85. Therefore, during nematode temporal patterning, an evolutionary rewiring of circadian clock components couples the timing of gene expression to the control of transcriptional dosage.
In brief
Kinney, Sahu, et al. report that genes implicated in mammalian circadian transcription are rewired in C. elegans to generate the oscillatory transcriptional patterns of miRNAs that program temporal patterning during post-embryonic development. This gene regulatory network directly controls lin-4 gene dosage in the heterochronic pathway, maintaining developmental robustness.
Graphical Abstract
INTRODUCTION
How cells within an organism sense and generate information to form and coordinate complex, reproducible patterns is a central question of developmental biology. While multicellular development inherently involves time-dependent changes in gene expression, many developmental systems incorporate gene regulatory networks (GRNs) that either directly encode timekeeping properties or measure time from environmentally- or internally-derived cues1. The animal kingdom contains numerous examples of organisms precisely timing gene expression programs and developmental milestones on vastly different timescales. For instance, human oocytes collectively arrest at the diplotene stage in Meiosis I for over a decade before a timely luteinizing hormone surge triggers meiotic resumption in sexually mature females2. During somitogenesis, reproducible 30min-to-5-hour cycles of gene expression generate metameric structures of epithelial tissues that will become vertebrae3–5. During D. melanogaster embryonic and larval development, neuroblasts sequentially express transcription factors over several days, specifying the temporal identity of their progeny6,7. On the extreme end of this spectrum, periodical cicadas synchronize their final molt into adulthood within hours after spending up to 17 years underground as larvae8,9. In each of the above cases, evolutionary pressure has selected GRNs that generate transcription at the correct relative time and maintain these expression patterns within specific ranges to ensure robust and reproducible developmental decisions. Most research has been devoted to either understanding how the timing of gene induction is established or, separately, how proper gene dosage is ensured. We only have a rudimentary understanding of how these two features may be coupled to organize robust developmental transitions.
Genetic analysis of the C. elegans model has illuminated many conserved principles of temporal patterning in animals10,11. This understanding is facilitated by the fact that post-embryonic maturation of C. elegans larvae is compartmentalized into four stages with distinct patterns of cell division, cell differentiation, and cuticle formation separated by molts12. Stage-specific temporal identity is controlled by the heterochronic gene regulatory network (GRN) composed of conserved transcription factors, RNA-binding proteins, and regulatory RNAs that transcriptionally and post-transcriptionally influence stage-specific gene expression patterns throughout the animal 13. Mutations in genes that enforce temporal identity result in the wholesale reiteration or skipping of stage-specific cell fate and gene expression patterns within the larval molting cycle, indicating that this GRN controls the sequence of developmental events in a modular fashion14. Importantly, transitions from one stage-specific pattern of cell division to the next and, therefore, the sequence of developmental events are mediated by the accumulation of multiple heterochronic microRNAs (miRNAs). These regulatory RNAs serially downregulate the expression of temporal identity genes at the post-transcriptional level10.
miRNA dosage is tightly regulated at the spatial and temporal levels to ensure developmental coordination across the organism. Defects in controlling heterochronic miRNA expression, where ectopic or abnormally higher or lower doses of miRNA transcription occur, can result in the wholesale skipping or reiteration of modular developmental programs15–19. Analysis of heterochronic miRNA expression during normal development indicates that the transcription of heterochronic miRNAs is highly periodic, peaking a single time during the molting cycle18–22. These patterns resemble the phased transcriptional cycles of a significant portion of the C. elegans protein-coding transcriptome20,23,24. While these observations suggest that a clock-like system produces the repetitive dynamics of heterochronic miRNA transcription, they raise the question of how these dynamics are programmed and coordinated with animal development and whether the systems that generate dynamic miRNA transcription share components with GRNs that control overall cyclical mRNA expression. Furthermore, whether the regulatory GRNs that produce these oscillatory expression patterns can modulate specific transcriptional features (e.g., phase, amplitude, or duration) to control gene dosage or if separate systems are in place to govern expression levels is unknown.
A single factor, LIN-42, is known to modulate miRNA transcriptional dosage in C. elegans. Lin-42 encodes the nematode ortholog of the circadian Period protein, is required for normal temporal patterning, and directly modulates the dynamic features of heterochronic miRNA transcription throughout post-embryonic development18,19,21. lin-42 mutations increase the amplitude and duration of oscillatory miRNA transcription, indicating that LIN-42, like its mammalian ortholog, functions as a transcriptional repressor18,19,21. Due to heterochronic miRNA over-expression, lin-42 mutants develop precociously18,25. In contrast to other protein-coding genes in the heterochronic GRN that are expressed in a graded fashion, lin-42 transcription is dynamic with a single peak of expression during each larval stage18,25,26. The similar molecular functions and expression dynamics between LIN-42 and its mammalian Period ortholog suggest that a regulatory architecture akin to the one that generates the once-daily circadian transcriptional patterns in mammals may play a role in temporal cell fate specification in nematodes. Intriguingly, C. elegans lacks orthologs of CLOCK and BMAL1, the central transcription factors that drive oscillatory circadian transcription in humans and mice and are the direct targets of mammalian Period repression. Thus, the mechanism by which LIN-42 modulates the transcription of C. elegans heterochronic miRNAs is currently unknown.
In this manuscript, we used the MS2/MCP-GFP tethering assay to directly image the transcription of the lin-4 heterochronic miRNA that promotes cell fate transitions during early larval stages and is dynamically expressed throughout post-embryonic development. lin-4 transcription is highly pulsatile, with a single approximately 90-minute pulse at each larval stage followed by 10–12 hours of quiescence. We then identify two highly conserved nuclear hormone receptors (NHRs), NHR-23 and NHR-85, that heterodimerize and bind cooperatively to lin-4 regulatory sequences to promote lin-4 transcription. Consistent with an essential role for these NHRs in integrating features of miRNA transcription and gene dosage within the heterochronic GRN, removing the conserved NHR-23 and NHR-85 binding sites within the lin-4 regulatory regions causes animals to display retarded temporal patterning defects. We demonstrate that the precise timing and duration of lin-4 transcriptional pulses are coordinated by the dynamic and partially overlapping expression patterns of these two NHRs within each molting cycle. Finally, we define the molecular mechanism by which LIN-42 modulates miRNA dosage within this GRN. Specifically, we show that LIN-42 binds to NHR-85 and modulates lin-4 transcription by limiting the temporal overlap in NHR-23 and NHR-85 expression patterns. We propose that the physical and regulatory interactions between NHR-23, NHR-85, and LIN-42 define the GRN that generates the cyclical transcription of lin-4 miRNAs and simultaneously modulates lin-4 dosage to ensure normal temporal patterning. Our results indicate that a common regulatory architecture used to control the timing of gene expression, namely a system similar to the circadian GRN, can also be employed to control gene dosage.
RESULTS
Oscillatory lin-4 transcription is pulsatile
Previous measurements of miRNA transcription used destabilized GFP reporters that must be transcribed, processed, and translated to visualize gene expression dynamics18,22. These features limit their temporal resolution and fail to capture direct transcriptional dynamics at the site of transcription. We used the MS2/MCP-GFP tethering system, where engineered RNA loops derived from MS2 bacteriophage and a co-expressed MS2 Coat Protein fused to Green Fluorescent Protein (MCP-GFP) can be concentrated and localized at a gene of interest by active transcription (Figure 1A)27. We measured the transcriptional dynamics of the lin-4 heterochronic miRNA that is expressed periodically throughout all larval stages and down-regulates lin-14 expression within the heterochronic gene regulatory network early in development22,28. We generated a transgene harboring 24 copies of a synthetic MS2 hairpin immediately downstream of the lin-4 pre-miRNA and within the defined transcriptional regions of all previously described lin-4 transcripts (Figure S1A)22,29. Because lin-4 is encoded within an intron of a host gene that is transcribed in the same orientation, we integrated the single-copy lin-4::24xMS2 transgene on chromosome I. This transgene rescues the adult gene expression, cell lineage, and cuticular phenotypes of a lin-4 null allele (Figure S1B and C). We also ubiquitously expressed MCP-GFP to detect MS2-tagged RNAs and a histone::mCherry fusion to locate nuclei (Figures 1A and B). Examination of transgenic animals revealed that transient nuclear MCP-GFP foci formation occurred at each larval stage in somatic tissue types known to transcribe lin-4 (Figure 1B)(Figure S1D–G). MCP-GFP foci were not observed in developing embryos (n > 50) or in starvation-arrested L1 larva (n = 23), consistent with the activation of lin-4 transcription after the initiation of larval development16.
Figure 1. lin-4 transcription is pulsatile at each larval stage and highly synchronous across the hypodermis.
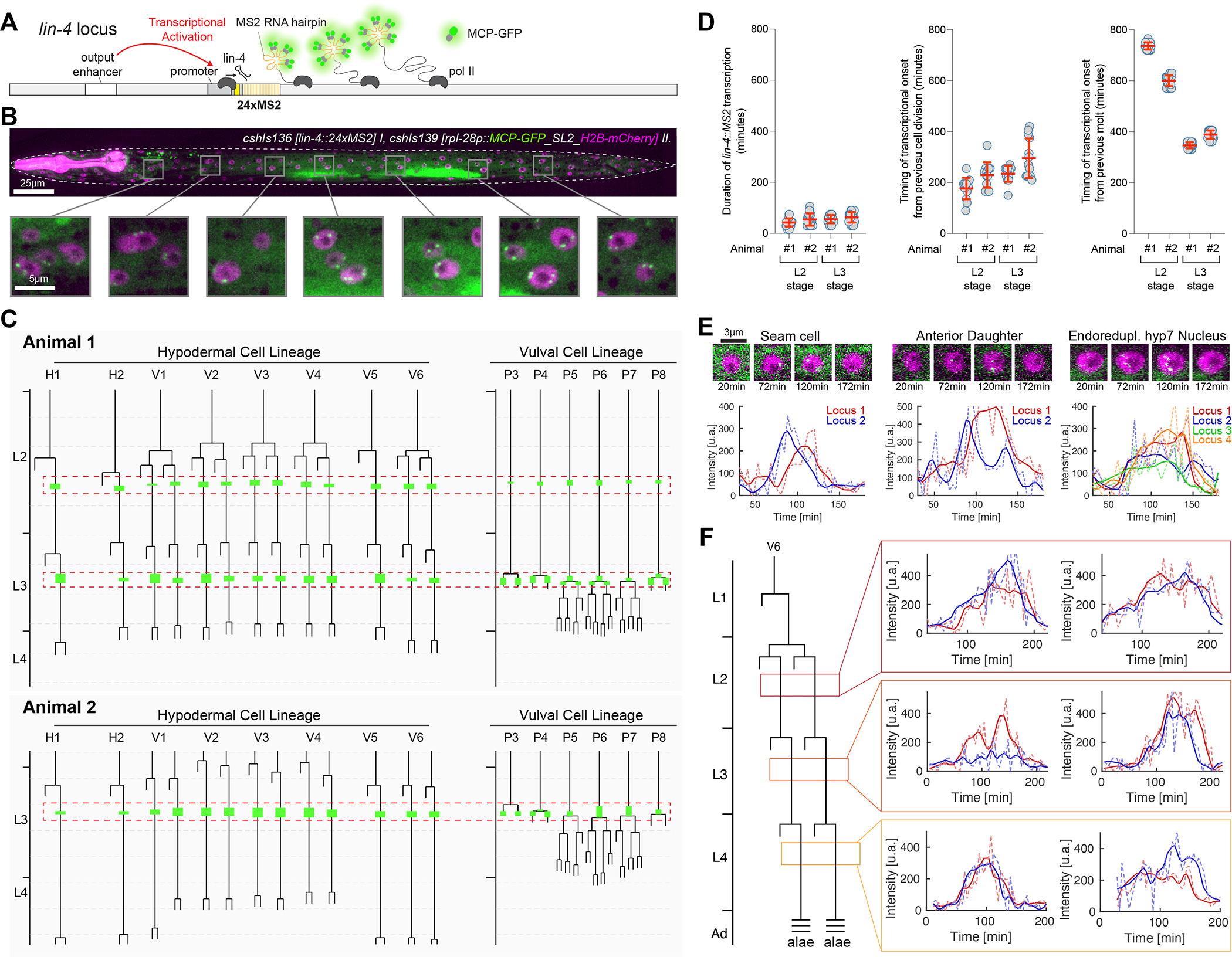
(A) The MS2/MCP system comprises an MS2 coat protein GFP fusion (MCP-GFP) which can bind to MS2 RNA hairpins engineered into primary miRNA transcripts.
(B) Magnified insets showing MCP-GFP spots (green) along the anteroposterior axis and H2B-mCherry (purple) expression localized in hypodermal nuclei.
(C) Examples of L2/L3 hypodermal and VPC division patterns in wild-type animals and annotations indicating when MCP-GFP foci were visible. Red boxes show the temporal region where synchronous lin-4 transcription is observed. Dashed horizontal grey bars indicate four hours of development at 20°C.
(D) Graphs representing the duration of MCP-GFP foci in the L2 and L3 stages of development and temporal relationships of these transcriptional epochs with the preceding cell division or molt cycle/ecdysis. Red bars indicate the mean with SD.
(E) Snapshots of individual seam cell and hyp7 cell MCP-GFP foci and the expression trajectories of these cells (L3-stage).
(F) Expression traces in pairs of V6.p seam cells from L2, L3, and L4-staged animals.
To determine the level of lin-4 transcriptional dynamics at single-cell resolution and across cell and tissue types in living animals, we used our microfluidics-based platform30 for long-term imaging of larvae harboring lin-4::24xMS2/MCP-GFP system. We quantified expression dynamics in hypodermal cells where the timing of transcriptional activity can be accurately assessed in relation to stage-specific cell division patterns and ecdysis from each larval molt12. We screened for periods of lin-4 transcriptional activity by imaging at 15min time intervals from the first larval stage (L1) to the mid-fourth larval stage (L4)(~60h) (n>10). lin-4::24xMS2 transcription was highly pulsatile, with a single transcriptional episode of ~40–105 minutes at each lin-4::24xMS2 loci during each larval stage, followed by long periods of inactivity (>10hrs at 20°C) (Figure 1B and C). Transcriptional activation across cells within the hypodermis was highly concordant, exhibiting similar transcriptional on and off times for lin-4::24xMS2 loci (Figures 1B and C). For instance, transcription began in hypodermal and vulval precursor cells (VPCs) after seam cell divisions in both L2 and L3 stages (Figure 1C). While lin-4 transcription was apparent in non-dividing, L2-staged VPCs, the appearance of lin-4::24xMS2 expression throughout the L3-staged skin cells generally occurred within minutes of the first VPC divisions (P3.p or P4.p)(Figure 1C). MCP-GFP foci became undetectable before the divisions of remaining VPCs (P5.p-P7.p) (n = 15) (Figure 1C).
