Abstract
Porous tissue-engineered 3D-printed scaffolds are a compelling alternative to autografts for the treatment of large periorbital bone defects. Matching the defect-specific geometry has long been considered an optimal strategy to restore pre-injury anatomy. However, studies in large animal models have revealed that biomaterial-induced bone formation largely occurs around the scaffold periphery. Such ectopic bone formation in the periorbital region can affect vision and cause disfigurement. To enhance anatomic reconstruction, we introduced geometric mismatches in the scaffolds used to treat full thickness zygomatic defects created bilaterally in adult Yucatan minipigs. We used 3D-printed, anatomically-mirrored scaffolds in combination with autologous stromal vascular fraction of cells (SVF) for treatment. We developed an advanced image-registration workflow to quantify the post-surgical geometric mismatch and correlate it with the spatial pattern of the regenerating bone. Osteoconductive bone growth on the dorsal and ventral aspect of the defect enhanced scaffold integration with the native bone while medio-lateral bone growth led to failure of the scaffolds to integrate. We found a strong positive correlation between geometric mismatch and orthotopic bone deposition at the defect site. The data suggested that strategic mismatch >20% could improve bone scaffold design to promote enhanced regeneration, osseointegration and long-term scaffold survivability.
Keywords: Yucatan pigs, midfacial, scaffolds, geometric mismatch, image registration, bone regeneration
Graphical Abstract
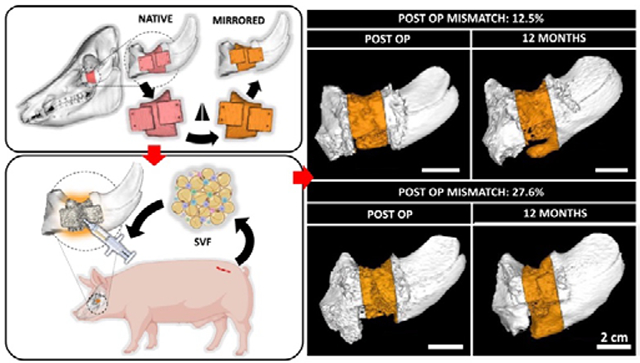
Using an advanced CT based imaging workflow, we demonstrate that post-operative geometric mismatch can promote orthotopic bone formation in clinically-sized periorbital bone defects treated with tissue-engineered scaffolds. Our findings suggest that strategic introduction of geometric mismatch (>20%) can potentially enhance bone regeneration in periorbital spaces and should be considered as a fundamental element of scaffold design.
INTRODUCTION:
Each year around 3 million cases of facial trauma (bony fractures requiring operative intervention) occur in the United States.[1] Developing successful biomaterial-based strategies to facilitate the anatomic reconstruction of midfacial bones is challenging due to their complex geometry and intimate association with sensory organs.[2,3] Clinical computed tomography (CT) imaging combined with 3D-printing technologies enable the manufacture of patient- and defect-specific porous scaffolds to promote bone regeneration. The premise of this strategy is that customized, macroscale architecture of the scaffold would adequately restore the pre-injury anatomy as new bone grows into and through the pore spaces to facilitate fracture healing and osseointegration. Consequently, significant effort has gone towards determining the optimum pore architecture[4,5] and pore size[6] conducive for new bone ingrowth. Although there are contradictory findings in the literature, the consensus is that optimum pore sizes to facilitate bone regeneration range from 200 – 1000 μm.[7–12] However, these findings are derived mostly from in vitro studies or small animal models and do not extrapolate well to large animal models of bone injury.
In large animal models,[13–18] there is considerable evidence demonstrating that new bone predominantly grows on the periphery of scaffolds extraneous to the defined macroscopic contours of the scaffolds. For instance, when critical-sized, segmental defects in sheep tibia were treated with the combination of porous polymeric scaffolds and autologous stem cells, bone growth was substantially higher outside the scaffold boundaries compared to within the interior.[13] Similarly, when multimodal porous titanium scaffolds (pore size: 80–1300 μm) designed to match the native bone stiffness were used to treat ovine femoral condylar defects, limited bone ingrowth into the scaffold pores was observed:[19] Fine woven bone was detected within the scaffold pores whereas lamellar bone was found pre-dominantly at the scaffold-to-native bone interface. Most recently, our group analyzed micro-computed tomography (μCT) data and confirmed two distinct patterns of bone regeneration into scaffolds implanted into 2 cm full-thickness, segmental, periorbital defects in Yucatan pigs. Specifically, the majority of newly deposited mineral appeared to originate from the adjacent native bone and grew around the scaffolds (osteoconductive bone) while smaller nodules accumulated by nucleating around the struts in the scaffold interior (osteoinductive bone) (Fig. S1).[20]
Understanding the spatial patterns of scaffold-mediated bone growth has significant implications for adequately and effectively treating midfacial bone injuries where the anatomic geometry is critical to function and aesthetics. Orthotopic bone regeneration is necessary for anatomically correct reconstruction as any major ectopic bony formation in the periorbital region may negatively affect vision. In the current study, we assessed the treatment of 2 cm full-thickness periorbital bilateral bone defects in adult, male Yucatan minipigs with geometrically mismatched scaffolds (comprised of polycaprolactone (PCL), 30% decellularized bone (DCB), and 20% polyvinyl alcohol (PVA) microtanks (μtanks)) infused with autologous stromal vascular fraction (SVF) harvested from adipose tissue of the animal. Half of the scaffolds were placed in chambers with pressurized oxygen (+O2) prior to implantation and compared to scaffolds of the identical composition that were not placed in chambers (−O2). We hypothesized that (1) deficits in the scaffold geometry relative to the native anatomy would facilitate increased osteoconductive bone infiltration and (2) local oxygen delivery from the μtanks would enhance bone regeneration at the defect site leading to better reconstructive outcomes.
RESULTS
Mirroring of native anatomy introduces geometric mismatch at defect site
The study utilized eight male Yucatan minipigs (Table S1). Each minipig was > 1 year old and castrated. CT images were taken of each skull prior to surgery. The native anatomical contours of the zygomatic arch were used as a template to design cutting guides and scaffolds. To introduce geometric mismatches, we mirrored the native bone anatomy around the midpoint in the coronal plane (Fig. 1A). The resulting geometries were used to 3D-print PCL-DCB-μtank scaffolds in a lattice structure with 800 μm isotropic pores. Full segmental 2 cm critical-sized periorbital defects were created bilaterally in the zygomatic bone of Yucatan minipigs and treated using geometrically mismatched scaffolds. Following implantation into the defects, the scaffolds were infused with SVF (2.25 ± 0.95 × 106 cells/ml) suspended in diluted fibrin hydrogel (TISSEEL)(Table S2). The surgical site and the SVF isolation site healed normally post-operatively (Fig. S2). CT scans of the animal skulls were acquired at 3, 6, and 12 months post-surgery (Fig. 1B). Scaffolds were often trimmed during surgery to facilitate their implantation within the defect site. Post-operative CT scans were used to quantify geometric mismatch. Analysis revealed that post-operative geometrical mismatch at the defect site varied a lot from defect to defect which might stem from a combination of factors like intraoperative sculpting of the scaffolds and difference in individual anatomy. Distinct spatial regenerative patterns were observed in defects with significantly different geometric mismatch values (Fig. 1C).
Figure 1. Design of geometrically mismatched scaffolds to treat critical-sized periorbital bone defects in a porcine model.
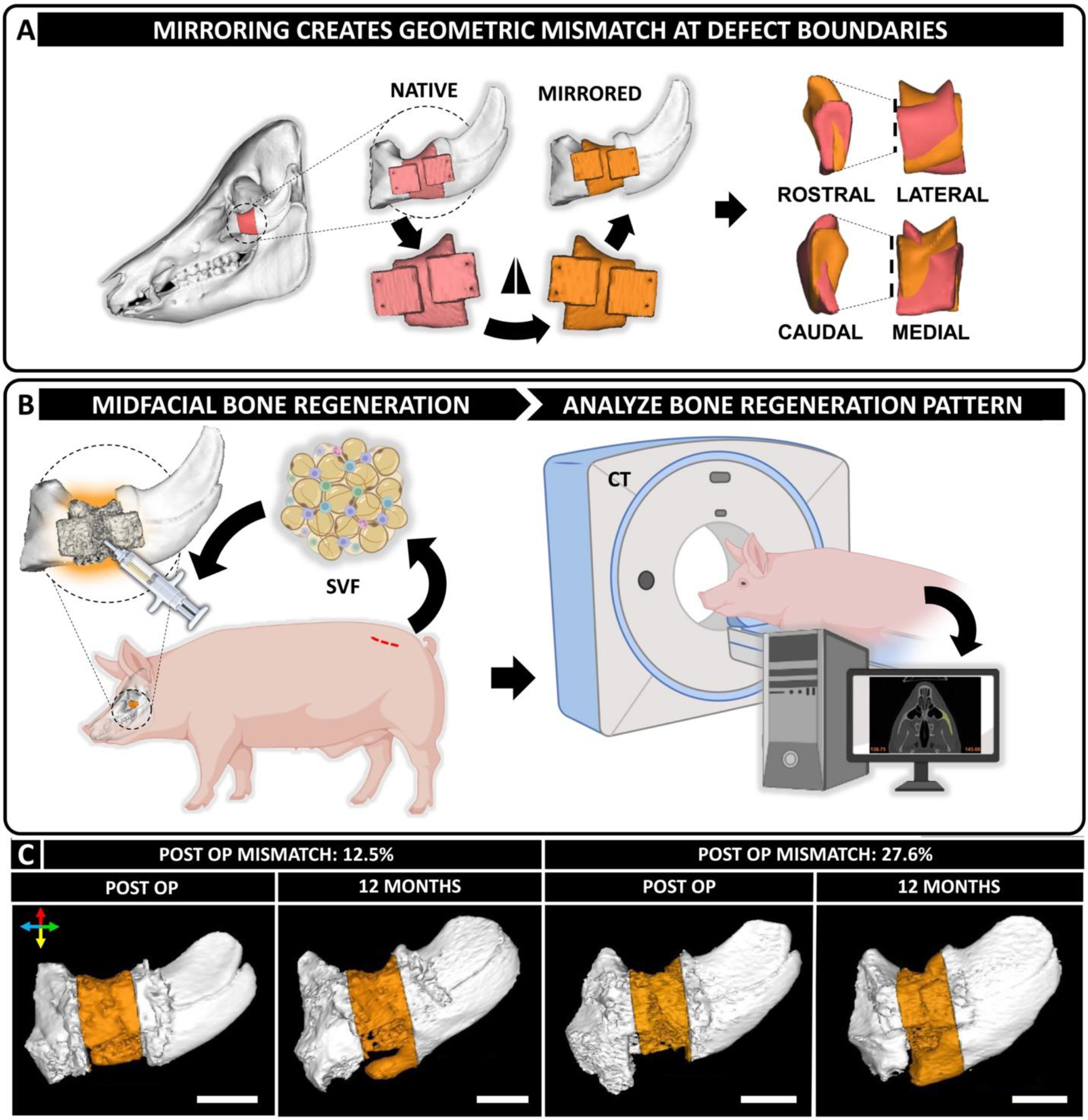
(A) CT scans of the porcine skulls were acquired pre-operatively and used as a template to design scaffolds. Native anatomy of the periorbital zygomatic bone was digitally mirrored along the coronal plane to introduce geometric mismatch at the defect site. (B) Treatment of 2 cm full thickness segmental defects in the zygomatic bone of Yucatan minipigs with geometrically mismatched scaffolds in combination with autologous SVF isolated from adipose tissue harvested from dorsal lumbar region of the animals. CT scans of the defect site and post-operative analysis are used to analyze and correlate bone regeneration pattern with anatomical accuracy of the reconstruction therapy. (C) Varying degree of post-operative geometric mismatch was obtained at the defect site post-reduction. Representative images demonstrating distinct spatial regenerative patterns of bone in defects with dissimilar post-operative geometric mismatch. Colored arrows convey orientation: dorsal (red), ventral (yellow), rostral (blue) and caudal (green). Scale bar = 2 cm. Created with BioRender.com.
Oxygen delivery enhances osteoinductive bone formation but not overall bone regeneration
Two experimental groups (−O2 and +O2) were considered for this study (Table S1). Large bone defects treated with tissue-engineered constructs could potentially develop a hypoxic environment which can negatively impact bone regeneration. We developed oxygen delivering scaffolds by incorporating hollow biodegradable microspheres called μtanks[21,22] within the PCL-DCB scaffold struts to store hyperbarically-loaded oxygen (Fig. S3). Scaffolds were made porous by incorporation of interconnected, isotropic 800 pores within the scaffold body (Fig. S4). Oxygen loaded (+O2) scaffolds demonstrated oxygen release for a period of ~2 hours (measured using a Seahorse XF Flux Analyzer as previously described[22]) (Fig. 2A). To perform quantitative CT analysis, bone volume/defect volume (BV/DV) at the defect site was calculated. Comparison of BV/DV at 12 months did not indicate any significant difference (-O2: 0.86 ± 0.12 and +O2: 0.92 ± 0.26) between the groups, however, bone growth was observed throughout (Fig. 2B, S6–9). Key hematological indicators suggested no systemic immune response in either of the groups (Fig. S5). Following euthanasia, mechanical testing was performed on harvested zygomas to assess osseointegration of the scaffolds with the native bone. No significant difference was observed in the fracture torque (-O2: 8.55 ± 3.31 Nm and +O2: 6.04 ± 1.01 Nm) between the two groups, indicating comparable osseointegration (Fig. 2C). Finally, to investigate how oxygen delivery affects bone formation within the scaffold pores, high resolution μCT scans of the defect sites were performed. Analysis of the μCT scans revealed the presence of bone nodules nucleating within the scaffold pores (Fig. 2D, Fig. S10). Quantification indicated higher bone nodule volume within the +O2 group, although not statistically significant (-O2: 33.11 ± 34.21 mm3 and +O2: 74.61 ± 63.13 mm3)(Fig. 2E).
Figure 2. Short-term oxygen delivery had minimal impact on bone regeneration in large midfacial bone defects.
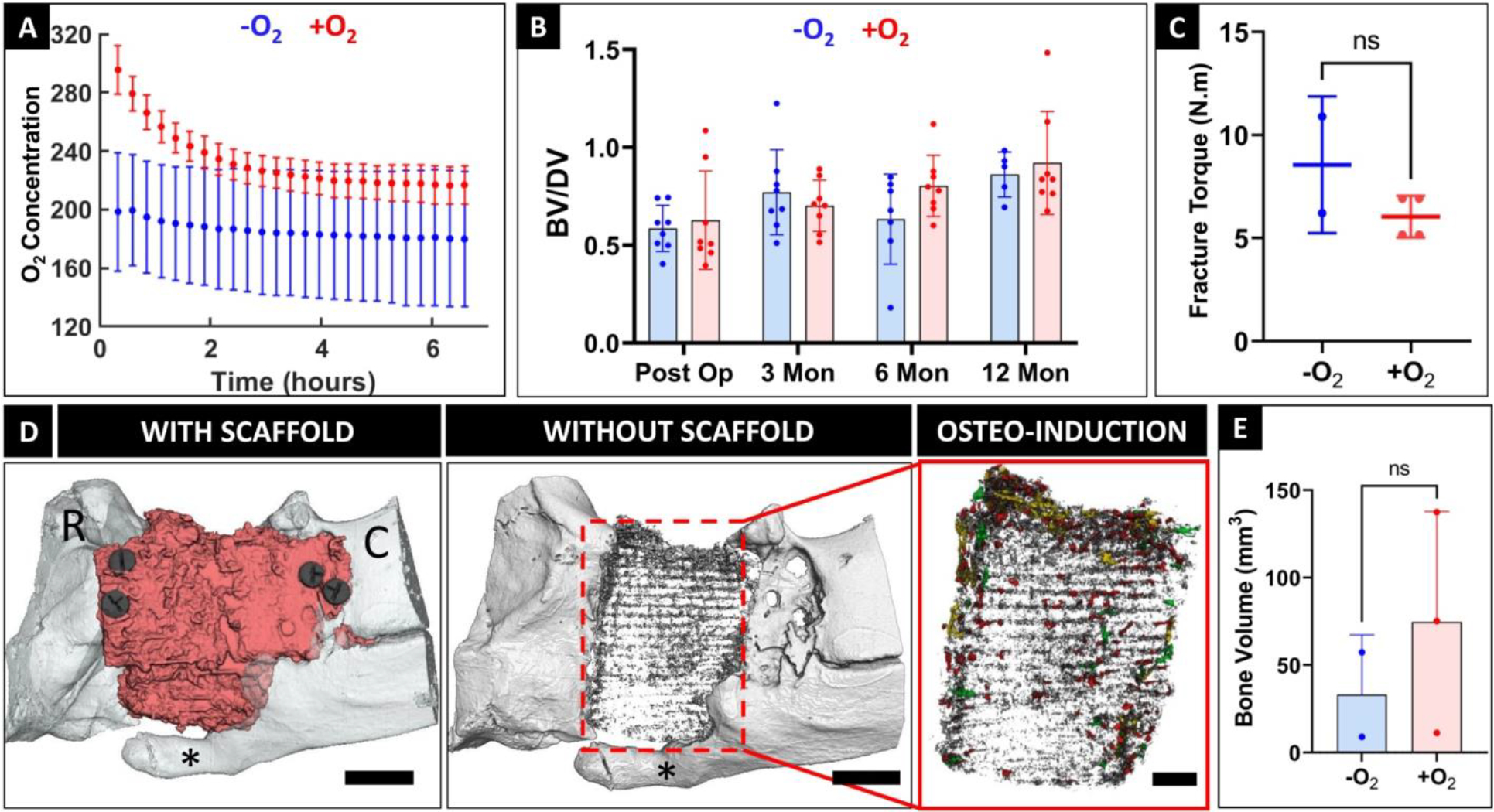
(A) Comparison of oxygen release profiles between -O2 and +O2 scaffolds demonstrate a therapeutic release window of approximately 2 hours. (B) Quantitative evaluation of bone volume/defect volume (BV/DV) at the defect site (n= 8 for both groups) revealed no significant differences. (C) Osseointegration of the scaffolds was assessed by mechanical testing (n=2 for -O2 and n=4 for +O2 groups) and showed statistically similar outcomes. (D) High resolution μCT scan of the defect site identified the scaffold (red), new osteoconductive bone (asterisk) growing from the surrounding native bone around the scaffold, and bone nodules growing within the scaffold pores as osteoinductive growth. Bone nodules were color coded based on their sizes namely fine (<0.0216 mm3, grey), small (0.0216–0.108 mm3, red), medium (0.108–0.216 mm3, green) and large (>0.216 mm3, yellow). R denotes rostral and C denotes caudal aspect of the zygomatic bone. Scale bar = 1 cm, 0.5 cm. (E) Comparison of osteoinductive bone nodule volume between the -O2 and +O2 groups. (n=2 for -O2 and 3 for +O2 groups).