We then correlated lin-4::24xMS2 transcriptional epochs with stage-specific cell division patterns and features of the larval molting cycle, two developmental milestones exhibiting similar temporal synchrony levels30,31. Transcriptional durations of lin-4::MS2 in each cell were similar across the population of lateral seam cells in both the L2 and L3 stages (Figures 1C and D). In contrast, the temporal variation between the completion of cell division and transcriptional activation within seam cells was more variable (coefficient of variation ranging from 13.5% to 25.3%)(Figure 1D). We then assessed the temporal relationships between transcriptional activation and the molting cycle by calculating the interval between transcriptional onset and prior ecdysis. Surprisingly, even though the completion of the preceding molt occurs hours before the programmed cell divisions of a given stage, we found a much smaller coefficient of variation between the previous ecdysis (shedding of the cuticle) and transcriptional activation (coefficient of variation from 1.9% to 4.1%)(Figure 1D). We also noted that transcriptional activation in non-dividing VPCs in L2-staged animals is tightly correlated with time from ecdysis (Figure 1B). This indicates that the timing of lin-4::24xMS2 transcriptional activation is likely coupled to the repetitive molting cycles of larval development and less so to individual cell division patterns that occur in each intermolt period.
To determine how similar the transcriptional dynamics are within individual hypodermis cells, we performed short-term imaging time courses (<6h) at 4min intervals in staged larvae. During transcriptional episodes, we detected near synchronous accumulation of MCP-GFP foci at each hypodermal lin-4::24xMS2 locus for 60–90 minutes (Figure 1E and F, Suppl. Movie 1 and 2) (>15 animals). We found no signs of “bursty” transcription32 as MCP-GFP foci were continuously maintained at each lin-4::24xMS2 transgene for the entire transcriptional episode (Suppl. Movie 1,2). These features were independent of the number of lin-4::24xMS2 loci per nucleus as cell types that undergo endoreduplication (i.e., hyp7 cells) exhibited MCP-GFP foci dynamics indistinguishable from diploid cells (Figure 1E). The dynamic features of lin-4::24xMS2 expression in hypodermal cells were similar across different developmental stages (Figure 1F), suggesting that the same regulatory programs controlling lin-4 transcription are repeated at each larval stage. Pulsatile transcription also occurs in additional cell types that normally express lin-4 (including the non-neuronal cells of the pharynx and intestinal cells) (Figure S1D–G). Therefore, the gene regulatory network that generates lin-4 transcriptional pulses at each stage of development organizes the timing, amplitude, and duration of transcription throughout the body in a highly reproducible manner.
Deletion of the lin-4 PCE sensitizes animals for L2-stage patterning defects
Full transcriptional activation of lin-4 requires a conserved upstream regulatory element, the Pulse Control Element (PCE), located ~2.8kb upstream of the lin-4 sequence (Figure 2A) 22. We aimed to determine if removing the PCE results in developmental timing defects. We integrated single-copy transgenes at a defined locus on chromosome I to accomplish this. Transgenes contained either a full-length 4023bp genomic fragment containing the lin-4 gene or variants of this transgene lacking individual conserved regions (Figure 2A). These transgenes were then crossed into a strain harboring a lin-4(0) allele, lin-4(e912), to determine if they could complement the absence of lin-4 activity. We found that the full-length lin-4 construct could rescue the cell lineage, adult alae, vulval, and gene misexpression phenotypes of lin-4(e912)(Figure 2A and Figure S2). In contrast, the 693bp genomic fragment used to clone the lin-4 gene that complements lin-4(e912) when expressed from high-copy extrachromosomal arrays fails to rescue lin-4(e912) developmental phenotypes from a single-copy transgene (Figure 2A and Figure S2). Surprisingly, transgenes lacking a large portion of the upstream sequence (Short fragment) or harboring deletions of the PCE or another conserved region of genomic DNA (D region) exhibit wild-type development under standard growth conditions.
Figure 2. lin-4 regulatory sequences control transcriptional output and buffer development against temporal patterning defects.
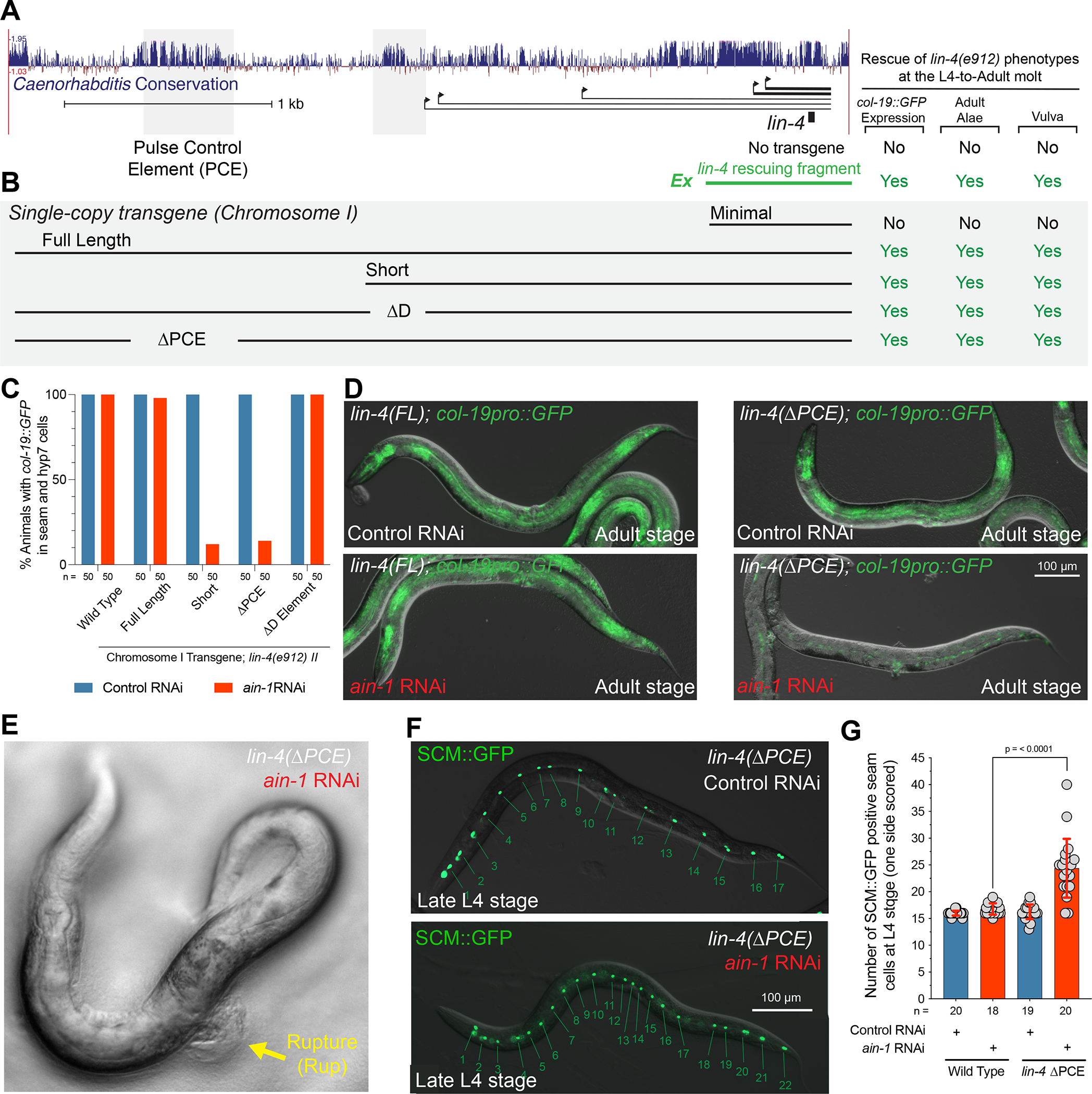
(A) A graphical depiction of the conservation of the lin-4 locus in multiple nematode species, the primary transcripts originating from C. elegans genomic region, and the fragment location of the lin-4 transgenes described in the main text.
(B) Graphical depiction of integrated lin-4 genomic rescue fragments and their ability to rescue temporal patterning phenotypes.
(C) Quantification phenotypes of various lin-4 mutants treated with control or ain-1 RNAi.
(D) Micrographs depicting the col-19::GFP expression phenotypes in wild-type or ΔPCE mutants treated with ain-1 or control dsRNAs.
(E) ΔPCE animal exposed to ain-1 dsRNA rupture from the vulva.
(F) Images of late L4-staged ΔPCE animals expressing the seam cell-specific GFP reporter exposed to control or ain-1 dsRNAs.
(G) Quantification of the supernumerary seam cells in ain-1-treated wild-type and ΔPCE animals.
We next aimed to determine whether mutations in these lin-4 cis-regulatory regions sensitize animals to heterochronic phenotypes by testing for genetic interactions with other genes that impact temporal gene expression. ain-1 encodes a C. elegans ortholog of GW182 and functions within the miRNA-induced silencing complex (miRISC) to repress miRNA targets33. Null mutations of ain-1 or RNA interference (RNAi) of ain-1 result in mild heterochronic phenotypes33,34. When ain-1 was depleted in wild-type animals or lin-4(e912) animals rescued with the full-length or ΔD region rescuing transgenes, animals exhibited normal col-19::GFP expression patterns and adult alae phenotypes (Figure 2C). In contrast, ain-1(RNAi) treatment of lin-4(e912) animals rescued with either the short promoter element or a transgene lacking only the PCE resulted in severe heterochronic phenotypes. These ain-1 treated animals only weakly express col-19::GFP in seam cells (Figures 2C and D). RNAi of ain-1 in these two genetic contexts also results in a vulval rupture (Rup) phenotypes where animals explode from the vulval opening at the L4 to adult (29%; n = 55)(Figure 2E). Consistent with ain-1 RNAi treatment altering temporal patterning throughout larval development in ΔPCE mutants, RNAi treatment of ΔPCE lin-4; lin-4(e912) animals exhibit a supernumerary seam cell phenotype where L2-specific lateral seam cell division patterns are inappropriately reiterated at later larval stages (Figures 2F and G).
NHR-23 and NHR-85 bind the PCE and function in lin-4 transcription and temporal development
Given that the PCE plays a role in maintaining the robustness of temporal patterning, we then performed a yeast one-hybrid screen to discover transcription factors (TFs) that bind the PCE to regulate lin-4 transcription using the entire 514bp PCE as bait. Using a yeast one-hybrid library that contains ~89% of C. elegans DNA binding proteins, we identified three TFs that specifically bound the PCE. These transcription factors were BLMP-1, NHR-23, and NHR-85 (Figure 3A). We have previously demonstrated that BLMP-1 promotes lin-4 expression and functions as a pioneer factor to decompact the lin-4 locus before transcriptional activation throughout development 22. NHR-85 and NHR-23 are two members of an expanded class of C. elegans nuclear hormone receptors (NHRs) that are the closest nematode orthologs of human circadian-related TFs Rev-Erb and ROR, respectively (Figure 3B). As such, NHR-85 and NHR-23 share significant sequence homology within their C4-type Zinc finger DNA-binding domains (Figure 3C)35. NHR-85 and NHR-23 are primarily expressed in hypodermal tissues, and not all cell types that express lin-4:24xMS2 (Figure S3)36,37. Disruption of nhr-85 expression results in egg-laying phenotypes (Egl) consistent with NHR-85 controlling hypodermal gene expression and vulval development36. NHR-23 expression cycles with the larval molts and has been implicated in spermatogenesis, molting, and the control of developmental pace during larval development38–41. In addition, nhr-23 and the terminal heterochronic miRNA, let-7, genetically interact to limit supernumerary molts during adulthood40,42. Analysis of publicly available ChIP-seq data indicated that all three TFs interact in vivo with lin-4 regulatory sequences (Figure 3A) and that their binding sites are enriched in the promoters of cyclically expressed mRNAs and other heterochronic genes (Figure S4) (Tables S1 and S2).
Figure 3. NHR-23 and NHR-85 bind the PCE and are required for normal temporal patterning and lin-4 expression.
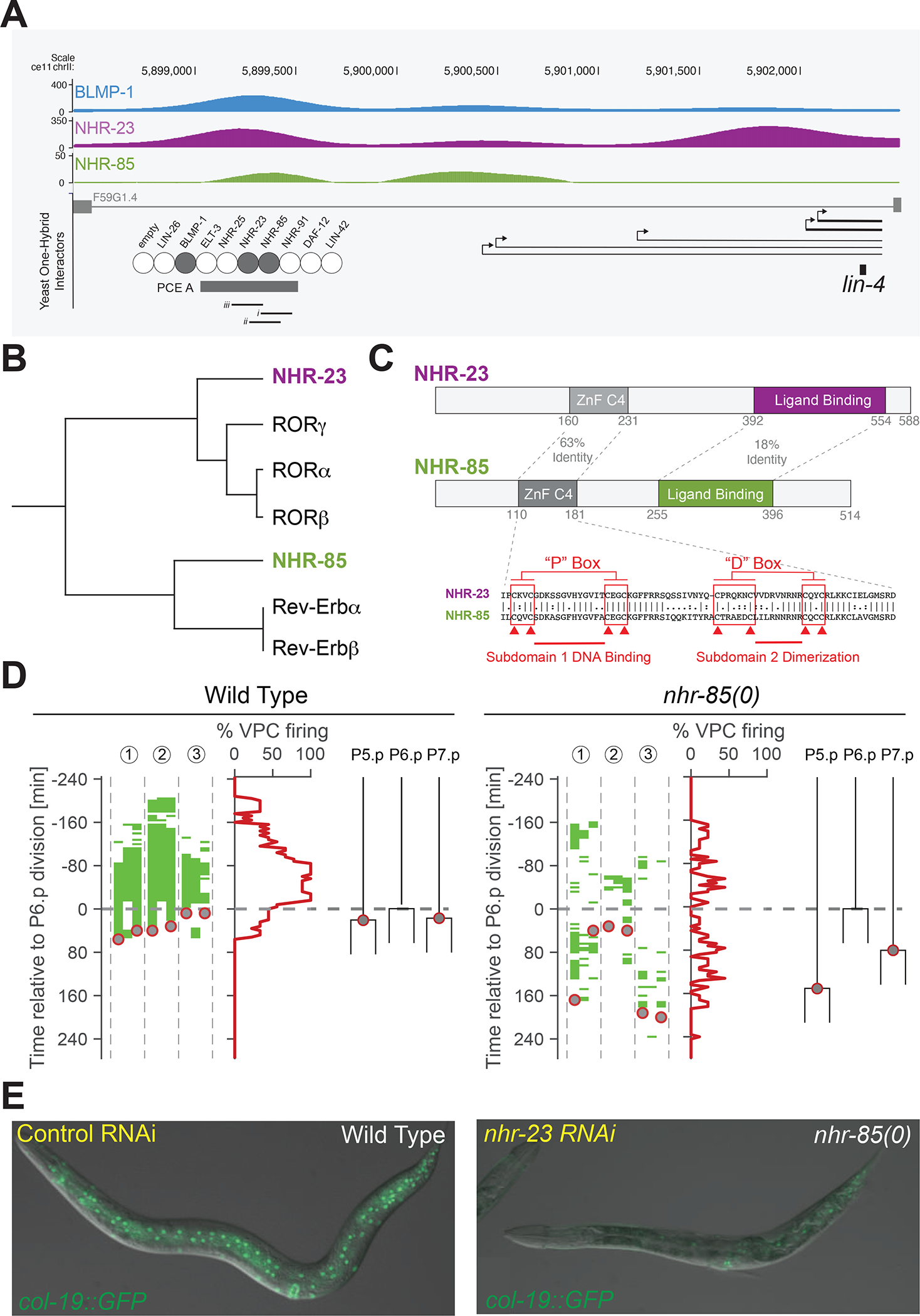
(A) Genome-Browser tracks showing BLMP-1, NHR-23, and NHR-85 bindings sites near the lin-4 locus.