Lamellar osteoconductive bone detected within geometrically mismatched scaffold pores
μCT analysis of the harvested zygoma specimens revealed osteoconductive bone ingrowth into the scaffold pores in 4 out of 9 samples (Fig. 3A). Histology was performed on undecalcified hard tissue specimens (n=5). Stevenel’s Blue staining confirmed the presence of mature, lamellar, osteoconductive bone ingrowth (Fig. 3B, C) while the bone nodules were identified as woven bone (Fig. 3D). H&E staining illustrated the presence of lymphocytes, macrophages, and multinucleated giant cells in intimate contact with the scaffold struts at 12 months (Fig. 3E). Semi-quantitative scoring indicated mild local inflammation around the scaffold struts with no major difference between the -O2 and +O2 groups. Given the similarities in bone formation and immune response between the two groups (Fig. 3F), we combined specimens belonging to both the groups in our subsequent assessments on the impact of geometric mismatch on bone regenerative outcome as oxygen was not considered as a confounding factor.
Figure 3: Histology at 12 months revealed the presence of lamellar bone and mild local inflammation within the scaffolds.
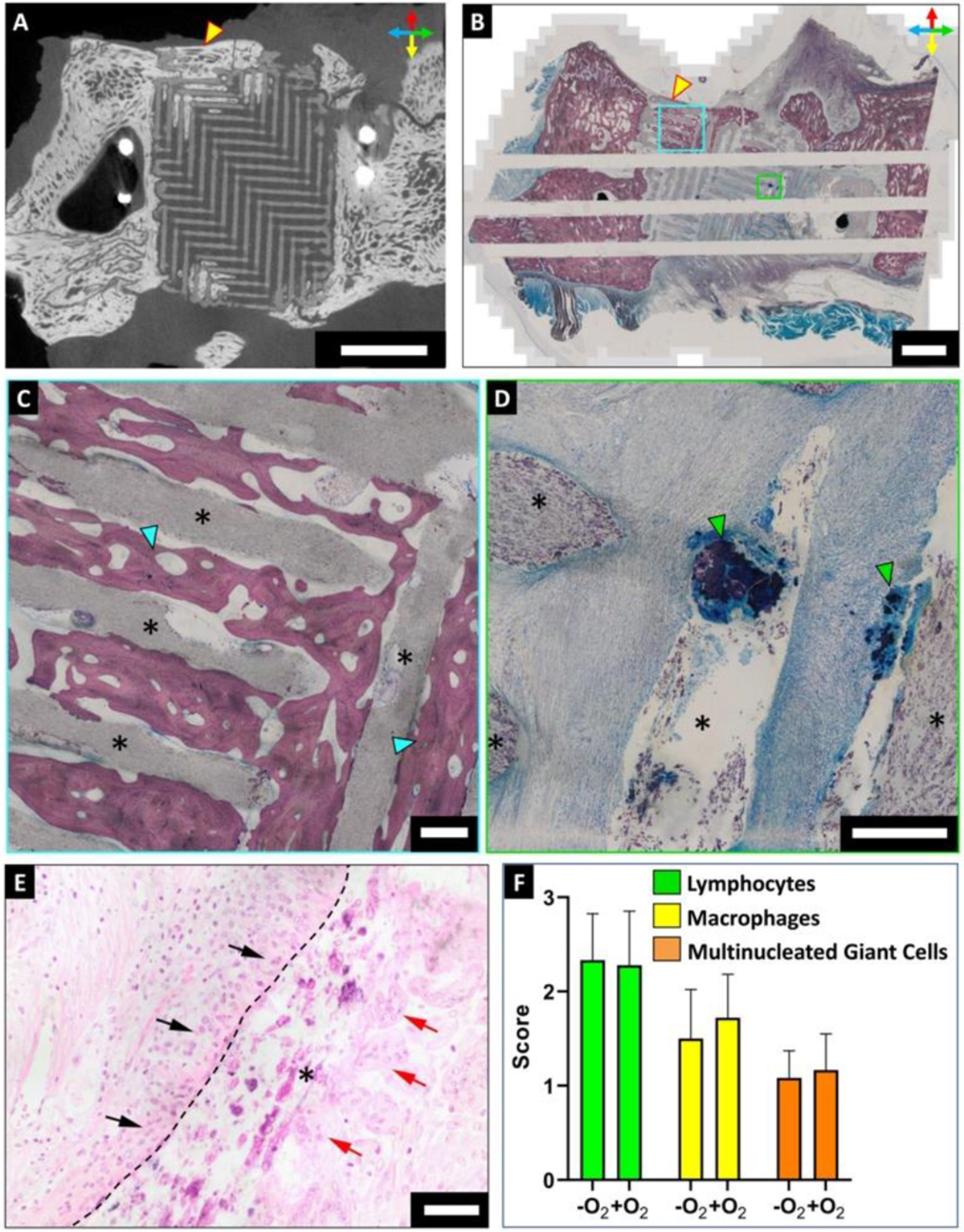
(A) μCT scan of harvested zygoma specimens confirmed osteoconductive bone infiltration into the scaffold. Scale bar = 10 mm. Orientation arrows: dorsal (red), ventral (yellow), rostral (blue) and caudal (green). (B-D) Stevenel’s Blue staining of undecalcified tissue specimens demonstrated osteoconductive bone growing on the periphery of scaffolds and infiltrating into the scaffold pores (cyan box; (C)). Bone nodules nucleating within the scaffold pores by osteoinduction were also observed (green box; (D)). Scale bar = 5 mm. (C, D) Magnified images of cyan and green boxes of (B) respectively, where asterisks denote scaffold struts. Scale bar = 500 μm. (E) H&E staining of undecalcified zygomatic specimens indicated the presence of immune cells within the scaffolds. Lymphocytes (black arrows) and multinucleated giant cells (red arrows) were found in close association with the scaffold strut (asterisk). Dashed line outlines the scaffold strut boundary. Scale bar = 50 μm. (F) Semi-quantitative scoring of H&E stained sections for assessing presence of lymphocytes, macrophages, and multinucleated giant cells within the scaffolds. Scores indicate: 0 = absent, 1 = rare, 2 = mild infiltrate, 3 =moderate/heavy infiltrate, 4 = packed infiltrate/sheets for MNGCs.
Imaging workflow facilitated characterization of post-operative geometric mismatch and spatial bone regeneration pattern
To identify areas of 3D geometric mismatch at the defect sites and quantitatively characterize them, we developed a CT based imaging workflow. First, post-operative and pre-operative CT scans of the porcine skulls were registered to segment out corresponding VOIs (Fig. 4A). Thereafter, the zygomatic bones were registered using independent iterative closest point (I-ICP10)[23] algorithm in MATLAB® to provide finer registration in the vicinity of the defect site (Fig. 4B). Post registration, areas of underfill (deficit areas where scaffold failed to recapitulate the native bone anatomy) were visualized and their respective magnitudes were quantified (Fig. 4C). Similarly, the process of segmentation and registration was repeated for post-operative and 3 months CT scans to identify regions of new bone growth. At later timepoints i.e., 6 and 12 months the porcine skull grows significantly[20] which makes skull registration challenging. Finally, we superimposed the underfill (deficit) regions with the regions of new bone growth. Overlapping of these two regions facilitated identification (and quantification) of orthotopic from ectopic bone. Moreover, underfill (deficit) regions filled by orthotopic bone were also identified and quantified (Fig. 4D).
Figure 4. Quantifying 3D geometric mismatch and spatial patterns of bone regeneration at defect sites.