(B) Sequence relationships between NHR-23 and NHR-85 and human Rev-Erb and ROR.
(C) A graphical representation of NHR-23 and NHR-85 domain organization.
(D) High-resolution time course analysis of lin-4::24xMS2 expression in wild-type and nhr-85(0) mutants, aligned to first P6.p cell division (t = 0). Green areas indicate detectable MCP-GFP foci in individual Pn.p cells (P5.p – P7.p) (n = 3 animals). Grey circles represent the timing of the P5.p and P7.p divisions.
(E) Depletion of nhr-23 in nhr-85(lf) mutants prevents the regular expression of the adult-stage specific col-19::GFP reporter.
We obtained mutants of nhr-23 and nhr-85 to determine whether the inactivation of these genes results in temporal patterning defects. Null mutants of nhr-23 exhibit variable developmental arrest phenotypes where most animals arrest during the L1 molt43. RNAi of nhr-23 mRNA or auxin-mediated depletion of AID-tagged NHR-23 alleles during post-embryonic development can result in developmental arrest at any of the four larval molts39,44,45. An nhr-85(lf) allele that removes the ZnF DNA-binding domain, nhr-85(ok2051), exhibits mild egg-laying (Egl) phenotypes consistent with previous studies examining nhr-85 function via RNAi36. To determine if NHR-85 contributes to lin-4 expression, we employed the MS2/MCP-GFP system in nhr-85(ok2051) animals. High-resolution imaging of VPC divisions and lin-4::24xMS2 expression dynamics indicates that two features of developmental timing were altered. In wild-type animals, transcriptional pulses of lin-4::24xMS2 are robust and concordant in adjacent wild-type VPCs (Figure 3D). In contrast, MCP-GFP foci in nhr-85(lf) mutants begin to accumulate at the same relative phase of L3-stage VPC development but are dimmer and only transiently observed (Figure 3D). Second, under identical imaging conditions, the rapid and highly coordinated VPC divisions observed in wild-type animals are altered in nhr-85(lf) mutants with some P5.p and P7.p dividing hours after the first P6.p division (Figure 3D). These results indicate that NHR-85 enhances the robustness of temporally regulated processes during development and that some level of lin-4 transcription occurs without NHR-85, perhaps driven by NHR-23 alone. RNAi-mediated depletion of nhr-23 activity in wild-type animals resulted in mild heterochronic phenotypes (Figure 3E). Consistent with the hypothesis that NHR-23 and NHR-85 function cooperatively to control temporal regulation, the penetrance of these phenotypes was enhanced when nhr-23 was also depleted in nhr-85(0) animals (Figure 3E)(nhr-23 RNAi in wild-type animals = 18% defective (n = 51) and nhr-23 RNAi in nhr-85(ok2051) animals = 47% defective (n = 51)).
NHR-23 and NHR-85 bind cooperatively to direct repeats found in the lin-4 PCE
Nuclear hormone receptors often bind cooperatively as homo- or hetero-dimeric complexes at closely spaced cis-regulatory DNA elements46–48. Several features of NHR-85, NHR-23, and the lin-4 PCE suggest this may also be the case for lin-4 transcription. First, NHR-85 and NHR-23 share significant sequence homology within their C4-type Zinc finger DNA-binding domains suggesting they may bind similar DNA sequences (Figure 3C). Second, we found that NHR-85 and NHR-23 bind to each other in yeast two-hybrid assays (Figure 4A). Furthermore, the recombinant proteins migrate through a gel filtration column at a rate consistent with a simple heterodimer (Figures 4B and C), and microscale thermophoresis analysis of recombinant NHR-85 and NHR-23 indicates high-affinity heterodimeric binding without DNA (5.8 +/− 2.2 nM KD)(Figure 4D). Third, sequences within the PCE element contain multiple GGTCA sequences found in the consensus binding motifs for NHR-85/Rev-Erb and NHR-23/ROR families of NHRs (Figure 4E)49.
Figure 4. NHR-23 and NHR-85 form heterodimers that bind cooperatively to sites within the PCE.
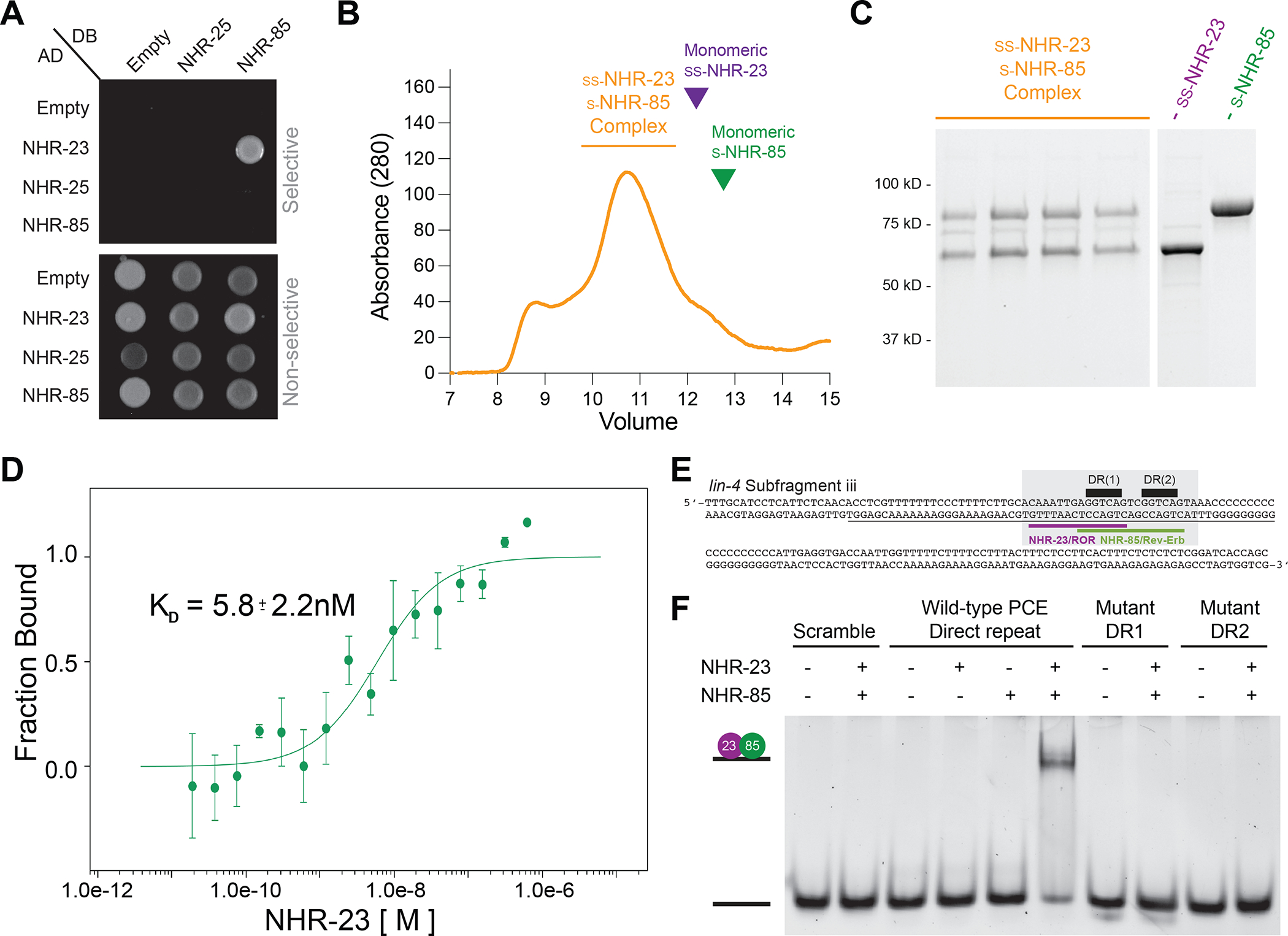
(A) NHR-23 and NHR-85 interact with each other in two-hybrid assays.
(B and C) Gel filtration of Strep2Sumo-NHR-23 and Strep2-NHR-85 complexes. Purple and Green carrots indicate where monomeric ss-NHR-23 and s-NHR-85 peaks occur with individual proteins run on identical gel filtration columns.
(D) Microscale thermophoresis analysis of NHR-23 and Alexa-647-labeled Strep2-NHR-85 indicates that NHR-85 and NHR-23 bind with high affinity (n = 3).
(E) The sequence of the PCEiii fragment identified in EMSAs (Figure S5) that can interact with NHR-23 and NHR-85. The grey box outlines the minimal DNA fragment bound by NHR-23/NHR-85 heterodimeric complexes (panel F). This fragment contains the GGTCA direct repeat (black bars) found in the overlapping consensus binding sites for NHR-23/ROR (purple) and NHR-85/Rev-Erb (green).
(F) EMSA experiments of wild-type and mutant target DNAs using recombinant NHR-85 and NHR-23.
To determine if NHR-23 and NHR-85 could directly bind to target sequences within the PCE, we performed electrophoretic mobility shift assays (EMSAs) using 161bp probes that span overlapping fragments of the 512bp PCE element and purified recombinant proteins. These assays identified a single sub-fragment, PCEiii (Figures S5A and B), that could be bound by NHR-23 when NHR-23 is in 5–10 molar excess to the DNA target (Figure S5B and C). We did not detect binding between NHR-85 and any PCE sub-fragments (Figure S5B). This PCEiii sub-fragment harbors two closely-spaced GGTCA sequences (Figure 4E). Importantly, we found that concentrations of NHR-23 that were insufficient to bind the PCE alone were dramatically stimulated by the addition of NHR-85, indicating that these NHRs bind cooperatively to the PCEiii fragment (Figures S5C, D, and E). These interactions could be recapitulated with a smaller 25nt probe that harbors the direct repeat element (Figure 4F). Cooperative binding of NHR-23 and NHR-85 to the minimal 25nt probe requires the presence of both closely-spaced direct repeats, as mutating either repeat prevents binding of the NHR-23/NHR-85 heterodimeric complex (Figure 4F).
Multiple NHR-23 and NHR-85 heterodimer binding sites redundantly control temporal patterning
Since NHR-23 and NHR-85 bind cooperatively presumably as heterodimer to closely spaced GGTCA binding sites within the PCE, we searched for additional candidate NHR-23- and NHR-85-binding elements within the full-length lin-4 rescuing transgene. These efforts identified six additional NHR-23/ROR and two additional NHR-85/Rev-Erb binding sites outside the PCE element (Figure 5A and S6B and C). Of these other binding sites, a region more proximal to the lin-4 gene, harboring adjacent NHR-23/ROR and NHR-85/Rev-Erb binding sites with a similar orientation and spacing (i.e., GGTCAnnnGGTCG) to the binding sites found in the PCE element was found (Figure 5B). This putative NHR-23/NHR-85 heterodimer binding site is located 266bp upstream of the encoded lin-4 pre-miRNA and immediately upstream of the transcriptional start site (TSS) of the two most abundant pri-lin-4 transcripts (Figure 5A)29. Furthermore, this sequence is present in the original genomic fragment used to rescue lin-4(0) developmental phenotypes when expressed from high-copy extrachromosomal arrays50. EMSAs using this proximal element demonstrate that NHR-23 and NHR-85 also bind cooperatively to this sequence (Figure 5C).
Figure 5. Conserved NHR-23 and NHR-85 binding sites in the lin-4 upstream regulatory sequence function redundantly to control temporal patterning.
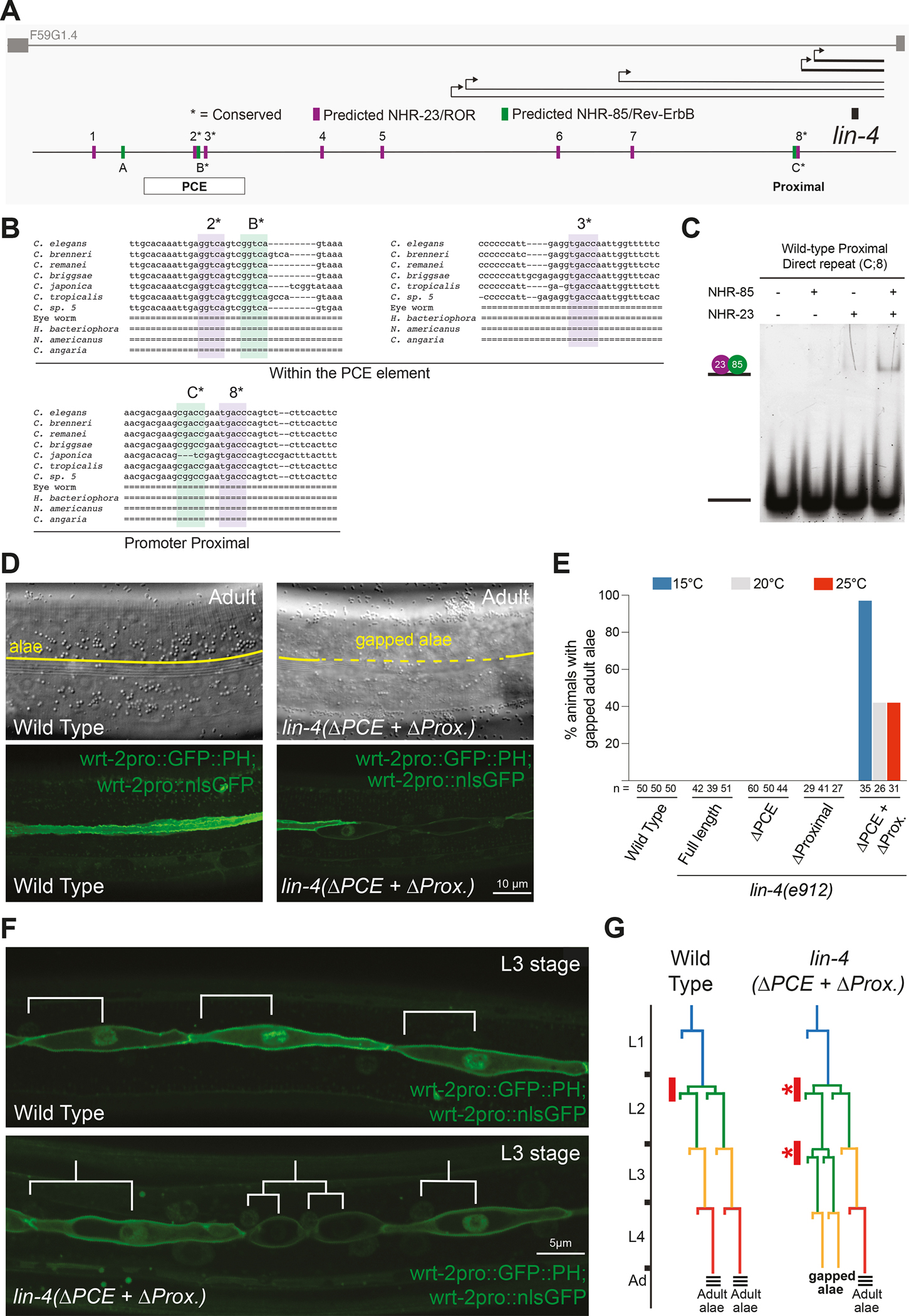
(A) The lin-4 locus and the predicted binding sites for NHR-23 (Purple) and NHR-85 (Green). Asterisks indicate predicted binding sites that are conserved in other nematode species.
(B) Alignment of indicated NHR-23 and NHR-85 binding sites in the lin-4 locus (see Figure S6 for complete alignments). Purple highlighted regions indicate conserved NHR-23 binding sites, and green indicates conserved NHR-85 binding sites.
(C) EMSA experiments demonstrate that recombinant NHR-23 and NHR-85 bind the two conserved direct repeat elements in the lin-4 locus proximal region.
(D) Micrographs showing the adult cuticle and lateral seam cell morphologies of wild-type animals and animals that harbor a deletion of the PCE and mutations in the proximal direct repeats.
(E) The retarded phenotypes of ΔPCE+ΔProximal animals are temperature sensitive. Asterisks indicate statistically significant differences from other conditions or genotypes.
(F) Analysis of the seam cell lineage of wild-type and ΔPCE+ΔProximal mutant animals show that L2 division patterns are inappropriately repeated in the L3 stages of development in PCE+ΔProximal mutants.
(G) Proposed seam cell lineage diagrams of wild-type and ΔPCE+ΔProximal mutant animals.
We next tested whether the proximal NHR-23/NHR-85 binding site is vital for normal development by mutating these sites using CRISPR/Cas9 genome editing. We found that altering the proximal site from AGCGACCGAATGACCCA to AGgctggGAAactggCA (ΔProximal) did not result in abnormal temporal patterning phenotypes (Figure 5E). We then edited the proximal sites in animals already harboring the PCE deletion (ΔPCE). Strikingly, almost half of the lin-4 ΔPCE+ΔProximal double mutants exhibit retarded terminal cell-fate specification phenotypes where the adult cuticular alae are gapped (Figures 5D and E). Lateral seam cells directly beneath alae gaps exhibit cell morphologies more similar to those of L4-staged seam cells (i.e., unfused with adjacent seam cells)(Figure 5D), suggesting that these phenotypes result from temporal patterning defects. The penetrance and expressivity of these phenotypes increase at lower temperatures (Figure 5E). Further examination of these cell lineage defects indicates that the retarded phenotypes of lin-4 ΔPCE+ΔProximal double mutants begin in the L3 stage for development, where L2 cell division programs are inappropriately reiterated a single time, leading to supernumerary seam cell numbers at each subsequent stage (Figures 5F and G). The expressivity and penetrance of this retarded L2-stage patterning defects are more severe when only one copy of the lin-4 ΔPCE+ΔProximal transgene is used to rescue lin-4(0) mutations. While a single copy of the full-length wild-type transgene fully rescues lin-4(0) phenotypes (n = 35), 89% of single-copy lin-4 ΔPCE+ΔProximal rescued animals lack adult-specific alae at the L4 molt (n = 46) and 76% of these animals burst from the vulva in early adulthood (n = 50). Importantly, the lin-4 ΔPCE+ΔProximal transgene fully rescues the defective stage-specific expression patterns of the neuropeptide NLP-45 in head ganglia cells of lin-4(0) animals (Figure S6C). These experiments indicate that the NHR-23/NHR-85 binding sites are dispensable for normal lin-4 activity in post-mitotic neurons but are required for normal lin-4 transcriptional dosage in the hypodermis.
lin-4 transcription in the hypodermis occurs during the brief overlap of NHR-85 and NHR-23 expression
We quantified mRNA and protein expression during post-embryonic development to determine how NHR-85, NHR-23, and LIN-42 expression patterns may contribute to the regulation of pulsatile lin-4 transcription. The expression of nhr-85 mRNAs begins from an L1-stage arrest with a pulse of transcription followed by a monotonic expression pattern for the remaining larval stages (Figure 6A). In contrast, nhr-23 and lin-42 mRNAs are expressed in phased, high-amplitude oscillatory patterns (Figure 6A). We next explored the temporal dynamics of the corresponding proteins by quantifying the expression of endogenously-tagged alleles during the L4 stage, where changes in vulval morphogenesis can be directly correlated with the developmental stage 51. NHR-23::mScarlet and LIN-42::YFP are dynamically expressed in all hypodermal cells, with a single peak of expression that matches the phased expression of their mRNAs (Figure 6B). We also found that NHR-85::GFP expression was highly dynamic during these periods, indicating substantial post-transcriptional regulation of expression in the L2-L4 stages of development. Specifically, expression of NHR-85::GFP peaks at ecdysis (shortly before NHR-23::mScarlet onset), is undetectable by the L4.3 stage of development and resumes at the L4.6 stage in an antiphasic manner to the expression pattern of LIN-42::YFP (Figure 6C). These proteins’ highly similar phased expression patterns are also maintained in L3- and L4-staged vulval cells (Figures 6D and E).
Figure 6. lin-4 transcription occurs during the brief periods where NHR-85::GFP and NHR-23::mScarlet are co-expressed.
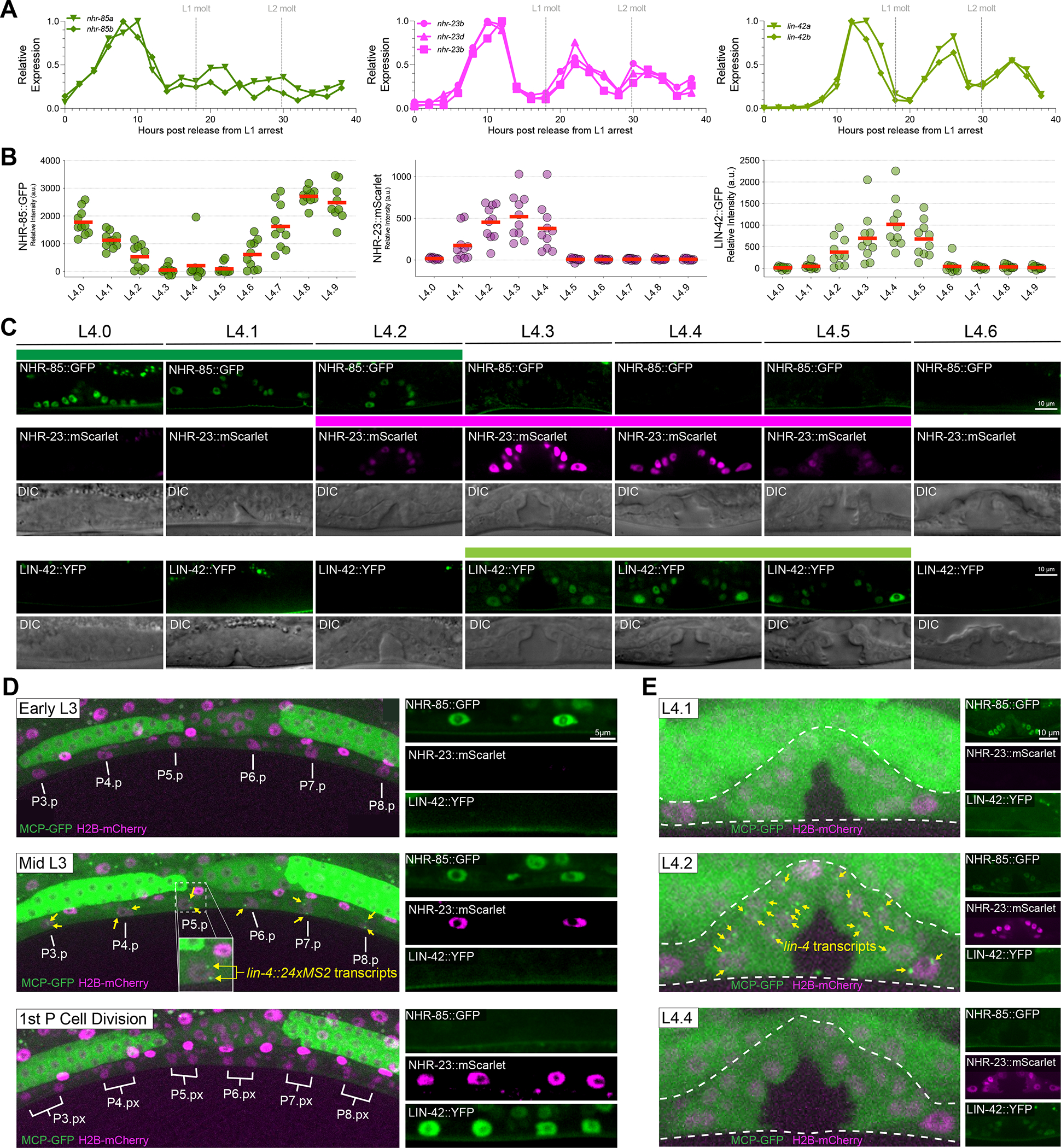
(A) RNA-seq time course data of nhr-85, nhr-23, and lin-42 mRNA expression patterns 20.
(B-C) Quantification and micrographs depicting NHR-85::GFP, NHR-23::mScarlet, and LIN-42::YFP expression in hypodermal seam cells and vulval cells, respectively, in each morphologically defined L4 substage 51. Circles in B represent average measurements from individual animals (3 cells sampled per animal); red bars indicate the mean. Colored bars indicate ranges of detectable expression.
(D) Time course experiments demonstrate that lin-4::24xMS2 expression occurs immediately before the first Pn.p cell divisions and not until NHR-85::GFP are NHR-23::mScarlet are co-expressed in the VPCs. In wild-type animals, lin-4::24xMS2 expression terminates when NHR-85::GFP expression is extinguished around the time of the first VPC division.
(E) Dynamic lin-4::24xMS2 transcription correlates with NHR-85::GFP/NHR-23::mScarlet expression in the L4 stages of vulval development.
Since NHR-85 and NHR-23 heterodimerize and bind cooperatively to regions of the lin-4 enhancer that control dynamic transcription, we hypothesized that the 60–90min pulses of lin-4 transcription might occur in the short window of each larval stage where NHR-85 and NHR-23 are co-expressed. To compare the timing of these events, we monitored the appearance of MCP-GFP foci in vulval cells during the L3 and L4 stages, where the rapid, stereotyped vulval cell division patterns30 and changes in morphology51 enable precise determination of the timing of lin-4::24xMS2 transcription and TF expression dynamics. We found a correspondence between NHR-85::GFP and NHR-23::mScarlet co-expression and lin-4::24xMS2 transcription in both vulval and hypodermal cells (Figures 6D and E). While the expression of both nuclear receptors is phased, the transient expression of the lin-4::24xMS2 transgene only occurs during the brief period when both NHRs are expressed (Figures 6D and E). The timing of NHR-85::GFP post-transcriptional downregulation is also concurrent with the onset of LIN-42::YFP expression in VPCs and hypodermal cells (Figures 6D and E). This suggests that the dynamic patterns of these three TFs control the timing and duration of lin-4 transcriptional pulses.
LIN-42 post-transcriptionally represses NHR-85 and controls the amplitude and duration of lin-4 transcription
Human Per2 (PERIOD2) protein interacts with multiple mammalian NHRs (including Rev-Erb) to modulate their transcriptional activity52. To test whether LIN-42 physically interacts with either NHR-85 or NHR-23, we used yeast two-hybrid assays53. We found that both major LIN-42 isoforms interact with NHR-85 but not NHR-23 (Figure 7A). We mapped the regions of LIN-42 that are required for NHR-85 binding and found that a minimal 51aa fragment present in both major LIN-42 isoforms is sufficient to mediate interactions (Figure 7A). This domain differs from the interaction motifs in mammalian Per2 and Rev-Erb binding52. To determine if LIN-42 binds additional NHRs, we performed two-hybrid experiments between LIN-42 isoforms and 241 of the remaining 282 encoded C. elegans NHRs. We identified 65 NHRs that physically interact with LIN-42 (Figure S7A). The additional interacting NHRs included DAF-12, which regulates the expression of the let-7-family of miRNAs and controls dauer development54–56, and NHR-14HNF4a, NHR-69HNF4a, and NHR-119 PPARα (Figure S7) whose orthologs are also bound by Per252. These findings suggest that many physical interactions between Period proteins and orthologous NHRs have been maintained since the divergence of nematodes and humans and are likely functional.
Figure 7. LIN-42 regulates the expression dynamics of NHR-85 to control the amplitude and duration of lin-4 transcription.
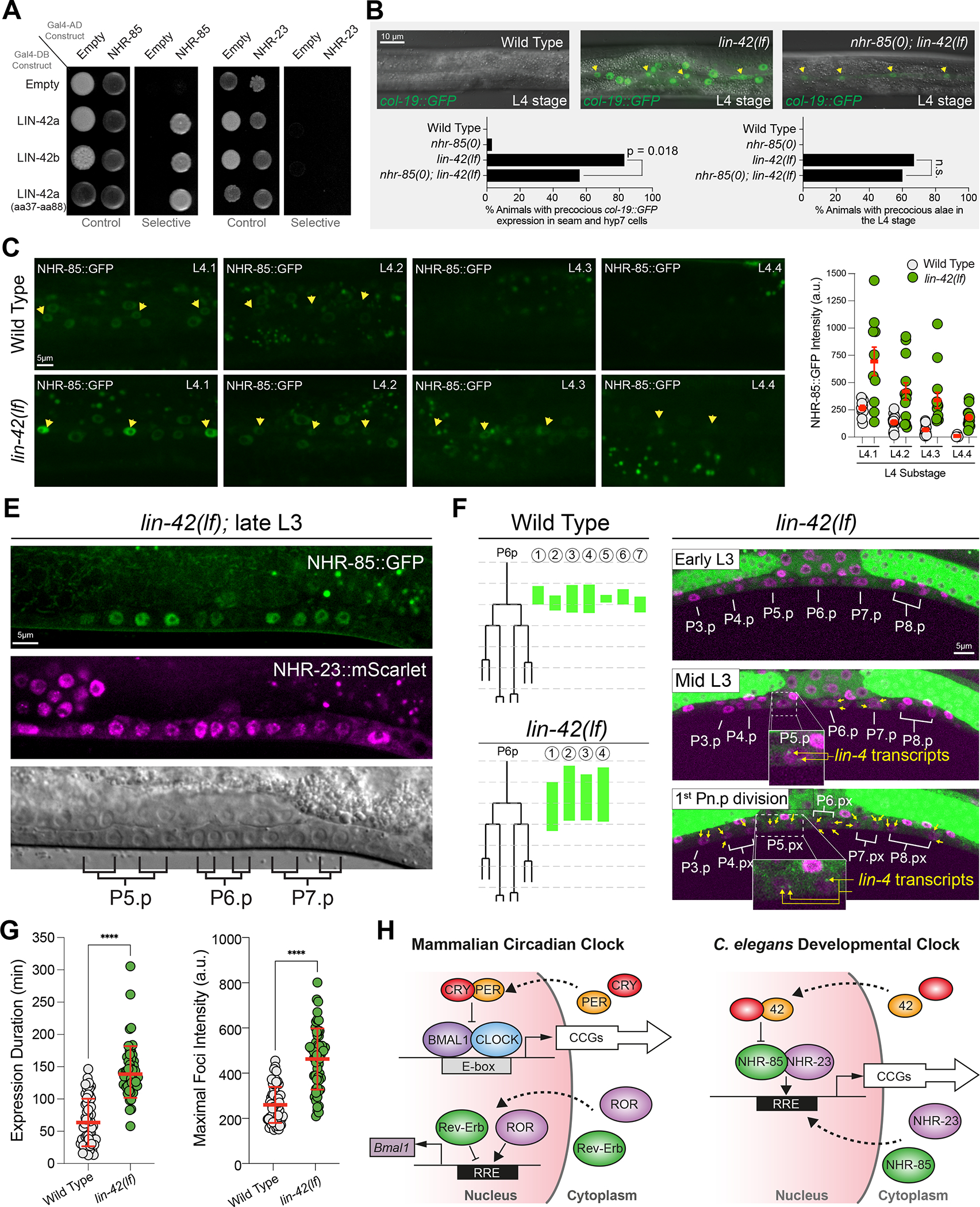
(A) LIN-42 interacts with NHR-85 but not NHR-23 in two-hybrid assays.