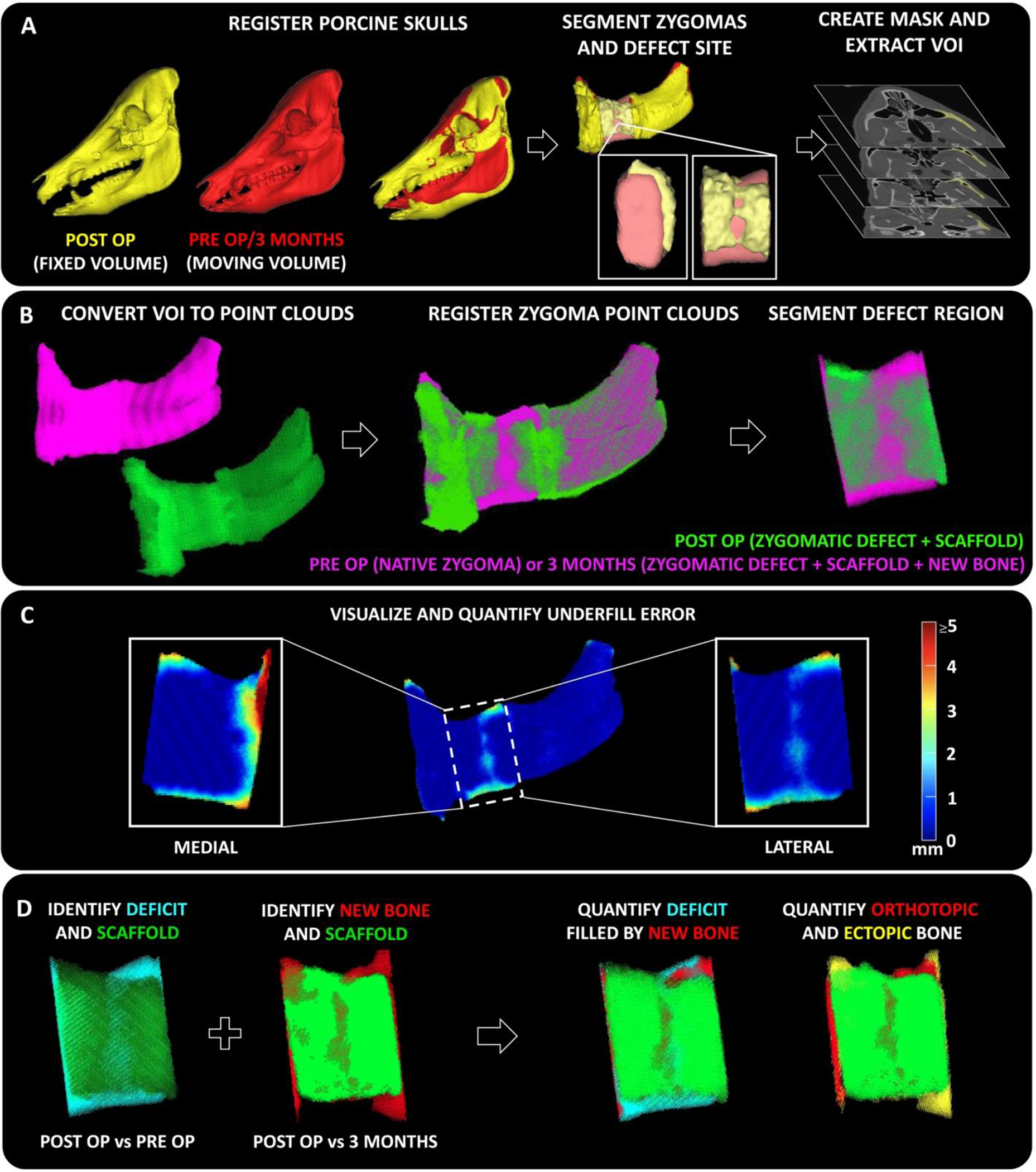
(A) Pre-operative and post-operative CT scans of the animal skulls were registered using Mimics software to segment out corresponding defect bone regions containing the scaffolds. Masks of the respective volume of interest (VOI) were created as bitmaps (.bmp). (B) The bitmaps were imported into MATLAB® and converted into point cloud objects and re-registered at a higher resolution using independent iterative closest point algorithm (I-ICP10)[23] (C) From the registered zygomatic point clouds, areas of anatomical mismatch (underfill) were visualized and their corresponding magnitudes were quantified at the defect site. Heatmap demonstrates magnitude of underfill at each point. (D) Regions of deficit (underfill, post op vs pre op) and new bone growth (overfill, post op vs 3 months) at the defect site were identified and superimposed spatially to quantify the amount of orthotopic or ectopic bone and calculate the amount of deficit region filled by new bone.
Higher geometric mismatch associated with higher orthotopic bone growth
We applied the imaging workflow to all 16 defects to quantitatively characterize post-operative geometric mismatch and spatial patterns of bone regeneration. Some defects exhibited very low levels of underfill whereas others demonstrated incidence of higher levels of underfill primarily located on the medial, dorsal, and ventral boundary of the defect (Fig. 5A). Anatomical mirroring introduced significant underfill (in magnitude and frequency) at the rostral aspect of the defect boundary due to the tapering of the maxillary sinus. However, some animals had relatively flat zygomas with less taper which gave rise to a wide range of geometric mismatch (Fig. 5B). At 3 months, ingrowth of new bone into the deficit volume was observed in some scaffolds while in others deficit volume remained unfilled (Fig. 5C). There was a strong positive Spearman correlation (r = 0.7098, p = 0.0021) observed between deficit volume and orthotopic bone volume (Fig. 5D). Finally, areas of new bone growth were identified and qualified as either orthotopic or ectopic depending on their spatial existence in relation to the native anatomical contour (Fig. 5E). Higher volume of orthotopic bone volume was observed in defects with high geometric mismatch which is reflected in the strong positive correlation between geometric mismatch % and orthotopic bone volume (Spearman correlation, r = 0.7147, p = 0.0026) (Fig. 5F). Moreover, new bone seemed to preferentially grow across the dorsal (superior) and ventral (inferior) margins of the scaffold as compared to lateral or medial aspects of the scaffold. Collectively, the observations indicate that higher geometric mismatch favors higher orthotopic bone formation at the defect site.
Figure 5. Quantification of post-operative geometric mismatch and spatial bone regenerative pattern.
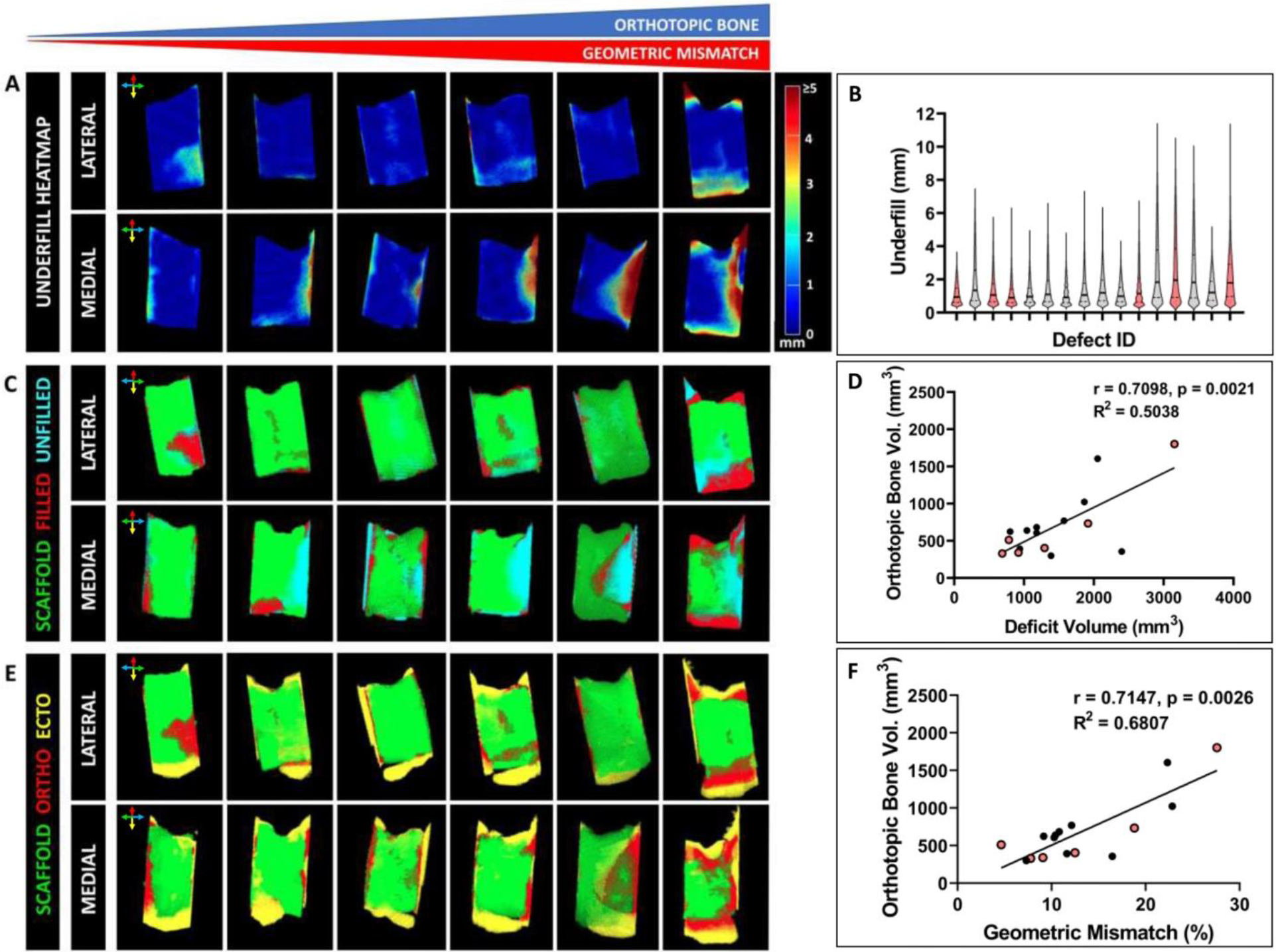
Six representative defect sites illustrated the range of observed geometric mismatch. (A) Heatmap demonstrated the spatial distribution of underfill on the medial and lateral aspects of the defect site. Color bar indicates the magnitude of underfill at a particular site (in mm). (B) Violin plots highlight the distribution underfill in all defects. Red colored plots correspond to the six defects depicted in (A). Median and quartiles are indicated by solid and dashed lines respectively. (C) Point cloud identifies sites of deficit (termed as “unfilled”) post scaffold implantation and bone infiltration (termed as “filled”) into the respective deficit areas at 3 months timepoint. (D) Strong positive correlation observed between post-operative deficit volume and orthotopic bone volume at 3 months (Spearman correlation, r = 0.7098, p = 0.0021). (E) Point cloud identifies the spatial distribution of new bone and facilitates their characterization as orthotopic or ectopic bone. (F) Strong positive correlation was observed between geometric mismatch % and orthotopic bone volume (Spearman correlation, r = 0.7147, p = 0.0026). Sense of orientation is conveyed by colored arrows: dorsal (red), ventral (yellow), rostral (blue) and caudal (green).