(B) Deletion of nhr-85(0) suppresses precocious col-19::GFP expression in lin-42(lf) mutants. Yellow arrows indicate the lateral seam cells of L4-staged animals. Error bars were calculated using two-tailed chi-square analysis.
(C and D) Images and quantification of NHR-85::GFP expression in hypodermal cells of L4-staged wild-type and lin-42(lf) animals. Yellow arrows indicate the lateral seam cells. Circles represent the average fluorescence in three seam cells of an individual animal. Error bars show mean and standard deviation. Significance was calculated using Welch’s t-test.
(E) NHR-85 expression perdures in developing VPC cells in lin-42(lf) mutants.
(F) Time course analysis of the onset/offset times for MCP-GFP foci in VPCs of wild-type and lin-42(lf). Green lines indicate the timing of lin-4::24xMS2 expression in P6.p cells of individual animals. Micrographs show representative images of the ventral surface of a single lin-42(lf) animal throughout the time course.
(G) Quantification of the duration and intensity of MCP-GFP foci in L3-staged animals. Error bars and significance are calculated as in D.
(H) Model of the mammalian circadian clock and the proposed C. elegans developmental clock.
Given the physical interaction between NHR-85 and LIN-42, we asked whether NHR-85 expression was required for the precocious phenotypes seen in lin-42(lf) mutants (lin-42(n1089)). We found that lin-42(lf) heterochronic phenotypes are partially ameliorated by removing nhr-85 function. Specifically, the precocious expression of adult-specific reporters (e.g., col-19::GFP) in both seam and hyp7 cells observed in lin-42(lf) mutants is suppressed by nhr-85 deletion, leaving weak expression in seam cells in double mutants. In contrast, precocious deposition of adult alae was not suppressed (Figure 7B). To examine whether LIN-42 regulates NHR-85 temporal expression patterns, we compared the dynamics of NHR-85::GFP and NHR-23::mScarlet abundance in wild-type and lin-42(lf) mutants over the course of the L4 stage. We found that the expression of NHR-85::GFP is altered in two ways by the lin-42 mutation. First, NHR-85::GFP signal is ~2.3x more abundant at the beginning of the L4 stage in lin-42 mutants than in equivalently-staged wild-type animals (Figures 7C and D). More importantly, the periodic dampening of NHR-85 expression usually occurs by the L4.2 stage of vulval morphogenesis (Figures 6B and C) and is altered in lin-42(lf) mutants. Specifically, NHR-85::GFP expression perdures into the L4.4 stage lin-42(lf) animals (Figures 7C and D). NHR-85 expression also perdures past the second VPC division in lin-42(lf) mutants (Figure 7E). Mutations in lin-42 do not alter the onset or duration of NHR-23::mScarlet accumulation in hypodermal or vulval cells. This suggests that LIN-42 regulates lin-4 transcriptional output by controlling the duration of NHR-85/NHR-23 heterodimeric complex formation in a manner that directly correlates with NHR-85 abundance.
We hypothesized that specific features of lin-4 transcriptional pulses would be altered in lin-42 mutants. Specifically, we anticipated that lin-4 transcriptional amplitude and duration would increase in lin-42 mutants because the perdurance of NHR-85 expression would enable a longer NHR-23/NHR-85 heterodimerization period. To determine if the increased duration of lin-4 transcription in lin-42 mutants is caused by an inappropriate perdurance of NHR-85/NHR-23 co-expression, we examined the appearance and duration of MCP-GFP foci in lin-42(lf) mutants. We found that MCP-GFP foci appear earlier in lin-42(lf) VPCs compared to the equivalent staged cells in wild-type (Figure 7F). Furthermore, they can continue past the second VPC division in lin-42 mutants, a developmental time when MCP-GFP foci have disappeared in wild-type animals (Figure 1C and Figure 7F). Quantification of MCP-GFP foci indicates that while most wild-type seam cells exhibit MCP-GFP foci during each larval stage (76%; n=66 L3 staged seam cell nuclei), the percentage of seam cells showing detectable lin-4 transcription is dramatically increased in lin-42 mutants (100%; n=58 L3 staged seam cell nuclei). This demonstrates that lin-42 normally dampens lin-4 transcriptional pulses in wild-type animals. In addition to elevating the likelihood that lin-4::24xMS2 transcription is above a threshold sufficient to generate measurable MCP-GFP foci, lin-42(lf) mutations increase the intensity of MCP-GFP foci indicating that LIN-42 also limits the rate of transcriptional activation of the lin-4 locus (Figure 7G). Time course experiments also revealed that the overall duration of transcription events in lateral seam cells was ~2.2 times longer in lin-42 mutants compared to wild-type (Figure 7G). These results demonstrate how LIN-42 regulates lin-4 transcription through direct interaction with NHR-85, preventing the persistence of transcriptionally activating NHR-23/NHR-85 heterodimers.
DISCUSSION
Dissecting post-embryonic gene expression dynamics with real-time in-vivo imaging
One-fifth of the C. elegans larval transcriptome exhibits reproducible, periodic expression patterns that are phase-locked with features of the molting cycle20,23,24. Oscillating genes are diverse (including transcription factors, cuticular collagens, proteases, signaling proteins, and miRNAs) with distinct phases of expression throughout each larval cycle. While this phenomenon was discovered based on population-based RNA-sequencing studies, understanding the origins and regulation of these patterns requires single-animal or single-cell techniques that can capture continuous expression dynamics within these contexts. Here, we describe an MS2/MCP-GFP tethering-based approach to study post-embryonic oscillatory gene expression dynamics in developing C. elegans larvae at single-cell resolution. We focused on measuring the dynamics of the lin-4 miRNA implicated in temporal cell fate specification across animal tissues and cell types. Having access to real-time transcriptional dynamics, we revealed striking synchrony in lin-4 transcription among cells within the same tissue (Figure 1C). These transcriptional episodes were surprisingly short compared to the duration of a larval stage. Quantification of these expression patterns during ongoing development allowed us to precisely characterize which dynamic transcriptional features (amplitude, phase, and duration) are affected in different genetic backgrounds. While low-intensity transcription from the lin-4 locus may occur outside of the periods of highly-dynamic transcription (as measured by the periodic formation of MCP-GFP foci), the dynamics we observe appear different from the stochastic and “bursty” patterns of transcriptional dynamics observed in the C. elegans germline and early drosophila embryos57–60.
The normal transcriptional dynamics of lin-4 require orthologs of the mammalian circadian GRN
The gene regulatory network controlling lin-4 transcriptional pulses we uncovered here shares integral components with the human circadian clock but exhibits essential differences in its regulatory architecture. In the circadian clock regulatory GRN, CLOCK and BMAL1 generate rhythmic expression patterns of clock control genes61–64 (Figure 7H), including two core circadian transcriptional repressors, Period and CRY, as well as Rev-Erb and ROR. Negative feedback on CLOCK/BMAL1 expression by Period/CRY heterodimers is essential for generating circadian rhythms. In contrast, Rev-Erb and ROR modulate CLOCK and BMAL1 expression levels through opposing transcriptional activities but are dispensable for the generation of circadian oscillations (Figure 7H)65–68. Genes for CLOCK and BMAL1 are absent in the C. elegans genome. We propose here that the worm orthologs of Rev-Erb and ROR replace CLOCK and BMAL1 as the central transcription factors of the hypodermal developmental clock (Figure 7H). In contrast to the antagonistic roles of Rev-Erb and ROR in the circadian clock, C. elegans NHR-85 and NHR-23 heterodimerize and bind cooperatively to lin-4 regulatory sequences to induce transcriptional pulses (Figure 4). We show here that the phased expression of NHR-85 and NHR-23 controls the precise timing of transcriptional induction within each developmental cycle. This physical interaction between NHR-23 and NHR-85 provides the mechanism by which the timing of lin-4 transcriptional and likely the transcriptional pulses of other heterochronic miRNAs (e.g., mir-241, mir-48, and mir-84; (Figure S4)) is initiated. In essence, this developmental clock functions as a coincidence detector. The overlapping expression of two transcription factors and specific biochemical interactions between them likely generates pulsatile transcriptional patterns of multiple miRNAs that program sequential cell fates. In addition, the developmental clock enables phased gene expression patterns of target genes to adaptively anticipate different aspects of the animal’s larval stage demands, similar to how the circadian clock adjusts gene expression patterns to anticipate regular, repeating environmental cues.
lin-4 transcriptional dosage is controlled by the duration of NHR-85/NHR-23 heterodimeric complex formation
In addition to controlling the timing of lin-4 transcriptional onset, the biochemical interactions (heterodimerization and cooperative binding to lin-4 regulatory elements) combined with the phased expression of these two NHRs also directly regulate lin-4 gene dosage. We demonstrate here that the duration and amplitude of lin-4 transcription are controlled by the NHR-85/NHR-23 heterodimeric complex (Figures 6D and 6E). Genetic alterations that alter the duration of NHR-85 and NHR-23 co-expression patterns (i.e., in lin-42(lf) backgrounds) result in predictable alterations in these transcriptional features, including an increase in transcriptional duration and amplitude (Figures 7E–G). In addition, we find that transcriptional dosage of lin-4 is controlled by the number of NHR-23/NHR-85 regulatory sites in lin-4 regulatory regions. We show that lin-4 dosage is controlled by separate GGTC direct repeats that function redundantly to control lin-4 activity (Figure 5). Deleting both GGTC repeat sequences (one within the PCE and the other in the lin-4 Proximal element) results in the inappropriate reiteration of L2 cell fate phenotypes in the L3 stage (Figures 5F and G). These defects are temperature-dependent developmental defects reminiscent of those associated with certain mutants with elevated lin-14 activity, such as the lin-14 3’ UTR deletion mutant lin-14(n536n540)69. It remains to be seen how deleting these elements alters the dynamic features of lin-4 transcription.
A potential role for hormonal coordination of temporal patterning
The core mammalian circadian clock is entrained by light/dark cycles through the direct connection between retinal projections into the suprachiasmatic nucleus, enabling light-activated transcription of the circadian Period gene within these tissues70. Concordant feeding/fasting rhythms can also be substantial entrainment factors for peripheral metabolic organs, indicating multiple mechanisms can shift the phase of circadian transcriptional programs71. While nutrition-mediated entrainment of peripheral circadian clocks likely requires the integration of numerous hormonal inputs, insulin/IGF-1-dependent signaling also promotes Period protein translation to alter the phase of a free-running clock72. NHR-85 and NHR-23 are orphan receptors, and transcriptional activation by these NHRs may depend on as yet unidentified ligands. We hypothesize that this activity may further refine transcriptional onset and offset times and enable the precise global coordination of lin-4 transcription in hypodermal cells we observe (Figure 1B and 1C).
Acute nutrient deprivation elicits checkpoints in developing C. elegans larvae where cell divisions and oscillatory gene expression are halted immediately after each larval molt22,73. Notably, these developmental arrests are controlled by the insulin/IGF signaling pathway, which generates several cholesterol-derived hormones73,74. Arrests occur at a similar phase of each larval stage during which nhr-23 expression peaks20,23,24, and resemble phenotypes associated with auxin-mediated depletion of NHR-2345. Cholesterol derivatives are agonists of RORs in mammals75. Heme is an endogenous ligand of Rev-Erb known to modulate Rev-Erb activity within the circadian pathway76,77. Remarkably, C. elegans is an auxotroph for both cholesterol and heme and developing larvae rely exclusively on the environmental uptake of both metabolites for continuous development78,79. As such, heme and modified cholesterol derivatives obtained from food may modulate developmental gene expression patterns and dosage by augmenting NHR-23 and NHR-85 activities. A direct connection between these or other metabolites and NHR-23 and NHR-85 transcription will require functional and structural studies.
LIN-42 functions to control gene dosage through interactions with NHR-85
We propose that NHR-85 and NHR-23 function within the C. elegans developmental clock in a manner analogous to their non-orthologous circadian counterparts, CLOCK and BMAL. However, we propose an evolutionarily conserved role for Period orthologs in the negative regulation of oscillatory transcriptional activity initiated by nuclear hormone receptors (Figure 7H). We demonstrate that interactions between Period orthologs and multiple NHRs are conserved (Figure S7)52. In mammalian systems, Period binds Rev-Erb when Rev-Erb is bound to its target DNA, and this association modulates the transcriptional output of Rev-Erb target genes52. This activity is thought to directly coordinate the transcriptional regulation of metabolic genes alongside the CLOCK and BMAL1 components of the circadian clock. We demonstrate that LIN-42 negatively regulates features of lin-4 transcriptional dynamics by directly binding to NHR-85 and dampening its expression during each larval stage (Figure 7). It is unknown if the similar physical interactions between the Period and Rev-Erb (and various conserved NHRs) alter the levels or turnover rates of these NHRs in mammalian systems.
Limitations of the study
This study uses the MS2/MCP-GFP imaging system to quantify lin-4 expression in various tissue types in developing larvae. While this imaging platform enabled us to accurately measure highly pulsatile lin-4 transcriptional patterns, we cannot exclude the possibility of a reduced but constitutive transcription of lin-4 RNAs throughout larval development. Further improvements to the signal-to-noise ratio of the MS2-tethering assay combined with in-situ hybridization experiments will be needed to assess transcription features such as the absolute number of actively transcribing polymerases or the rate of miRNA or mRNA production for specific loci80,81. Nevertheless, our results demonstrate the power of the MS2/MCP-GFP approach, which enabled us to assign roles for particular transcription factors in controlling transcriptional features and gene dosage.
This manuscript also identifies the GRN that programs lin-4 transcriptional pulses in hypodermal cells and defines the basic regulatory architecture between two NHRs and the C. elegans Period ortholog (LIN-42) that mediate these transcriptional patterns. Because pulsatile lin-4 transcription occurs in cell types that don’t express NHR-23 and NHR-85, we hypothesize that additional NHRs play related roles in similarly-structured GRNs in other tissues. Consistent with this hypothesis, we demonstrate that LIN-42 interacts with many conserved NHRs. Whether LIN-42 regulates the post-transcriptional expression patterns of the 65 additional NHRs it interacts with (Figure S7) remains to be seen. If so, this would suggest that LIN-42, whose tissue expression pattern is more diverse than NHR-23 and NHR-85 (Figure S3), may coordinate other NHR-centric developmental clocks globally.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher M. Hammell (chammell@cshl.edu).
Materials Availability
Plasmids generated in this study are available upon request made to the Lead Contact. C. elegans strains generated in this study are available upon request to the Lead Contact.
Data and Code Availability
The published article includes all the datasets generated or analyzed during this study.
A public repository containing MATLAB scripts used for figures and statistical analyses in this paper can be found at https://github.com/wolfgangkeil/Kinney_Sahu_et_al_2023_code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
C. elegans maintenance and genetics
C. elegans strains were maintained on standard media at 20°C and fed E. coli OP50 82. A list of strains used in this study is provided in the Key Resources Table. Some strains were provided by the CGC, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| MAX Efficiency DH5alpha (E. coli) | Agilent | Cat#230280 |
| DH10MultiBac (E. coli) | Geneva Biotech | Cat#DH10MultiBac |
| NA22 | C. elegans Genome Center | NA22 |
| OP50 | C. elegans Genome Center | OP50 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor™ 647 NHS Ester (Succinimidyl Ester) | A20006 | ThermoFisher |
| D-desthiobiotin | Cat#D1411 | Sigma-Aldrich |
| HyClone Insect Cell Culture Media (CCM3) | Cat#16777–272 | Cytiva / VWR |
| IPTG | Cat#I2481C50 | Gold Biotechnology |
| Levamisole hydrochloride | 16595-80-5 | Sigma |
| Poly(2'-deoxyinosinic-2'-deoxycytidylic acid) sodium salt | 118578-37-3 | Sigma |
| Polyethylenimine, linear | 9002-98-6 | Sigma |
| Recombinant Protein: NHR-23 | This study | N/A |
| Recombinant Protein: strep-NHR-85 | This study | N/A |
| Recombinant Protein: ULP1* protease | This study | N/A |
| Sodium azide | 26628-22-8 | Sigma |
| Strep-Tactin superflow resin (IBA) | 2-1206-025 | IBA Technologies |
| Suprerdex200 Increase 10/300GL | 28990944 | Cytiva Life Sciences |
| Critical commercial assays | ||
| Monolith NT.115 capillaries | Nanotemper Tech | MO-K022 |
| Qiagen miniprep kit | Qiagen | Cat#27104 |
| Experimental models: Cell lines | ||
| S. cerevisiae Strain Y1HaS2: MATa, ura3-52, his3-Δ1, ade2-101, ade5, lys2-801, leu2-3,112, trpl-901, tyr1-501, gal4Δ, gal80Δ, ade5::hisG | Reece-Hoyes et al. 201185 | N/A |
| S. cerevisiae Strain PJ69-4A: MATa trp1-901 leu2- 3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. 199686 | N/A |
| Experimental models: Organisms/strains | ||
| C. elegans Strain HML1019: cshIs136[lin-4::24xMS2] I; mnCI-mCherry/cshIs139[rpl-28pro::MCP-GFP::SL2 Histone mCherry] | This study | N/A |
| C. elegans Strain HML1065: cshIs145[(ΔPCEA)lin- 4::24XMS2] I; mnCI-mCherry/cshIs139[rpl-28pro::MCP-GFP::SL2 Histone mCherry] | This study | N/A |
| C. elegans Strain VT1367: maIs105 [col-19pro::GFP] V. | Hammell et al. 20 0934 | N/A |
| C. elegans Strain HML1005: cshIs130[lin-4(FL)] I; lin-4(e912); maIs105 [col-19pro::GFP] V. | This study | N/A |
| C. elegans Strain HML979: cshIs129[minimal lin-4] I; lin-4(e912) II; maIs105 [col-19pro::GFP] V. | This study | N/A |
| C. elegans Strain HML995: cshIs131[lin-4 ΔPCE]; lin-4(e912); maIs105 [col-19pro::GFP] V. | This study | N/A |
| C. elegans Strain HML1010: cshIs134[lin-4_short]I; lin-4(e912); maIs105 [col-19pro::GFP] V. | This study | N/A |
| C. elegans Strain HML1007: cshIs133[lin-4ΔD]; lin-4(e912); maIs105 [col-19pro::GFP] V. | This study | N/A |
| C. elegans Strain HML243: wIs51 [scm::GFP} | This study | N/A |
| C. elegans Strain HML1051: cshIs131[lin-4 ΔPCE]; lin-4(e912); wIs51 [scm::GFP]] V. | This study | N/A |
| C. elegans Strain OP539: unc-119(tm4063) III; wgIs539[nhr-85::TY1::EGFP::3xFLAG + unc-119(+)]. | Caenorhabditis Genetics Center | CGC: OP539 RRID:WB-STRAIN WBCnstr00021773 |
| C. elegans Strain OP109: unc-119(ed3) III; wgIs 109[blmp-1::TY1::EGFP::3xFLAG + unc-119(+)]. | Caenorhabditis Genetics Center | CGC: OP109 RRID:WB-STRAIN WBCnstr00007730 |
| C. elegans Strain OP43: unc-119(ed3) III; wgIs43[nhr-23::TY1::EGFP::3xFLAG(92C12) + unc-119(+)]. | Caenorhabditis Genetics Center | CGC: OP43 RRID:WB-STRAIN WBCnstr00016818 |
| C. elegans Strain N2 Bristol | Brenner 197482 | CGC: N2, RRID:WB-STRAIN: WBStrain0000000 1 |
| C. elegans Strain HML990: cshIs130 [lin-4(FL)] I; lin-4(e912) | This study | N/A |
| C. elegans Strain HML994: cshIs131 [lin-4(ΔPCE)] I; lin-4(e912) | This study | N/A |
| C. elegans Strain HML1209: cshIs206 [lin-4(Δproximal)] l; lin-4(e912) | This study | N/A |
| C. elegans Strain HML1210 cshIs207[lin-4(ΔPCEΔproximal)] l; lin-4(e912) | This study | N/A |
| C. elegans Strain SV1009: heIs63 [wrt-2p::GFP::PH + wrt-2p::GFP::H2B + lin-48p::mCherry] V. | Wildwater 201183 | N/A |
| C. elegans Strain HML1217: cshls207[lin-4(ΔPCEΔpmximal)] I; lin-4(e912); heIs63 [wrt-2p::GFP::PH + wrt-2p::GFP::H2B + lin-48p::mCherry] V | This study | N/A |
| C. elegans Strain HML1163: nhr-23::mScarlet(wrd33) nhr-85::GFP(wrd25) I. | This study | N/A |
| C. elegans Strain HML445: cshIs45 [lin-42::YFP] II. | This study | N/A |
| C. elegans Strain OH16380: nlp-45(ot1032[nlp-45::T2A::GFP::H2B]) X | Sun et al, 20 2084 | N/A |
| C. elegans Strain HML1245: lin-4(e912) II; nlp-45(ot1032[nlp-45::T2A::GFP::H2B]) X | This study | N/A |
| C. elegans Strain HML1244: cshIs130 [lin-4(FL)] I; lin-4(e912) II; nlp-45(ot1032[nlp-45::T2A::GFP::H2B]) X | This study | N/A |
| C. elegans Strain HML1243: cshIs206 [lin-4(ΔPCE+Δproximal)] I; lin-4(e912) II; nlp-45(ot1032[nlp-45::T2A::GFP::H2B]) X | This study | N/A |
| Oligonucleotides | ||
| Oligo lin-4 PCE: TTTGCATCCTCATTCTCAACACCTCGTTTTTTTCCCTTTTCTTGCACAAATTGAGGTCAGTCGGTCAGTAAACCCCCCCCCCCCCCCCCCCATTGAGGTGACCAATTGGTTTTTCTTTTCCTTTACTTTCTCCTTCACTTTCTCTCTCTCGGATCACCAGC | This study | Synbio Technologies |
| Oligo C_ROR(1)REV: TTTGCATCCTCATTCTCAACACCTCGTTTTTTTCCCTTTTCTTGCACAAATTGAcctqtGTCGGTCAGTAAACCCCCCCCCCCCCCCCCCCATTGAGGTGACCAATTGGTTTTTCTTTTCCTTTACTTTCTCCTTCACTTTCTCTCTCTCGGATCACCAGC | This study | Synbio Technologies |
| Oligo C_REV: TTTGCATCCTCATTCTCAACACCTCGTTTTTTTCCCTTTTCTTGCACAAATTGAGGTCAGTCcctgtcTAAACCCCCCCCCCCCCCCCCCCATTGAGGTGACCAATTGGTTTTTCTTTTCCTTTACTTTCTCCTTCACTTTCTCTCTCTCGGATCACCAGC | This study | Synbio Technologies |
| Oligo C_ROR(2): TTTGCATCCTCATTCTCAACACCTCGTTTTTTTCCCTTTTCTTGCACAAATTGAGGTCAGTCGGTCAGTAAACCCCCCCCCCCCCCCCCCCATTGAccactCCAATTGGTTTTTCTTTTCCTTTACTTTCTCCTTCACTTTCTCTCTCTCGGATCACCAGC | This study | Synbio Technologies |
| Oligo Scramble_top: GGTCGCGATATAGGTATAACATCGA | This study | Sigma Aldrich |
| Oligo Scramble_bot: TCGAT GTTATACCT AT ATCGCGACC | This study | Sigma Aldrich |
| Oligo wtLIN-4_23_85top: ACAAATTGAGGTCAGTCGGTCAGTA | This study | Sigma Aldrich |
| Oligo wtLIN-4_23_85bot: TACTGACCGACT GACCTCAATTT GT | This study | Sigma Aldrich |
| Oligo Mut_DR1_top: ACAAATTGAcctgtGTCGGTCAGT A | This study | Sigma Aldrich |
| Oligo Mut_DR1_bot: ACTGACCGACacaggTCAATTTGT | This study | Sigma Aldrich |
| Oligo Mut_DR2_top: ACAAATTGAGGTCAGTCcctgtcTA | This study | Sigma Aldrich |
| Oligo Mut DR2 bot: TAgacaggGACTGACCTCAATTTGT | This study | Sigma Aldrich |
| Oligo Proximal_top: CGAAGCGACCGAATGACCCAGTCTC | This study | Sigma Aldrich |
| Oligo Proximal_bot: GAGACTGGGTCATTCGGTCGCTTCG | This study | Sigma Aldrich |
| Oligo Proximal mutations: GGGACCGCGGCAAAAAAGAATAACGACGAAGgctggGAAactggCAGTCTCTTCACTTCTCTACTTTCGATCCTCCTCCTTC-3’ | This study | Sigma Aldrich |
| Recombinant DNA | ||
| Plasmid pCMH2177: (lin-4(FL)24xMS2v6_Cb_unc-119) | This study | N/A |
| Plasmid pCMH2171: rpl-28pro::MCP-GFP_SL2_H2B::mCherry | This study | N/A |
| Plasmid pCMH2127: lin4(Sall3_Cb_unc-119) | This study | N/A |
| Plasmid pCMH2138: lin-4(FL)_Cb_unc-119) | This study | N/A |
| Plasmid pCMH2152: lin-4(FL)(ΔPCE)_Cb_unc-119) | This study | N/A |
| Plasmid pCMH2149: lin-4(FL)(ΔD)_Cb_unc-119) | This study | N/A |
| Plasmid pCMH2058: pMW#2 + PCE element for on-hybrid | This study | N/A |
| Plasmid pCMH1986: pAD-empty | This study | N/A |
| Plasmid pCMH1987: pDB-empty | This study | N/A |
| Plasmid pCMH1966: pAD-NHR-23 | This study | N/A |
| Plasmid pCMH2122: pAD-NHR-85 | This study | N/A |
| Plasmid pCMH1865: pAD-LIN-42b | This study | N/A |
| Plasmid pCMH1864: pAD-LIN-42a | This study | N/A |
| Plasmid pCMH1968: pDB-LIN-42b | This study | N/A |
| Plasmid pCMH1967: pDB-LIN-42a | This study | N/A |
| Plasmid pCMH2322: pDB-LIN-42a(aa37-88) | This study | N/A |
| Plasmid pJW135: pAD-NHR-25 | Ward et al. 2013 87 | N/A |
| Plasmid pJW136: pDB-NHR-25 | Ward et al. 2013 87 | N/A |
| Plasmid pCMH1434: lin-42 sgRNA in pDD122 backbone | This study | N/A |
| Plasmid pPD129.35: control RNAi | Timmons et al. 200188 | |
| Plasmid pCMH1629: nhr-23 RNAi | This study | N/A |
| Plasmid pCMH2206: ph-trep-NHR-85_p10-eGFP | This study | N/A |
| Plasmid pCMH1662: ph-strep-Sumo-NHR-23-P10-eGFP | This study | N/A |
| Software and algorithms | ||
| GraphPab Prism | Graphpad Software | V10.0.1 |
| MP Affinity Analysis | Nanotemper Technologies | v2.1.3 |
| Zen Imaging software | Zeiss | V2.5 |
| Metamorph software | Molecular Devices | V7.8 |
| Original Code | This study | https://github.com/wolfgangkeil/Kinney_Sahu_et_al_2023_code |
Yeast strains
Yeast strains were maintained on standard media as previously outlined 53,85,89.
METHOD DETAILS
RNAi Feeding
RNAi by feeding was performed using E. coli HT115 expressing double-stranded RNA corresponding to the indicated target gene or containing a control dsRNA expression plasmid that does not contain sequences corresponding to any C. elegans gene90,91. To prevent contamination by E. coli OP50, L4-staged animals were added to RNAi plates individually after removing co-transferred bacteria. For RNAi against nhr-23, bacterial cultures were diluted with control RNAi cultures at the indicated levels before experimental onset. In experiments in Fig. 2, starved L1 animals of the indicated genotypes were used. Unless otherwise noted, F1 progeny were analyzed for RNAi-induced phenotypes 48 to 60 hours later (20°C). Plasmids used for RNAi are outlined in Key Resources Table.
CRISPR genome editing
Genome editing/transgene insertion was accomplished using standard CRISPR/Cas9-mediated genomic editing to the ttTi5605 or ttTi4348 landing site following standard protocols 92. For CRISPR/Cas9 editing of the endogenous lin-42 gene, pCMH1434 (expressing Cas9 and a synthetic CRISPR guide RNA targeting a genomic region encoding the LIN-42 C-terminus) and pCMH1439 (encoding a LIN-42::YFP fragment) were injected into N2 animals and screened by PCR to identify transgene insertion at the lin-42 gene.
For CRISPR editing of the proximal NHR-23/NHR-85 binding sites, a single sgRNA with the sequence ttgcacaaattgaggtcagt (Synthgo ) and a ssDNA repair oligonucleotide of the following sequence was used:
5’-GGGACCGCGGCAAAAAAGAATAACGACGAAGgctggGAAactggCAGTCTCTTCACTTCTCTACTTTCGATCCTCCTCCTTC- 3’
Multiple independent clones were isolated, validated by sequencing, and outcrossed two times to parental strain. Each clone expressed identical phenotypes.
Yeast one-hybrid assays
Yeast one-hyid assays were performed using the wTF2.2 gal4 AD library of C. elegans transcription factors93.
Yeast two-hybrid assays
Plasmids containing target proteins fused to GAL4 DNA-binding-domain (pBD) and GAL-4 Activation Domain (pAD) were co-transformed into the pJ69–4a Y2H yeast strain86 as previously described53,89. Transformed yeast was plated on SC-TRP-LEU plates for three days. Three colonies from each transformation plate were streaked onto SC-HIS-TRP-LEU plates containing 3-AT at the indicted concentrations. Protein interactions were determined by visible growth on 3-AT conditions with negative growth in empty vector controls after three days. For the large-scale LIN-42 screen, pBD containing LIN-42a, LIN-42b, and the empty vector control were individually mated to each pAD construct from the WTF2.2 yeast library85. For visualization of results, individual colonies were grown overnight in YPD in 96-well plates. Overnight cultures were diluted 1/200 in ddH20, and 3μL was pipetted onto selective 3-AT and control plates. After three days of growth, plates were imaged on a Fotodyne FOTO/Analyst Investigator/FX darkroom imaging station.