Medio-lateral osteoconductive bone growth compromised long term scaffold survivability in periorbital bone defects
We observed scaffold failure (i.e. significant displacement of the scaffolds from the initial implantation site) in 5 out of 16 defects. All the scaffolds failed in a similar way i.e., new bone originated from the caudo-ventral aspect of the defect boundary at around 3 months (Fig. 6, S11). This bone grew in a medio-lateral direction and pushed the scaffolds out of the defect site laterally causing scaffold failure around ~5–6 months (3 defects) or closer to endpoint (2 defects). Interestingly, even after the failed scaffolds were explanted, the regenerating bone continued growing medio-laterally until the animals were sacrificed. However, complete bridging at the defect site was not observed in any of these cases.
Figure 6. Medio-lateral bone growth was associated with scaffold failure.
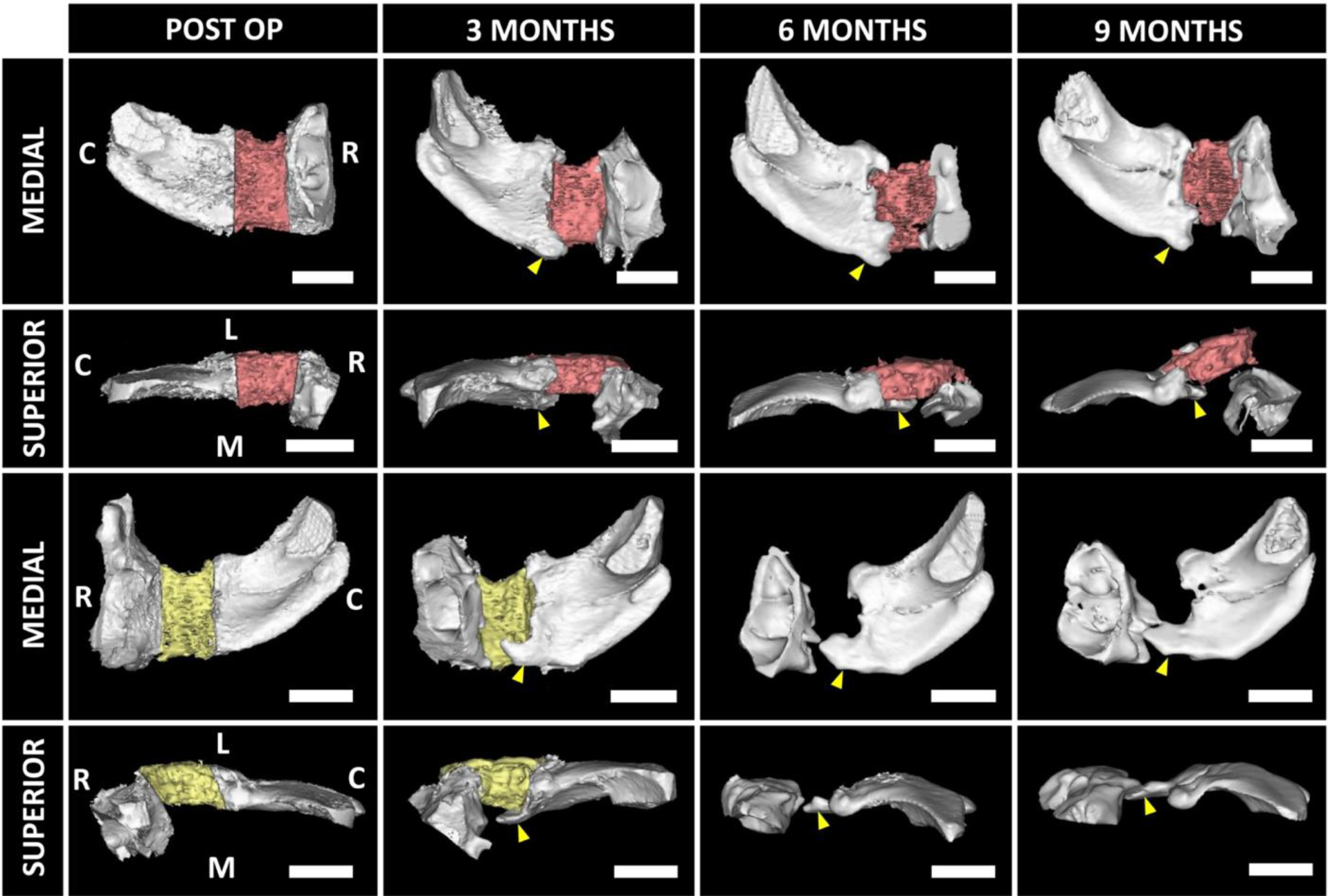
Representative images of two zygomas demonstrated medio-lateral growth of native bone from the caudo-ventral aspect (yellow arrowheads) of the defect boundary initiates around 3 months timepoint. The growth became prominent around 5–6 months and pushed the scaffolds (red and yellow indicate left and right zygoma, respectively) out of the defect sites. C: Caudal; R: Rostral; L: Lateral; M: Medial. Scale bar = 2 cm.
DISCUSSION
Autologous bone grafts are the standard clinical care for treatment of large human-sized craniofacial bone defects but are associated with several limitations, most notably, donor-site morbidity. Consequently, point-of-care strategies employing porous tissue-engineered 3D-printed scaffolds in conjunction with autologously sourced stem cells are considered as desirable alternatives. The midfacial region, especially the periorbital bones, have complex anatomies which are difficult to reconstruct using bone grafts. This makes them compelling indications for treatment with tissue-engineered scaffolds. Such scaffolds are customized to match the anatomy of lost bony tissue and have a porous framework to facilitate new bone ingrowth and osseointegration. However, our pre-clinical study[20] involving treatment of large bone defects with such constructs demonstrate that new bone mostly grows on the periphery of the scaffolds by osteoconduction with limited infiltration into the pores of the scaffolds. In periorbital bone defects, this ectopic bone formation could have significant negative impacts such as impaired vision or facial disfiguration. The incorporation of large pores (> 800 μm) into the scaffold framework did not adequately facilitate robust bone ingrowth.[19] In contrast, critical-sized bone defects in small animals treated with hydrogels demonstrated bone formation throughout the defect volume.[24,25] Consequently, there might be some benefit to designing tissue-engineered scaffolds that underfill the defect volume and there remains a need to improve our understanding of how surrounding native bone responds and how new bone grows spatially in relation to the scaffold implanted at the defect site. In this study, we developed a CT-based imaging workflow to provide insight into the spatial relationship and regenerative pattern of bone in periorbital defects especially around the scaffold implanted at the defect site.
We hypothesized that inducing controlled geometric mismatch at the zygomatic defect would promote dense, lamellar bone formation within orthotopic regions, enhanced bridging across the defects, and improved integration of the native and regenerated bone. The zygoma has a complex geometry and varies from animal to animal. Therefore, to provide a simple yet unbiased and standard approach to introducing geometric mismatches, we digitally mirrored the native bone anatomy around the defect midpoint in the coronal axis. The fixation tabs ensured that the external surface of the scaffold remained flush with the native bone and mirroring the scaffold maintained the native bone’s curvature. The inherent limitation with this approach is that we only explored a narrow range of geometric mismatches (4.65% - 27.6%). In some cases, the extent of the anatomic mismatch was further reduced by the need to trim the scaffolds during surgery to fit them into the defects. Despite these drawbacks and the small number of samples, we observed clear correlations demonstrating the potential benefit of mismatching on bone regeneration.
In most of the scaffolds, osteoconductive bone growth was observed primarily at the dorsal and ventral margins irrespective of the degree of geometric mismatch. However, when osteoconductive bone grew along the inner surface of the scaffolds in a medio-lateral direction, it resulted in premature scaffold failure around 5–6 months post-surgery. Taken together, these data have clinical significance and suggest that establishing a database in which patterns of bone growth in midfacial/periorbital clinical injuries could be observed might be a crucial step in geometric design of tissue-engineered scaffolds to maximize bone regeneration. Many in silico studies[26–28] which resort to radiological or histological data from longitudinal pre-clinical studies to computationally simulate bone regeneration can significantly benefit from these findings to streamline and enhance their models.
The CT imaging workflow developed to analyze geometric mismatch and patterns of de novo bone growth provides a robust, unbiased platform to assess surgical accuracy, monitor and identify areas of orthotopic or ectopic bone regeneration. CT based image registration methodologies have been previously used for pre-operative surgical planning[29], intraoperative surgical navigation[30], and studying bone healing and remodeling[31,32]. However, the use of such techniques to study spatial bone regeneration patterns around tissue-engineered scaffolds has been limited. Most clinical studies[33–35] involving periorbital surgical reconstruction using tissue-engineered scaffolds resort to CT for post-operative follow up and, if applied clinically, this method would provide a means of determining the need for appropriate and timely medical intervention.