Protein preparation
Full-length C. elegans protein NHR-23 was cloned as an N-terminal Strep-SUMO fusion protein in a pFL vector of the MultiBac Baculovirus expression system to create pCMH1662 94. This construct was expressed in insect Sf9 cells grown in CCM3 media (HyClone) at 27°C for 60 h. Cells were harvested by spinning at 2200 rpm for 20 min and resuspended in lysis/wash buffer (20 mM Tris pH 8.0, 200 mM NaCl, 5 mM BME) and a protease inhibitor cocktail before flash freezing in liquid N2. Cell pellets were stored at −80°C. Cell pellets were thawed and sonicated once. Polyethylene imine (PEI) was added at 0.2% to the lysate after cell pellets were thawed and sonicated. The lysate was then spun by ultracentrifuge at 38,000 rpm for 45 min, at 4°C. The lysate supernatant was then used for batch binding with Strep-Tactin superflow resin (IBA) for 1 hour while on a rolling shaker at 4°C. The affinity beads were harvested by spinning at 1000 rcf for 5 minutes, then resuspended in lysis/wash buffer and applied to a gravity column. The column was washed with 30 column volumes of lysis/wash buffer and 5 column volumes of ATP wash buffer (20 mM Tris pH 8.0, 200 mM NaCl, 5 mM BME, 2 mM ATP). The protein was eluted from the affinity column in two column volumes of elution buffer (20 mM Tris pH 8.0, 200 mM NaCl, 5 mM BME, 2 mM desthiobiotin). The Strep-SUMO tag was cleaved from NHR-23 by ULP1* protease overnight at 4°C. The protein was then concentrated and loaded onto a 10/300 Superdex200 Increase gel filtration column (Cytiva Life Sciences), running in lysis/wash buffer, chromatographed for ~30 mL at 0.6 mL min−1. SDS-PAGE was used to assess protein purity and cleavage efficiency.
Full-length C. elegans protein NHR-85 was cloned as an N-terminal Strep-fusion protein in a pFL vector of the MultiBac Baculovirus expression system to create pCMH2206. NHR-85 was purified using the same method as above, with the exception of the overnight N-terminal tag cleavage step.
Microscale thermophoresis analysis
Binding assays of purified NHR23 or strep-NHR85 was measured using a Monolith NT.115 Pico running MO Control version 1.6 (NanoTemper Technologies). Assays were performed in 100 mM NaCl, 20 mM Tris pH 8.0, 0.05% Tween-20. AlexaFluor647 NHS Ester (ThermoFisher Scientific) labeled NHR-23 (200 pM) was mixed with 16 serial dilutions of strep-NHR-85 starting at 31.5 uM and loaded into microscale thermophoresis premium coated capillaries (NanoTemper Technologies). MST measurements were recorded at 25°C using 30% excitation power and 60% MST power. Measurements were performed in duplicate. Determination of the binding constant was performed using MO Affinity Analysis v.2.3.
AlexaFluor647 NHS Ester (ThermoFisher Scientific) labeled strep-NHR-85 (400 pM) was mixed with 16 serial dilutions of NHR-23 starting at 625 nM. MST measurements were recorded at 25°C using 15% excitation power and 40% MST power. Measurements were performed in triplicate. Determination of the binding constant was performed using MO Affinity Analysis v.2.3.
Electrophoretic Mobility Shift Assays (EMSAs)
For larger fragment gel shifts, PCR products with free DNA, 5’ IRDye (IRDye700 or IRDye800)-labeled and unlabeled oligos were obtained from IDT (Coralville, Iowa) and used to amplify DNA probes of the indicated sequences. For wild-type probes, the indicated PCE fragments were amplified pCMH1954. For mutant probes that harbor mutations in either GGTCA repeat, synthetic DNAs were obtained from Synbio Technologies (Manmouth Junction, NJ, USA) and used to amplify the corresponding mutant DNA fragments (Table above with mutation lowercase). For binding reactions containing 161bp probes (Figure S5, recombinant proteins were incubated with gel-purified DNA probes in 10 mM Tris pH 7.5, 50 mM KCl, 1 mM DTT, 0.1mg/mL poly (dIdC) (Sigma-Aldrich), and 0.25% Tween 20 for 30 minutes at 20°C (in dark chamber). Samples were then run in a 4% native polyacrylamide gel containing 50mM Tris pH 7.5, 0.38 M glycine, and 2mM EDTA in 1x TBE buffer. Gels were imaged and quantified using a Li-Cor Odyssey Imager (Lincoln, Nebraska). For small, 25bp EMSAs, dsDNA probes (65nM) were incubated in 75mM NaCl, 20mM Tris pH 8.0, 2mM BME, 10% glycerol, and recombinant NHR-85 or recombinant NHR-23 at a 1.5 stoichiometric excess for 30 minutes at room temperature. Binding reactions were then resolved on a 5% TBE native gel, 30 min, 100V, in 0.5x TBE running buffer.
Microfluidics and long-term imaging
For microfluidics experiments, early to mid-L1-staged animals were isolated 6h after starvation-induced L1 arrest at 20°C before an experimental time course. Other stages were individually isolated by observing defined cellular and morphological features indicative of animals’ developmental stage 12. Animals were mounted into the microfluidic device as previously described 30. During imaging, animals were constantly fed NA22 E. coli suspended in S medium. The temperature was kept constant at 20°C both at the objective and the microfluidic device using a custom-built water-cooled aluminum ring (for the objective) and a custom-built aluminum stage inset directly coupled to a thermal Peltier device.
Image acquisition
MS2-MCP-GFP live imaging.
Live imaging was performed with a 60x, 1.2NA objective on a Nikon Ti2 Eclipse microscope equipped with a V3 CREST spinning disk confocal module. To ensure fast multichannel acquisition, hardware triggering was implemented between a MadCityLabs NANO Z200-N piezo z-stage, a Photometrics Prime 95B sCMOS camera with 25mm field of view (2048×2048 pixels, pixel size 11um corresponding to 183nm), and a Lumencor© Celesta solid-state laser source via a National Instruments (NI) PCIe-6323 card. Laser wavelengths of 488nm and 545nm were used to excite MCP-GFP and histone-mCherry, respectively. Acquiring a dual-color z-stack with 51 slices and 50ms exposure times takes approximately 3.2 seconds with this setup.
Confocal Microscopy:
Images were acquired using a Hamamatsu Orca EM-CCD camera and a Borealis-modified Yokagawa CSU-10 spinning disk confocal microscope (Nobska Imaging, Inc.) with a Plan-APOCHROMAT x 100/1.4 or 40/1.4 oil DIC objective controlled by MetaMorph software (version: 7.8.12.0). Animals were anesthetized on 5% agarose pads containing 10mM sodium azide and secured with a coverslip. Imaging on the microfluidic device was performed on a Zeiss AXIO Observer.Z7 inverted microscope using a 40X glycerol immersion objective and DIC and GFP filters controlled by ZEN software (version 2.5). Images were captured using a Hamamatsu C11440 digital camera. For scoring plate-level phenotypes, images were acquired using a Moticam CMOS (Motic) camera attached to a Zeiss dissecting microscope.
Wide-field Fluorescence microscopy:
Images were acquired with a Zeiss Axio Observer microscope equipped with Nomarski and fluorescence optics as well as a Hamamatsu Orca Flash 4.0 FL Plus camera. An LED lamp emitting at 470 nm was used for fluorophore excitation. For single images, animals were immobilized on 2% agarose pads supplemented with 100mM Levamisole (Sigma). For single images, animals were immobilized on 2% agarose pads supplemented with 100mM Levamisole (Sigma). For long-term imaging methods, see Microfluidics and Long-term Imaging section.
Fluorescent Reporter Quantification
Reporter lines were imaged using wide-field fluorescence or confocal microscopy, as described above. The average intensity (arbitrary units) per seam cell was measured using ImageJ. The measurement of the fluorescent intensity of the nucleus minus the intensity of a background sample determined each seam cell intensity. The average of three seam cells determined the fluorescent intensity of each animal. Ten animals per developmental stage were imaged unless otherwise noted.
MCP-GFP live imaging
Long-term imaging:
For long-term live imaging across several larval stages, animals were mounted in a microfluidic chamber 6h after L1 arrest and grown on NA22 bacteria suspended in S-medium until mid-L4 as previously described 30. At each time point, animals were reversibly immobilized using microfluidic pressures and flows. A z-stack of 51 images separated by 0.5um was acquired at four overlapping positions, covering the entire microfluidic chamber. Thereafter, the animal was released from immobilization and left to roam and feed freely until the next time point.
Short term imaging:
For short-term live imaging (Figs. 1, 4 & 5) developmentally staged animals were mounted into the microfluidic chamber as previously described 30, a few hours before the expected appearance of MS2 spots. Minutes before the appearance of MS2 spots, animals were immobilized using microfluidic pressures and flows and kept immobilized for the entire experiment. This enabled automated analyses to maintain a stable worm position (see below). At each time point (every 4min), a stack of 21 images separated by 0.5um was acquired at four overlapping positions, covering the entire microfluidic chamber. Occasionally, animals arrested development upon prolonged immobilizations, as evidenced by the cessation of germline divisions (L2-L4 larvae), the cell-cycle arrest of vulval precursor cells (VCPs) (L3 larvae), or by failure to advance through vulval morphogenesis (L4 larvae). These animals were excluded from further analysis. As opposed to all other genotypes imaged, nhr-85(0) mutants animals exhibited a pronounced tendency to roll under these imaging conditions, precluding MS2 spot tracking within nuclei of the lateral hypodermis.
MCP-GFP live imaging data analysis
Short-term imaging:
All events (cell divisions, onset, and offset of MCP-GFP spots) were scored manually in the time series. For short-term live imaging, all analysis was performed using custom-written FIJI macros, and pixel classification with random forest trees in Ilastik and MATLAB© scripts. The main challenge in this analysis is residual animal movement between time points and the low signal-to-noise ratio of the MCP-GFP signal.
Long-term imaging and 3D tracking:
First, the worm backbone was detected in each frame by skeletonization, using a thresholded probability map obtained by processing a maximum z-projection of the MCP-GFP channel through a custom-trained Ilastik pixel classifier. Next, computational straightening was performed along this backbone 30 to obtain a time series of straightened worm z-stacks. This straightened time series has the advantage that residual movement is primarily along the anteroposterior animal axis. Next, we divided the straightened worm z-stack along the worm axis into overlapping (20%) segments of 500 pixels (~91um) in length. Each of these segments was then manually registered to obtain z-stacks in which tracking of almost all hypodermal nuclei could automatically performed with minimal user corrections. The histone-mCherry signal was augmented using a custom-trained Ilastik pixel classifier to improve the nuclear signal for segmentation. 3D tracking and a manual correction were performed on the resulting Ilastik probability maps using the FIJI TrackMate plugin 95 with LoG detector and LAP tracker. For each segment, frames with substantial animal movement between z-slices were excluded from subsequent analysis. Whenever nuclei were tracked twice (due to overlapping worm segments), the nucleus with the most tracked time points was chosen for subsequent MCP-GFP spot analysis.
MCP-GFP spot tracking and intensity quantification
MS2 spots were tracked in each tracked nucleus, using the FIJI TrackMate plugin with LoG detector and LAP tracker and the Ilastik nucleus probability maps as a mask to include only spots inside nuclei. Each MCP-GFP spot track was manually corrected, and spots were added to frames occasionally, in which the TrackMate LoG detector failed to detect them. For quantification of MCP-GFP spot intensities, a 2D Gaussian fit to the maximum z-projection of three z-slices around the peak slice determines the position of the spot. The background was calculated as the average intensity in a ring between 3 and 5 pixels away from the spot position. The spot intensity was calculated by integrating the fluorescence over a circle with a radius of 2 pixels around the spot position and subtracting the estimated background.
MS2-MCP-GFP trace analysis
MCP-GFP traces were smoothed with a Gaussian width of 1.5 frames (6 minutes). Using this filtered trace, duration and maximum intensity during the transcriptional pulse were determined for each tracked locus. Loci were considered “ON” if spot intensity was above 150 counts and “OFF” if below. The duration of transcription for each locus was determined as the interval between the first “ON” time point and the last. Typically, loci stayed “ON” for the entire duration of the transcriptional pulse within a given larval stage.
Bioinformatic analysis of BLMP-1, NHR-82, and NHR-23 ChIP-seq data
ChIP-seq short reads were first clipped off adaptor sequences. Reads of a minimum of 22bp were mapped to the UCSC C. elegans genome (ce10) using bowtie program76 96, looking for unique alignments with no more than two mismatches. MACS program (v1.4)97 was used for peak calling with a significant p-value cutoff equaling 1e-5. Target annotations were based on WormBase (version 220) using customized R scripts and Bioconductor packages. ModeEncode data sets98 (BLMP-1(ENCFF108AEB and ENCFF615AMZ), NHR-23(ENCFF019QMH) and NHR-85(ENCFF018KHA)) were used in the initial analysis, and we defined the promoter region as upstream 3kbp to downstream 300bp around the transcription start site (TSS). Peaks located in promoter regions were annotated to their closest TSS sites for coding and non-coding genes (Table S1). These potential targets were then overlapped with sets of oscillatory genes identified in a previous mRNA-seq-based study 23. Overlapping target gene sets organized in Figure 2D are from postembryonic ChIP-seq samples for each TF. Raw data for each of the 265 ModEncode data sets (annotation numbers listed individually in Table S1) used in Figure 2E were downloaded from modENCODE98 and processed in the same way as BLMP-1, NHR-23, and NHR-85 datasets22. ModEncode-derived peaks from each TF ChIP-seq dataset were compared to identify common sites with at least one base pair overlapping using BEDTools. All ChIP-seq mapping graphs and images were produced in R by customized scripts.
QUANTIFICATION AND STATISTICAL ANALYSIS. QUANTIFICATION AND STATISTICAL ANALYSIS
Plots and diagrams were generated using GraphPad Prism v9 (GraphPad Software, San Diego, Ca) or custom-written MATLAB® scripts. Statistical significance was determined using a two-tailed unpaired Student’s t-test. P<0.05 was considered statistically significant. **** indicates P < 0.0001.
Supplementary Material
Video S1: Time-laps video depicting the formation of MCP-GFP/lin-4::24xMS2 transcriptional foci in hypodermal cells of a transgenic L3-staged animal, related to Figure 1.
Video S2: Time-laps video depicting the formation of MCP-GFP/lin-4::24xMS2 transcriptional foci in vulval precursor cells of a transgenic L3-staged animal, related to Figure 1.
Table S1: Consolidation of NHR-23-GFP, NHR-85-GFP, and BLMP-1::GFP ChIP-seq data set analysis from ModEncode data, related to Figure 3.
Table S2: Analysis of ModEncode ChIP-seq data set analysis parsing transcription factor binding sites to target genes and expression classes, related to Figure 3.
Highlights.
lin-4 miRNA transcription is highly dynamic with ~100 min pulses per larval stage.
NHR-85/NHR-23 heterodimers bind cooperatively to the upstream lin-4 enhancers.
The duration of NHR-85/NHR-23 dimerization controls lin-4 transcriptional dynamics.
LIN-42 binds directly to NHR-85 to dynamically modulate lin-4 transcriptional dosage.