This study also briefly explored the role of oxygen release on bone regeneration. We employed PCL-DCB-μtank scaffolds which differed in composition from the PCL-μtank scaffolds used in our previous study[22], where oxygen releasing scaffolds significantly enhanced bone regeneration in critical-sized murine calvarial defects. Based on our μCT analysis, we measured roughly twice as much volume of mineralized nodules (osteoinductive bone) in the scaffold interior in groups containing oxygen. However, in keeping with our observations that most of the new bone volume is comprised of lamellar bone formed through osteoconduction, we did not observe similar increases in the overall bone regenerative outcome in the large periorbital bone defects. Another potential reason for the lack of overall differences between the two groups is likely that the short oxygen release window. Oxygen released during the period between scaffold removal from hyperbaric O2 chamber to implantation at defect site could limit its therapeutic benefit, and the study was designed to minimize that window by opening the hyperbaric chamber within the surgical suite after the defect had been created. Interestingly, the elution time for the PCL-DCB-μtank scaffolds was significantly shorter than what was measured for PCL-μtank (without DCB) scaffolds, which raises speculation that changes in the microporosity in the scaffold from the inclusion of DCB might have affected the release kinetics.
Overall, this study employed an advanced imaging workflow to investigate the impact of geometric mismatch on bone formation in a challenging model of bone regeneration in pigs. Our findings demonstrated that new osteoconductive bone primarily grew largely along the dorsal and ventral aspects of the scaffolds and underfill facilitates orthotopic bone growth. Thus, strategic introduction of geometric mismatch at the dorsal and ventral aspects of the scaffold enhanced the regenerative outcomes. One limitation of our study is the limited range of geometric mismatch achieved by anatomically mirroring scaffolds. Interestingly, the correlation between geometric mismatch and orthotopic bone volume became much stronger within the geometric mismatch range of 15–30% (r = −0.914, p = 0.0299, n=5) compared to 0-<15% (r = −.3473, p = 0.1206, n=11). Future studies might introduce geometric mismatch by deliberate removal of scaffold at strategic sites (for e.g., dorsal and ventral aspects of the periorbital defect boundary) to maximize bone regeneration while providing adequate implant stability at the defect site. Adequate apposition between the scaffold boundary (rostral and caudal) and native bone should be provided to avoid micromotion[36]. The zygomatic bone is subjected to different forces varying in magnitude and direction due to its association with the masseter muscle[37]. Incorporating geometric mismatch into porous scaffolds might alter its mechanical properties and load bearing ability. Hence, care should be taken to ensure the scaffold is suited to sustain physiological forces at the defect site. Collectively, we propose that strategic incorporation of geometric mismatch into the scaffold design strategy has the potential to enhance bone regeneration in large periorbital and midfacial bone defects in human patients.
MATERIALS AND METHODS
Biodegradable microtank (μtank) fabrication
μtanks were fabricated as previously described.[22] Briefly, a water/oil/water double emulsion of 10% (w v−1) polyvinyl alcohol (Sigma Aldrich), 7% (w v−1) poly(lactic-co-glycolic acid) (Polyscitech) in dichloromethane (Sigma Aldrich) were mixed at 500 rpm for 1 hour to create a primary emulsion. The primary emulsion was mixed with 1% gelatin (Sigma Aldrich) and stirred at 500 rpm overnight to allow dichloromethane (DCM) to evaporate. μtanks were collected, washed, filtered, lyophilized, and stored at room temperature until use in 3D-printed scaffolds.
Custom PCL-DCB-μtank filament fabrication
Custom filaments incorporated with μtanks were fabricated from a blend of clinical grade polycaprolactone (PCL) and decellularized bone (DCB) for 3D-printing custom scaffolds as follows. Polycaprolactone (PURASORB® PC 12, Corbion, The Netherlands) and Bio-Oss® (Geistlich Biomaterials) (i.e., DCB) powder as received from the vendor were mixed in a ratio of 62.5:37.5 (w w−1) and stored at −80°C overnight. Thereafter, the mixture was cryo-milled to obtain a fine homogeneous powdered mix using a SPEX cryo-mill (45, 1 minute cycles, at a frequency of 10 Hz). μtanks were mixed with cryomilled PCL-DCB powder to obtain a final ratio of 5:3:2 (PCL:DCB:μtank) The mixture was fed into in a polymer extruder (Dynisco LME-120, Franklin, MA) at 85–90°C and were drawn into filaments with an average diameter of 2.85 mm.
Design and fabrication of anatomically mirrored scaffolds
All experiments were conducted with skeletally mature, castrated, male Yucatán pigs at Johns Hopkins University under an approved Institutional Animal Care and Use Committee protocol (SW17M378). Scaffolds were designed and fabricated as previously described.[20] Briefly, pre-operative CT scans of animals were acquired, and the native anatomy was used as a template to design the scaffolds. 3D medical image processing software Mimics (Materialise Mimics 14.0®, Leuven, Belgium) was used for segmentation. First the scan was mirrored in the rostro-caudal direction to provide a mirrored anatomy to facilitate design of anatomically mirrored scaffolds. Thereafter, 2 cm wide wedge shaped solid (.stl) object was used as a surgical guide to define the defect boundaries and virtual ostectomy of the zygoma was performed. The virtually resected bone was rendered solid by wrapping the hollow sinus. The scaffold design was completed by addition of fixation tabs digitally. Masks were created from the solid scaffold volumes. Next, masks for surgical guides were created by segmenting out the lateral aspect of corresponding zygomatic bones. The masks were converted into bitmap (.bmp) slices and exported out of Mimics.
To incorporate isotropic porosity into the solid scaffold volume and generate the corresponding print path (g-code file), the exported bitmap slices were imported into a MATLAB® based in-house slicer program (ScafSLICR[38]). The slicer programm introduced isotropic pores of width 800 μm and struts of width 500 μm (which translates to a theoretical porosity of 62%)[38] into the scaffold body whereas the fixation tabs were kept solid except for screw holes. A Lulzbot TAZ 5 3D-printer (Aleph Objects) was provided with the sliced g-code file and PCL-DCB-μtank filaments to fabricate custom scaffolds. All scaffolds were subjected to PBS wash and incubation in an ethanol ladder. Later they were dried, packed in sterilization pouches and treated with ethylene oxide gas overnight, before being hyperbarically loaded (+O2) in a custom chamber[22] at 500 psi for 7 days prior to surgery.
Longitudinal CT acquisition
Longitudinal imaging (time points: pre-operative, post-operative, 3, 6, 12 months) was performed on an O-arm™ intraoperative Cone Beam Computed Tomography (CBCT) (Medtronic, Littleton MA) operated in a High Definition (HD) mode. The scan consisted of 720 views over a 360°rotation; the x-ray tube voltage was 120 kV; the total exposure was 240 mAs. The flat-panel x-ray detector (PaxScan 4030D, Varex Imaging, Salt Lake City, UT) was read out in 2×4 binning with 1024×384 pixels measuring 0.388×0.776 mm2. A 12:1 anti-scatter grid was placed on the detector. The images were reconstructed on a 0.415 × 0.415 × 0.833 mm3 voxel grid.
Pre-operative preparation and survival surgery
All animals were fasted a minimum of 12 hours prior to surgery while water was continued to be provided ad libitum. Pigs were sedated using a combination of ketamine 20–30mg kg−1 and xylazine 2–3mg kg−1 given intramuscularly followed by placement of an intravenous catheter in a marginal ear vein for delivery of fluids (Lactated Ringers Solution (LRS) 5 –10 mL kg−1 hr−1) and anesthesia throughout the procedure. Propofol 0.83–1.66 mg kg−1 was given intravenously followed by placement of an endotracheal tube for delivery of inhalational anesthesia (0.5–2.5% isoflurane). Sterile ophthalmic lubrication was applied to the corneas. Cefazolin (22 mg kg−1 IV) was administered approximately 15 minutes prior to the start of surgery and repeated every 90 minutes intraoperatively. Pantoprazole (Protonix ®) (1.0 mg kg−1 IV) was also administered once at the start of surgery. All surgical sites were aseptically prepped alternating between chlorhexidine and 70% ethanol at least three times before finishing with the application of an ionophore to the skin’s surface for a contact time of no less than 10 minutes. Specifically, for this surgery, the entire face was shaven. Heart rate, blood pressure (NIBP), pulse oximetry, capnography, electrocardiography, respiratory rate, and rectal temperature were monitored continuously and recorded every 15 minutes. Mechanical ventilation was initiated if indicated based on respiratory rate and end tidal CO2 values. Hypothermia was prevented using a Bair Hugger warm-air blanket.
Isolation of SVF from subcutaneous porcine fat
As previously described[20] under anesthesia, subcutaneous adipose tissue was excised from the lumbar paravertebral area. Briefly, following aseptic preparation a skin incision was made over the area superficial to L3 - L6 using a #10 blade and subcutaneous adipose tissue was sharply excised. Thereafter, using a combination of interrupted and continuous patterns with absorbable suture subcutaneous and skin layers were closed. To remove blood the harvested adipose tissue was washed multiple times with phosphate buffered saline. Therafter to remove blood clots, the tissue was minced for 5 minutes into <2–3 mm pieces, and washed in RBC (red blood cell) lysis buffer (0.155 M ammonium chloride, 0.01 M potassium bicarbonate, 0.1 mM EDTA, dissolved in sterile deionized water) for 1 minute. Centrifugation was performed to separate the minced tissue from RBC lysis buffer. Agitation for 30 minutes with collagenase solution (4 mg ml−1 collagenase type 1, Worthington Biochemical; 25 mg ml−1 bovine serum albumin, Sigma Aldrich; dissolved in 1 g l−1 glucose Dulbecco’s modified eagle medium) at 37°C helped to digest the tissue. SVF pellet was collected by centrifuging at 300 g for 5 minutes at 37°C following digestion. After removing collagenase, oil, and residual undigested fat layers the remaining cell pellet was resuspended in RBC lysis buffer. After filtering through a 800 μm mesh and a 100 μm cell strainer, the cell suspension was centrifuged to obtain a pellet. After resuspending in PBS the cells were split into three groups: 45% of the volume for each defect and 10% for cell characterization. The cell suspension was centrifuged again. Pellets for each defect were homogenously resuspended in the fibrinogen component of a TISSEEL™ packet (diluted 1:5 in phosphate buffered saline). For in situ injection of SVF into the created defect, the cell-fibrinogen mixture was drawn into 5 ml double-barreled syringes, along with the thrombin component contained in the other side. For future studies, the remaining 10% of cells were plated in expansion media.
Zygomatic defect creation
Zygomatic defects were created as previously described.[20] Once the animal was sedated and subcutaneous fat was harvested, a transcutaneous 5 cm subtarsal incision was made with a #15 blade over the zygoma. A dissection was performed with a cautery down to the inferior orbital rim and then down to the zygomatic bone. With a periosteal elevator, the dissection proceeded in a sub-periosteal fashion over the anterior, lateral portions of the zygomatic bone. To facilitate complete release of the zygomatic defect, careful attention was placed to the dissection of the masseter off the zygoma on its inferior margin. The proximal and distal osteotomy sites were marked with a pencil while placing a 3D-printed, animal-specific 2 cm osteotomy cutting guide onto the anterior wall of the zygoma/orbital rim. To complete the cuts, a 27 mm reciprocating saw tip (Johnson & Johnson – De Puy Synthes) was then used to make the proximal and distal osteotomies followed by using a 4 mm straight osteotome. To create the defect the cut bone was removed and for removal of any residual bone or sharp bone edges, a thorough exploration of the defect and zygoma was performed. Using two molt elevators, the scaffold was then inserted into the zygoma defect and held in place. To fixate the implant onto the defect, two 8 (or 10) × 1.8 mm screws were placed bilaterally on the built-in fixation tabs. The construct was copiously irrigated and injected with SVF. Using absorbable stitches, the incision was closed in layers starting at the periosteal level, followed by a deep subcutaneous layer, and lastly in a subcuticular fashion. Finally, to achieve immediate closure of the wound dermal skin glue was placed over the incision. No surgical site dressing was utilized.
Blood collection and complete blood count
Blood draws were performed immediately post-operatively and every 3 months until endpoint at 12 months. Animals were sedated using ketamine 20–30 mg kg−1 and xylazine 2–3 mg kg−1 given intramuscularly. A maximum of 5 ml of whole blood was obtained from each animal from the cranial vena cava and complete blood counts and blood chemistry panels were performed at an outside laboratory (IDEXX Reference Laboratories).
Scaffold explantation
Scaffolds were explanted as previously described.[20] Upon reaching the endpoint (12 months), all animals were euthanized and the zygomas (zygomatic (arch + flange) + squamosal bone) containing the scaffolds were harvested. Briefly, using a #10 blade a transcutaneous 8 cm incision with 1 cm vertical extensions proximally and distally were made over the malar region of the animal. This was followed by sharp dissection down to the anterior and posterior borders of the zygoma. Until the entire zygoma was exposed, dissection proceeded in a subperiosteal fashion, including the former zygomatic defect region. The zygomas were dissected free and osteotomized using a reciprocating saw followed by a 4 mm osteotome (at approximately 2 cm proximal and distal to the defect margin). The entire zygomatic complex including the implanted construct was then removed en bloc.
Micro-CT (μCT) scan
μCT scans were acquired as previously described[20]. Bruker Sky-Scan 1173 system was used to visualize the selected harvested zygomas. During the scan, each sample was rotated 360° and projections were captured using the following scan settings, source voltage: 130 kV, source current: 61 μA, exposure time: 1200 ms, rotation step: 0.25°, 0.25 mm brass filter. Nrecon software was used to reconstruct the projection images and to generate computed axial cross-section slice images with 30 μm pixel resolution once all projections were obtained. Bruker’s CTVox software was used to render these axial sliced images, whereas Avizo (FEI Visualization Sciences Group, Burlington, MA) was used for performing segmentation of the images. Labeling and analysis of the zygomatic bones, scaffolds, the bone formation on the periphery and within the scaffold, and the metal pins were performed using the watershed tool, volume edit function, and label analysis functions in Avizo.
Mechanical testing
Mechanical testing was performed on the harvested zygomas to assess osseointegration as previously described[20]. To remove excess tissue, all the harvested zygoma specimens were scraped with care taken to not damage the soft tissue coverage around the PCL-DCB-μtank scaffolds. To serve as pots for the zygomas, rectangular cross-sectional plastic tubes (McMaster-Carr) were cut into small sections of approximately 1 inch depth. Polymethylmethacrylate (PMMA, Keystone Bosworth Fastray) was poured to uniformly fill the entire volume around the zygoma, while they were held vertically upright within the pot. The slurry was allowed to cure for approximately 20 minutes. The defect boundaries and the midpoint of the scaffold were identified and marked as reference points. On a servo hydraulic testing machine (Model 858, MTS, Eden Prairie, MN), the potted zygomas were clamped into a multiaxial vise such that the medial aspect of the zygoma faced the load applicator of the testing machine. At the midpoint of the scaffold, the load was applied. At a rate of 1 mm s−1, the load applicator was displaced until failure occurred at the interface between the scaffold and the posterior (caudal) aspect of the native bone. Measurement of the distance between the load application site and the fixed end of the specimen was performed and used to calculate the applied bending moment as follows:
Applied bending moment = Load (N) × distance (m).
Hard tissue histology
Hard tissue histology was performed as previously described[20]. To primarily distinguish new bone from pre-existing bone and identify sites of osteoid deposition or mineralization within the extra-cellular matrix, a special bone stain namely Stevenel’s Blue (SB)[39] with Van Gieson’s picrofuchsin counterstain was employed while H&E stain was used to gain insight about the cell types present at the defect site. To aid in trimming each specimen in order to capture the implant area with adjacent pre-existing bone, all zygoma specimens were radiographed (Bruker Digital Radiograph System, Bruker, Billerica, MA, USA). Dehydration of tissues was done through a graded series of ethanol and then they were infiltrated and embedded with methyl methacrylate (MMA). At three levels each in the rostro-caudal orientation and superior-inferior orientation respectively, the resultant MMA blocks from each zygoma were then sectioned. Each level was ground and polished to approximately 100 μm thickness (Exakt® Systems, Norderstedt, Germany). H&E stain was performed on the middle level section from each orientation whereas SB stain was used for rest of the sections. Olympus VS120 scanner (Olympus Corporation of America, Center Valley, PA, USA) was used for scanning all slides. H&E-stained slides were scored semi-quantitatively by pathologists for overall inflammation, polymorphonuclear cells, lymphocytes, macrophages, and multinucleated giant cells (MNGC) where scores indicate 0 = absent, 1 = rare, 2 = mild infiltrate, 3 = moderate/heavy infiltrate, 4 = packed infiltrate/sheets for MNGC.
Imaging workflow for characterizing geometric mismatch and spatial bone regenerative pattern
A custom imaging workflow was developed to register defect sites and characterize post-operative geometric mismatch and spatial bone regeneration pattern. Briefly, post-operative CT scans of the animals were first registered with the respective pre-operative skulls in Materialise Mimics 14.0® using the inbuilt image registration tool. After registration of the skulls, boundaries of corresponding zygomatic bone region and defect sites from both the timepoints were identified, segmented and converted into masks. The hollow sinus region of the pre-operative mask co-localized with the post-operative defect site was rendered solid using the wrap tool in Mimics. Finally, masks were exported out of Mimics in the form of bitmap (.bmp) slices. The bitmaps were imported into MATLAB and converted to point clouds. The point clouds of the post-operative and pre-operative zygomas were registered again using our previously developed autonomous I-ICP10 algorithm[23] to provide more accurate local registration at the defect site. Thereafter, the registered defect site and the corresponding pre-operative point clouds were segmented out and areas of underfill were identified using knnsearch tool in MATLAB and their magnitude was determined by calculating the Euclidian distance between the closest neighboring points belonging to both the point clouds. (Note: For post-operative vs pre-operative registration, underfill is defined as deficit of scaffold failing to recapitulate the native bone anatomy). The same process of registration and segmentation was repeated for post-operative and 3 months CT scans. However, in this case overfill was determined which is defined as new bone growth extraneous to the scaffold volume (post-operative). Since, corresponding pre-operative and 3 months CT scans were registered to the same post-operative zygoma volumes they were spatially registered too. Applying knnsearch tool on registered pre-operative and 3 months point-clouds in MATLAB identified orthotopic bone from ectopic bone and their respective volumes could be quantified. Geometric mismatch was calculated using alphaShape tool in Matlab. Briefly, an alphaShape object (a bounding volume that envelops all points in the corresponding point cloud) of the post-operative point cloud was created using alphaShape tool. Using, inShape tool the percentage of pre-operative points (native bone) lying outside the post-operative alphaShape object was determined and has been termed as geometric mismatch (%). All scripts were executed on a 32 GB RAM Windows 10 system with 6th generation Intel Core i7 processor and AMD Radeon R5 340X graphics card.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 software. Normality of data was tested using D’Agastino-Pearson omnibus test and Spearman correlation (r) was evaluated. Simple linear regression was performed to determine goodness of fit i.e., coefficient of determination (R2). Quantitative data were expressed as the mean ± standard deviation. Statistical analysis for bone volume was carried out by two-way ANOVA followed by a Bonferroni post-hoc test. Comparison of mechanical testing data was done using an unpaired, two-tailed Mann Whitney t-test. For complete blood work data (dot plot), each dot represents an individual animal. Statistical significance has been denoted as ns (p>0.05), *(p < 0.05), **(p < 0.01) and ***(p < 0.001).