ACKNOWLEDGEMENTS:
C.M.H is funded by the National Institutes of Health R01GM117406, National Science Foundation 2217560, and Cold Spring Harbor Laboratory. The CSHL Cancer Training Grant T32CA148056 supported B.K.. W.K. was funded by the CNRS ATIP/Avenir program and the Conseil Regional d’Île de France (DIM ELICIT-AAP-2020, 20002719). S.S. was supported by the Fondation pour la Recherche Médicale FDT202204015083. L.J. is an investigator of the Howard Hughes Medical Institute. We want to thank M. Walhout, J. Ward, J. Kimble, and M Barkoulas for strains and reagents. We would like to also thank G. Sumati for recombinant protein expression.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ebisuya M, and Briscoe J (2018). What does time mean in development? Development 145. 10.1242/dev.164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filicori M (1999). The role of luteinizing hormone in folliculogenesis and ovulation induction. Fertil Steril 71, 405–414. 10.1016/s0015-0282(98)00482-8. [DOI] [PubMed] [Google Scholar]
- 3.Palmeirim I, Henrique D, Ish-Horowicz D, and Pourquie O (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 4.Pourquie O (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663. 10.1016/j.cell.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuda M, Hayashi H, Garcia-Ojalvo J, Yoshioka-Kobayashi K, Kageyama R, Yamanaka Y, Ikeya M, Toguchida J, Alev C, and Ebisuya M (2020). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455. 10.1126/science.aba7668. [DOI] [PubMed] [Google Scholar]
- 6.Isshiki T, Pearson B, Holbrook S, and Doe CQ (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521. 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 7.Maurange C, Cheng L, and Gould AP (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891–902. 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 8.Ito H, Kakishima S, Uehara T, Morita S, Koyama T, Sota T, Cooley JR, and Yoshimura J (2015). Evolution of periodicity in periodical cicadas. Sci Rep 5, 14094. 10.1038/srep14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon C, Cooley JR, Karban R, and Sota T (2022). Advances in the Evolution and Ecology of 13- and 17-Year Periodical Cicadas. Annual Review of Entomology 67, 457–482. 10.1146/annurev-ento-072121-061108. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V (2011). MicroRNAs and developmental timing. Current opinion in genetics & development 21, 511–517. 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57. [DOI] [PubMed] [Google Scholar]
- 12.Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental biology 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 13.Rougvie AE, and Moss EG (2013). Developmental Transitions in C. elegans Larval Stages. Curr Top Dev Biol 105, 153–180. 10.1016/B978-0-12-396968-2.00006-3. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V, and Horvitz HR (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- 15.Rougvie AE (2001). Control of developmental timing in animals. Nature reviews. Genetics 2, 690–701. [DOI] [PubMed] [Google Scholar]
- 16.Feinbaum R, and Ambros V (1999). The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Developmental biology 210, 87–95. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Jones-Rhoades MW, Lau NC, Bartel DP, and Rougvie AE (2005). Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Developmental cell 9, 415–422. 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Perales R, King DM, Aguirre-Chen C, and Hammell CM (2014). LIN-42, the Caenorhabditis elegans PERIOD homolog, Negatively Regulates MicroRNA Transcription. PLoS genetics 10, e1004486. 10.1371/journal.pgen.1004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Wynsberghe PM, and Pasquinelli AE (2014). Period homolog LIN-42 regulates miRNA transcription to impact developmental timing. Worm 3, e974453. 10.4161/21624054.2014.974453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D.h., Grün D, and van Oudenaarden A (2013). Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet 45, 1337–1344. 10.1038/ng.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCulloch KA, and Rougvie AE (2014). Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression. Proceedings of the National Academy of Sciences of the United States of America 111, 15450–15455. 10.1073/pnas.1414856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stec N, Doerfel K, Hills-Muckey K, Ettorre VM, Ercan S, Keil W, and Hammell CM (2021). An Epigenetic Priming Mechanism Mediated by Nutrient Sensing Regulates Transcriptional Output during C. elegans Development. Curr Biol 31, 809–826 e806. 10.1016/j.cub.2020.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendriks G-J, Gaidatzis D, Aeschimann F, and Großhans H (2014). Extensive Oscillatory Gene Expression during C. elegans Larval Development. Mol Cell 53, 380–392. 10.1016/j.molcel.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Meeuse MW, Hauser YP, Morales Moya LJ, Hendriks GJ, Eglinger J, Bogaarts G, Tsiairis C, and Grosshans H (2020). Developmental function and state transitions of a gene expression oscillator in Caenorhabditis elegans. Mol Syst Biol 16, e9498. 10.15252/msb.20209498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon M, Gardner HF, Miller EA, Deshler J, and Rougvie AE (1999). Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141–1146. [DOI] [PubMed] [Google Scholar]
- 26.Monsalve GC, Van Buskirk C, and Frand AR (2011). LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Current biology : CB 21, 2033–2045. 10.1016/j.cub.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 27.Tutucci E, Vera M, Biswas J, Garcia J, Parker R, and Singer RH (2018). An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods 15, 81–89. 10.1038/nmeth.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wightman B, Ha I, and Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- 29.Bracht JR, Van Wynsberghe PM, Mondol V, and Pasquinelli AE (2010). Regulation of lin-4 miRNA expression, organismal growth and development by a conserved RNA binding protein in C. elegans. Developmental biology 348, 210–221. 10.1016/j.ydbio.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil W, Kutscher LM, Shaham S, and Siggia ED (2016). Long-Term High-Resolution Imaging of Developing C. elegans Larvae with Microfluidics. Developmental cell. 10.1016/j.devcel.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filina O, Demirbas B, Haagmans R, and van Zon JS (2022). Temporal scaling in C. elegans larval development. Proc Natl Acad Sci U S A 119, e2123110119. 10.1073/pnas.2123110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson DR, Singer RH, and Zenklusen D (2009). A single molecule view of gene expression. Trends in cell biology 19, 630–637. 10.1016/j.tcb.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding L, Spencer A, Morita K, and Han M (2005). The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19, 437–447. [DOI] [PubMed] [Google Scholar]
- 34.Hammell CM, Lubin I, Boag PR, Blackwell TK, and Ambros V (2009). nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burris TP (2008). Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol 22, 1509–1520. 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, and Sluder AE (2004). Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol 266, 399–416. 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragle JM, Aita AL, Morrison KN, Martinez-Mendez R, Saeger HN, Ashley GA, Johnson LC, Schubert KA, Shakes DC, and Ward JD (2020). The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis. Development 147. 10.1242/dev.193862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostrouchova M, Krause M, Kostrouch Z, and Rall JE (2001). Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 98, 7360–7365. 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R, Galagali H, Kim JK, and Frand AR (2022). Feedback between a retinoid-related nuclear receptor and the let-7 microRNAs controls the pace and number of molting cycles in C. elegans. Elife 11. 10.7554/eLife.80010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacNeil LT, Watson E, Arda HE, Zhu LJ, and Walhout AJM (2013). Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252. 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes GD, Frand AR, and Ruvkun G (2006). The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133, 4631–4641. 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 43.Kouns NA, Nakielna J, Behensky F, Krause MW, Kostrouch Z, and Kostrouchova M (2011). NHR-23 dependent collagen and hedgehog-related genes required for molting. Biochemical and biophysical research communications 413, 515–520. 10.1016/j.bbrc.2011.08.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meeuse MWM, Hauser YP, Nahar S, Smith AAT, Braun K, Azzi C, Rempfler M, and Grosshans H (2023). C. elegans molting requires rhythmic accumulation of the Grainyhead/LSF transcription factor GRH-1. EMBO J 42, e111895. 10.15252/embj.2022111895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson LC, Vo AA, Clancy JC, Aguilera J, Levenson MT, Wohlenberg C, Rechtsteiner A, Ragle JM, and Ward JD (2022). NHR-23 activity is necessary for developmental progression and apical extracellular matrix structure and function. bioRxiv, 2021.2010.2027.465992. 10.1101/2021.10.27.465992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forman BM, Umesono K, Chen J, and Evans RM (1995). Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81, 541–550. 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 47.Perlmann T, Umesono K, Rangarajan PN, Forman BM, and Evans RM (1996). Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol 10, 958–966. 10.1210/mend.10.8.8843412. [DOI] [PubMed] [Google Scholar]
- 48.Rastinejad F, Perlmann T, Evans RM, and Sigler PB (1995). Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375, 203–211. 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 49.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Perez N, et al. (2022). JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 50, D165–D173. 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee RC, Feinbaum RL, and Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 51.Mok DZL, Sternberg PW, and Inoue T (2015). Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC developmental biology 15, 26. 10.1186/s12861-015-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, and Albrecht U (2010). The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24, 345–357. 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walhout AJ, Boulton SJ, and Vidal M (2000). Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast 17, 88–94. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tennessen JM, Opperman KJ, and Rougvie AE (2010). The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development 137, 3501–3511. 10.1242/dev.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammell CM, Karp X, and Ambros V (2009). A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 106, 18668–18673. 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, and Antebi A (2009). Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324, 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee C, Shin H, and Kimble J (2019). Dynamics of Notch-Dependent Transcriptional Bursting in Its Native Context. Dev Cell 50, 426–435 e424. 10.1016/j.devcel.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lammers NC, Galstyan V, Reimer A, Medin SA, Wiggins CH, and Garcia HG (2020). Multimodal transcriptional control of pattern formation in embryonic development. Proc Natl Acad Sci U S A 117, 836–847. 10.1073/pnas.1912500117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falo-Sanjuan J, Lammers NC, Garcia HG, and Bray SJ (2019). Enhancer Priming Enables Fast and Sustained Transcriptional Responses to Notch Signaling. Dev Cell 50, 411–425 e418. 10.1016/j.devcel.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada S, Whitney PH, Huang SK, Eck EC, Garcia HG, and Rushlow CA (2019). The Drosophila Pioneer Factor Zelda Modulates the Nuclear Microenvironment of a Dorsal Target Enhancer to Potentiate Transcriptional Output. Curr Biol 29, 1387–1393 e1385. 10.1016/j.cub.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, and Bradfield CA (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hogenesch JB, Gu YZ, Jain S, and Bradfield CA (1998). The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A 95, 5474–5479. 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. (1997). Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reick M, Garcia JA, Dudley C, and McKnight SL (2001). NPAS2: an analog of clock operative in the mammalian forebrain. Science 293, 506–509. 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 65.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, and Schibler U (2002). The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 66.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. (2002). A transcription factor response element for gene expression during circadian night. Nature 418, 534–539. 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 67.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, and Hogenesch JB (2004). A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537. 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 68.Francois P, Despierre N, and Siggia ED (2012). Adaptive temperature compensation in circadian oscillations. PLoS Comput Biol 8, e1002585. 10.1371/journal.pcbi.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambros V, and Horvitz HR (1987). The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes & development 1, 398–414. [DOI] [PubMed] [Google Scholar]
- 70.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, and Okamura H (1997). Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053. 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 71.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, and Menaker M (2001). Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493. 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 72.Crosby P, Hamnett R, Putker M, Hoyle NP, Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA, et al. (2019). Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell 177, 896–909 e820. 10.1016/j.cell.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindler AJ, Baugh LR, and Sherwood DR (2014). Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS genetics 10, e1004426. 10.1371/journal.pgen.1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baugh LR, and Hu PJ (2020). Starvation Responses Throughout the Caenorhabditis elegans Life Cycle. Genetics 216, 837–878. 10.1534/genetics.120.303565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santori FR, Huang P, van de Pavert SA, Douglass EF, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, et al. (2015). Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell metabolism 21, 286–298. 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, and Rastinejad F (2007). Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14, 1207–1213. 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, and Lazar MA (2007). Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789. 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 78.Chitwood DJ, and Lusby WR (1991). Metabolism of plant sterols by nematodes. Lipids 26, 619–627. 10.1007/BF02536426. [DOI] [PubMed] [Google Scholar]
- 79.Rao AU, Carta LK, Lesuisse E, and Hamza I (2005). Lack of heme synthesis in a free-living eukaryote. Proc Natl Acad Sci U S A 102, 4270–4275. 10.1073/pnas.0500877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stavreva DA, Garcia DA, Fettweis G, Gudla PR, Zaki GF, Soni V, McGowan A, Williams G, Huynh A, Palangat M, et al. (2019). Transcriptional Bursting and Co-bursting Regulation by Steroid Hormone Release Pattern and Transcription Factor Mobility. Mol Cell 75, 1161–1177 e1111. 10.1016/j.molcel.2019.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia HG, Tikhonov M, Lin A, and Gregor T (2013). Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol 23, 2140–2145. 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wildwater M, Sander N, de Vreede G, and van den Heuvel S (2011). Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 138, 4375–4385. 10.1242/dev.066431. [DOI] [PubMed] [Google Scholar]
- 84.Sun H, and Hobert O (2021). Temporal transitions in the post-mitotic nervous system of Caenorhabditis elegans. Nature 600, 93–99. 10.1038/s41586-021-04071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, and Walhout AJ (2011). Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat Methods 8, 1059–1064. 10.1038/nmeth.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.James P, Halladay J, and Craig EA (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward JD, Bojanala N, Bernal T, Ashrafi K, Asahina M, and Yamamoto KR (2013). Sumoylated NHR-25/NR5A regulates cell fate during C. elegans vulval development. PLoS Genet 9, e1003992. 10.1371/journal.pgen.1003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Timmons L, Court DL, and Fire A (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- 89.Walhout AJ, and Vidal M (2001). High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24, 297–306. 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 90.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, and Ahringer J (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 91.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 92.Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature methods 10, 1028–1034. 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. (2006). A gene-centered C. elegans protein-DNA interaction network. Cell 125, 1193–1205. 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 94.Bieniossek C, Richmond TJ, and Berger I (2008). MultiBac: multigene baculovirus-based eukaryotic protein complex production. Current protocols in protein science Chapter 5, Unit 5.20–25.20.26. 10.1002/0471140864.ps0520s51. [DOI] [PubMed] [Google Scholar]
- 95.Ershov D, Phan MS, Pylvanainen JW, Rigaud SU, Le Blanc L, Charles-Orszag A, Conway JRW, Laine RF, Roy NH, Bonazzi D, et al. (2022). TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat Methods 19, 829–832. 10.1038/s41592-022-01507-1. [DOI] [PubMed] [Google Scholar]
- 96.Borden KL (1998). RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochemistry and cell biology = Biochimie et biologie cellulaire 76, 351–358. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137–139. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Araya CL, Kawli T, Kundaje A, Jiang L, Wu B, Vafeados D, Terrell R, Weissdepp P, Gevirtzman L, Mace D, et al. (2014). Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512, 400–405. 10.1038/nature13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Time-laps video depicting the formation of MCP-GFP/lin-4::24xMS2 transcriptional foci in hypodermal cells of a transgenic L3-staged animal, related to Figure 1.
Video S2: Time-laps video depicting the formation of MCP-GFP/lin-4::24xMS2 transcriptional foci in vulval precursor cells of a transgenic L3-staged animal, related to Figure 1.
Table S1: Consolidation of NHR-23-GFP, NHR-85-GFP, and BLMP-1::GFP ChIP-seq data set analysis from ModEncode data, related to Figure 3.
Table S2: Analysis of ModEncode ChIP-seq data set analysis parsing transcription factor binding sites to target genes and expression classes, related to Figure 3.
Data Availability Statement
The published article includes all the datasets generated or analyzed during this study.
A public repository containing MATLAB scripts used for figures and statistical analyses in this paper can be found at https://github.com/wolfgangkeil/Kinney_Sahu_et_al_2023_code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