Supplementary Material
Acknowledgements
We thank Stephen Belkoff and Parker Treadway for help in performing the mechanical testing procedure on the harvested zygoma specimens, and Derick Vollmer for help with acquisition and analysis of the μCT scans. We thank the staff at Department of Molecular and Comparative Pathobiology, Johns Hopkins University for veterinary assistance and Geistlich Pharma North America, Inc. for generously providing us with Bio-Oss® for conducting the studies.
Funding
We acknowledge the funding support of this work by the Maryland Stem Cell Research Fund (2018-MSCRFV-4314) and the National Institutes of Health (R01DE027957) to W.L.G. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the funders.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Credit Author Statement
SS: Designed research, performed research, analyzed data, wrote the paper. YZ: Analyzed data. ALF: Designed research, performed research, analyzed data. ECW: Analyzed data. ELN: Designed research, performed research, analyzed data. ANO: Designed research, performed research, analyzed data. NYZ: Performed research, analyzed data. ANR: Performed research. CCW: Performed research. WZ: Provided resources. WG: Performed research. DG: Performed research, Analyzed data. NDJ: Analyzed data. DK: Designed Research. JMI: Performed research. JL: Performed research. MPG: Designed research, performed research, analyzed data, WLG: Designed research, analyzed data, wrote the paper.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Su P, Paquet C, O’Dell K, Reinstadler D, Kokot N, Granzow J, Chambers TN, Kochhar A, Laryngoscope 2021, 131, 1985. [DOI] [PubMed] [Google Scholar]
- [2].Salentijn EG, van den Bergh B, Forouzanfar T, Journal of Cranio-Maxillofacial Surgery 2013, 41, 630. [DOI] [PubMed] [Google Scholar]
- [3].Kochhar A, Byrne PJ, Otolaryngol Clin North Am 2013, 46, 759. [DOI] [PubMed] [Google Scholar]
- [4].Zhang M, Lin R, Wang X, Xue J, Deng C, Feng C, Zhuang H, Ma J, Qin C, Wan L, et al. , Sci Adv 2020, 6, eaaz6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hollister SJ, Nat Mater 2006, 5, 590. [Google Scholar]
- [6].Karageorgiou V, Kaplan D, Biomaterials 2005, 26, 5474. [DOI] [PubMed] [Google Scholar]
- [7].Mantila Roosa SM, Kemppainen JM, Moffitt EN, Krebsbach PH, Hollister SJ, J Biomed Mater Res A 2010, 92, 359. [DOI] [PubMed] [Google Scholar]
- [8].Sicchieri LG, Crippa GE, de Oliveira PT, Beloti MM, Rosa AL, J Tissue Eng Regen Med 2012, 6, 155. [DOI] [PubMed] [Google Scholar]
- [9].Zhao YN, Fan JJ, Li ZQ, Liu YW, Wu YP, Liu J, Artif Organs 2017, 41, 199. [DOI] [PubMed] [Google Scholar]
- [10].Chen Z, Yan X, Yin S, Liu L, Liu X, Zhao G, Ma W, Qi W, Ren Z, Liao H, et al. , Materials Science and Engineering C 2020, 106, 110289. [DOI] [PubMed] [Google Scholar]
- [11].Huri PY, Arda Ozilgen B, Hutton DL, Grayson WL, Biomedical Materials 2014, 9, 045003. [DOI] [PubMed] [Google Scholar]
- [12].Li W, Dai F, Zhang S, Xu F, Xu Z, Liao S, Zeng L, Song L, Ai F, ACS Appl Mater Interfaces 2022, 14, 20693. [DOI] [PubMed] [Google Scholar]
- [13].Reichert JC, Cipitria A, Epari DR, Saifzadeh S, Krishnakanth P, Berner A, Woodruff MA, Schell H, Mehta M, Schuetz MA, et al. , Sci Transl Med 2012, 4, 141ra93. [DOI] [PubMed] [Google Scholar]
- [14].Weisgerber DW, Milner DJ, Lopez-Lake H, Rubessa M, Lotti S, Polkoff K, Hortensius RA, Flanagan CL, Hollister SJ, Wheeler MB, et al. , Tissue Eng Part A 2018, 24, 943. [DOI] [PubMed] [Google Scholar]
- [15].Diloksumpan P, Bolaños RV, Cokelaere S, Pouran B, de Grauw J, van Rijen M, van Weeren R, Levato R, Malda J, Adv Healthc Mater 2020, 9, DOI 10.1002/adhm.201901807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li JJ, Roohani-Esfahani SI, Dunstan CR, Quach T, Steck R, Saifzadeh S, Pivonka P, Zreiqat H, Biomedical Materials (Bristol) 2016, 11, 015016. [DOI] [PubMed] [Google Scholar]
- [17].Vidal L, Kampleitner C, Krissian S, Brennan M, Hoffmann O, Raymond Y, Maazouz Y, Ginebra MP, Rosset P, Layrolle P, Sci Rep 2020, 10, 7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Henkel J, Medeiros Savi F, Berner A, Fountain S, Saifzadeh S, Steck R, Epari DR, Woodruff MA, Knackstedt M, Schuetz MA, et al. , Bone 2021, 153, 116163. [DOI] [PubMed] [Google Scholar]
- [19].Ghouse S, Reznikov N, Boughton OR, Babu S, Ng KCG, Blunn G, Cobb JP, Stevens MM, Jeffers JRT, Appl Mater Today 2019, 15, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Singh S, Nyberg EL, O’Sullivan AN, Farris A, Rindone AN, Zhang N, Whitehead EC, Zhou Y, Mihaly E, Achebe CC, et al. , Biomaterials 2022, 282, 121392. [DOI] [PubMed] [Google Scholar]
- [21].Cook CA, Hahn KC, Morrissette-McAlmon JBF, Grayson WL, Biomaterials 2015, 52, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Farris AL, Lambrechts D, Zhou Y, Zhang NY, Sarkar N, Moorer MC, Rindone AN, Nyberg EL, Perdomo-Pantoja A, Burris SJ, et al. , Biomaterials 2022, 280, 121318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang N, Singh S, Liu S, Zbijewski W, Grayson WL, 3D Print Med 2022, 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Suvarnapathaki S, Wu X, Zhang T, Nguyen MA, Goulopoulos AA, Wu B, Camci-Unal G, Bioact Mater 2022, 13, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chamieh F, Collignon A-M, Coyac BR, Lesieur J, Ribes S, Sadoine J, Llorens A, Nicoletti A, Letourneur D, Colombier M-L, et al. , Sci Rep 2016, 6, 38814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Perier-Metz C, Duda GN, Checa S, Front Bioeng Biotechnol 2022, 10, 980727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jaber M, Poh PSP, Duda GN, Checa S, Front Bioeng Biotechnol 2022, 10, 995266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu C, Entezari A, Zheng K, Fang J, Zreiqat H, Steven GP, Swain M. v., Li Q, Nat Comput Sci 2021, 1, 532. [DOI] [PubMed] [Google Scholar]
- [29].Bartling SH, Peldschus K, Rodt T, Kral F, Matthies H, Kikinis R, Becker H, J Comput Assist Tomogr 2005, 29, 305. [DOI] [PubMed] [Google Scholar]
- [30].Wang J, Shen Y, Yang S, Int J Comput Assist Radiol Surg 2019, 14, 763. [DOI] [PubMed] [Google Scholar]
- [31].Nicolielo LFP, van Dessel J, Shaheen E, Letelier C, Codari M, Politis C, Lambrichts I, Jacobs R, Int J Oral Sci 2017, 9, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lynch JA, Grigoryan M, Fierlinger A, Guermazi A, Zaim S, MacLean DB, Genant HK, Journal of Orthopaedic Research 2004, 22, 362. [DOI] [PubMed] [Google Scholar]
- [33].Han HH, Shim JH, Lee H, Kim BY, Lee JS, Jung JW, Yun WS, Baek CH, Rhie JW, Cho DW, Plast Reconstr Surg Glob Open 2018, 6, e1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xue R, Lai Q, Sun S, Lai L, Tang X, Ci J, Zhang Z, Wang Y, Journal of Craniofacial Surgery 2019, 30, e127. [DOI] [PubMed] [Google Scholar]
- [35].Oh TS, Jeong WS, Chang TJ, Koh KS, Choi JW, Journal of Craniofacial Surgery 2016, 27, 2020. [DOI] [PubMed] [Google Scholar]
- [36].Kohli N, Stoddart JC, van Arkel RJ, Sci Rep 2021, 11, 10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rafferty KL, Herring SW, Artese F, Journal of Experimental Biology 2000, 203, 2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nyberg E, O’Sullivan A, Grayson W, PLoS One 2019, 14, e0225007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jackson N, Assad M, Vollmer D, Stanley J, Chagnon M, Toxicol Pathol 2019, 47, 280. [DOI] [PubMed] [Google Scholar]
- [40].“Hematologic Reference Ranges - Special Subjects - Merck Veterinary Manual,” can be found under https://www.merckvetmanual.com/special-subjects/reference-guides/hematologic-reference-ranges, n.d. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


